Abstract
The N-end rule pathway is conserved from bacteria to man and determines the half-life of a protein based on its N-terminal amino acid. In Escherichia coli, model substrates bearing an N-degron are recognised by ClpS and degraded by ClpAP in an ATP-dependent manner. Here, we report the isolation of 23 ClpS-interacting proteins from E. coli. Our data show that at least one of these interacting proteins—putrescine aminotransferase (PATase)—is post-translationally modified to generate a primary N-degron. Remarkably, the N-terminal modification of PATase is generated by a new specificity of leucyl/phenylalanyl-tRNA-protein transferase (LFTR), in which various combinations of primary destabilising residues (Leu and Phe) are attached to the N-terminal Met. This modification (of PATase), by LFTR, is essential not only for its recognition by ClpS, but also determines the stability of the protein in vivo. Thus, the N-end rule pathway, through the ClpAPS-mediated turnover of PATase may have an important function in putrescine homeostasis. In addition, we have identified a new element within the N-degron, which is required for substrate delivery to ClpA.
Keywords: ClpS, LFTR, N-degron, N-end rule pathway, substrate binding
Introduction
Regulated protein degradation is essential for the proper function of both pro- and eukaryotic cells. It contributes to a vast array of processes, including general protein quality control, cell growth, differentiation, signal transduction and response to stress (Wickner et al, 1999; Dougan et al, 2002a; Jenal and Hengge-Aronis, 2003; Varshavsky, 2005; Ravid and Hochstrasser, 2008). One example of regulated proteolysis, common to both prokaryotes and eukaryotes, is the N-end rule pathway (Varshavsky, 1996; Mogk et al, 2007; Tasaki and Kwon, 2007). This pathway selects proteins for destruction based on the identity of their N-terminal amino acid, which may be either stabilising or destabilising (Varshavsky, 1996). In eukaryotes, destabilising residues are recognised by at least one member of the N-recognins, which result in ubiquitylation of the target protein, culminating in its ATP-dependent turnover by the 26S proteasome. The eukaryotic N-end rule is hierarchical, and destabilising residues can be separated into three tiers (primary, secondary and tertiary). Primary destabilising residues can be further divided into two categories: type 1 (basic; Arg, Lys and His) and type 2 (bulky hydrophobic; Trp, Tyr, Phe, Leu and Ile), which are recognised directly by N-recognins. In contrast, secondary (Asp, Glu and oxidised Cys) and tertiary (Asn, Gln and Cys) destabilising residues must first be converted to a primary destabilising residue, by specific cellular enzymes, for N-recognin binding to occur.
Although the N-end rule pathway controls a number of important events in eukaryotes, such as peptide import, chromosome stability, cardiovascular development and nitric oxide detection (Varshavsky, 2005; Tasaki and Kwon, 2007), the functions and physiological substrates of the N-end rule pathway in bacteria remain elusive. Despite this, many of the components of the N-end rule pathway have been identified using model substrates (Tobias et al, 1991; Shrader et al, 1993; Erbse et al, 2006). These findings show that the N-end rule pathway in Escherichia coli is also hierarchical. Primary destabilising residues are bulky and hydrophobic (Phe, Tyr, Trp and Leu), whereas secondary destabilising residues are basic (Arg and Lys). The conjugation of a primary destabilising residue onto a secondary destabilising residue is catalysed by leucyl/phenylalanyl-tRNA-protein transferase (LFTR). Biochemical and structural studies of this enzyme indicate that its catalytic activity is largely limited to the addition of Leu and Phe to a target protein containing an N-terminal Arg or Lys (Soffer, 1974; Tobias et al, 1991; Watanabe et al, 2007). Regardless of the method used to generate an N-degron, the recognition of all primary N-degrons in E. coli is mediated by ClpS, a distant relative of eukaryotic N-recognin (Lupas and Koretke, 2003; Erbse et al, 2006). The ClpS–substrate complex is delivered directly to the ClpA unfoldase (Weber-Ban et al, 1999) through a specific interaction between ClpS and the N-terminal domain of ClpA (Zeth et al, 2002), resulting in the ATP-dependent degradation of the substrate by the ClpAP protease (Erbse et al, 2006). Despite this detailed knowledge of the N-end rule pathway in E. coli, no natural N-end rule substrates have been defined, and hence, it still remains unclear how natural N-degrons are produced in vivo.
Here, we report the affinity isolation of 23 ClpS-interacting proteins from E. coli. Our preliminary ex vivo degradation assay, of the ClpS-interacting proteins, identified two potential N-end rule substrates—DNA protection during starvation (Dps) and putrescine aminotransferase (PATase). Further characterisation of these affinity-isolated proteins indicated that both contained a primary destabilising residue at the N-terminus, essential for their ClpS-dependent degradation by ClpAP in vitro. Further detailed analysis of PATase revealed that the N-degron was created by the unexpected conjugation of Leu or Phe to an N-terminal Met by LFTR. Our data show that the ClpAP-mediated turnover of PATase is strictly dependent on LFTR and ClpS both in vitro and in vivo. These data identify PATase, not only as the first physiological substrate of LFTR, but also as the first substrate of the bacterial N-end rule pathway. Importantly, this modification of the N-terminal Met of PATase, by LFTR, represents a new destabilising residue of the N-end rule pathway in E. coli. In addition, we have identified a hydrophobic element within these substrates, which is also present in all other bacterial model N-end rule substrates studied to date. Mutational analysis of this hydrophobic element has little effect on ClpS binding, and yet, dramatically alters the ClpAP-mediated degradation of the substrate, suggesting a role in substrate delivery. Finally, a model for N-degron generation and delivery to its cognate protease in E. coli is presented, which incorporates the unique findings revealed in this study.
Results
N-end rule dipeptides specifically compete with model N-end rule substrates for binding to ClpS, and thereby inhibit their ClpAP-mediated degradation
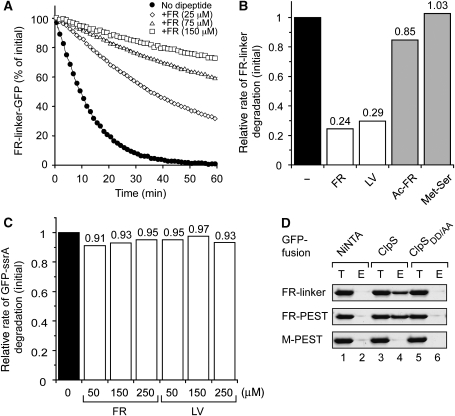
Previously, it was shown that ClpS binds to model N-end rule proteins with high affinity, and that binding was abolished in a subtrate-binding mutant of ClpS bearing a double mutation D35A/D36A referred to here as ClpSDD/AA (Erbse et al, 2006). Therefore, with the aim of developing a method for the affinity purification of natural N-end rule substrates, we tested the ability of various dipeptides to inhibit the ClpS-mediated delivery of a model N-end rule substrate to ClpAP. Initially, we monitored the ClpAPS-mediated degradation of FR-linker-GFP (Erbse et al, 2006) in the absence and presence of the Phe-Arg (FR) dipeptide (Figure 1A). In the absence of dipeptide, FR-linker-GFP was rapidly degraded by ClpAPS (Figure 1A, filled circles), whereas the addition of FR resulted in a concentration-dependent inhibition of FR-linker-GFP degradation (Figure 1A, open symbols). To ensure that the FR dipeptide effect was specific to N-end rule dipeptides, we compared the ability of two N-end rule dipeptides (FR and Leu-Val (LV)) and two control dipeptides (acetylated-FR (Ac-FR) and Met-Ser) to inhibit the degradation of FR-linker-GFP by ClpAPS (Figure 1B). Importantly, only the N-end rule dipeptides (FR or LV) reduced the rate of FR-linker-GFP degradation (Figure 1B, white bars). In contrast, the non-N-end rule dipeptides (Ac-FR or Met-Ser) had little to no effect on the degradation of FR-linker-GFP (Figure 1B, grey bars). To ensure that the N-end rule dipeptide specifically inhibited ClpS binding to model N-end rule substrates, and did not disrupt ClpAP function, we also monitored the ClpAP-mediated degradation of GFP-ssrA in the absence of ClpS (Figure 1C). In contrast to the ClpS-specific inhibition of FR-linker-GFP degradation, N-end rule dipeptides did not inhibit the ClpAP-mediated degradation of GFP-ssrA (Figure 1C, white bars). These data suggest that an N-end rule dipeptide can compete for binding to ClpS, with a protein containing an N-degron. To determine whether an N-end rule dipeptide could dissociate a preformed ClpS–substrate complex, we performed a series of ‘pull-down' experiments using the immobilised ClpS and various GFP-fusion proteins (FR-linker-GFP, FR-PEST-GFP and M-PEST-GFP, Figure 1D). After application of the different GFP-fusion proteins to immobilised ClpS or ClpSDD/AA, only the GFP-fusion proteins containing an N-degron (FR-linker-GFP or FR-PEST-GFP), but not M-PEST-GFP, were specifically eluted from the Ni-NTA agarose beads containing ClpS (Figure 1D, lane 4). Importantly, none of the GFP-fusion proteins was eluted from the control columns, in which the substrate-binding mutant ClpSDD/AA was immobilised (Figure 1D, lane 6) or that lacked an immobilised protein (Figure 1D, lane 2). Collectively, these data reveal that an N-end rule dipeptide can dissociate a pre-bound ClpS–substrate complex, thereby permitting the use of immobilised ClpS to affinity purify natural N-end substrates from an E. coli lysate.
Figure 1.
N-degron dipeptides compete with model N-end rule substrates for binding to ClpS. (A) The ClpAPS-mediated degradation of FR-linker-GFP (1 μM) was monitored by fluorescence in the absence (filled circles) or presence of 25 μM (open diamonds), 75 μM (open triangles) and 150 μM (open squares) of FR. (B) The ClpAPS-mediated degradation of FR-linker-GFP was monitored in the absence of dipeptide (black bar), the presence of 150 μM N-degron dipeptide (white bars) or 150 μM non-N-end rule dipeptide (grey bars). The rate of FR-linker-GFP degradation by ClpAPS in the presence of dipeptide was determined relative to the rate in the absence of dipeptide. (C) The ClpAP-mediated degradation of GFP-ssrA (1 μM) was monitored either in the absence of dipeptide (black bar) or in the presence of increasing concentration of N-degron dipeptides (white bars). The rate of GFP-ssrA degradation by ClpAP in the presence of dipeptide was determined relative to the rate in the absence of dipeptide. (D) GFP-fusions FR-linker-GFP, FR-PEST-GFP and M-PEST-GFP were applied (T) to Ni-NTA agarose beads (lane 1), beads with immobilised ClpS (lane 3) and beads with immobilised ClpSDD/AA (lane 5). After a wash step, columns were treated with FR dipeptide and eluted samples (lanes 2, 4 and 6) were separated by SDS–PAGE and visualised by Coomassie blue staining; 4% of the total (T) and 10% of the eluted (E) samples were analysed.
Affinity isolation of putative N-end rule substrates from ΔclpA E. coli
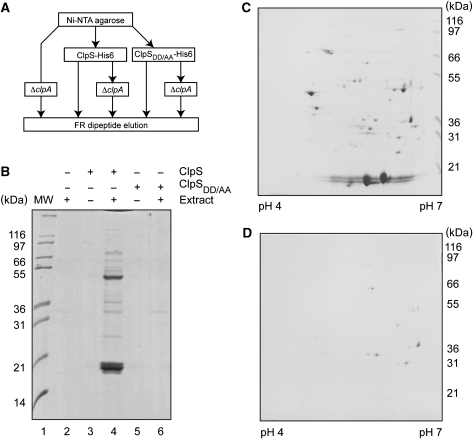
To identify natural N-end rule substrates from E. coli, we reasoned that they would accumulate in strains lacking a component of the ClpAPS machinery. To avoid substrate overlap with the ClpXP protease, we excluded the use of ΔclpP cells, likewise ΔclpS cells were also excluded as these cells contain ClpA and it has been proposed that the degradation of N-end rule substrates can occur independent of ClpS (Wang et al, 2007). Therefore, we chose to isolate putative N-end rule substrates from ΔclpA cells using the affinity purification (pull-down) method illustrated in Figure 2A. Initial experiments comparing wild-type and ΔclpA cells indicated that ClpS-interacting proteins did indeed accumulate in cells lacking ClpA. Consequently, all subsequent experiments were performed with ΔclpA cell extracts harvested during stationary phase (Figure 2B). Under these conditions, approximately 30 different proteins were specifically eluted from immobilised ClpS, using an N-end rule dipeptide (Figure 2B, lane 4 and Figure 2C). In contrast, very few proteins were eluted from Ni-NTA agarose beads, in the absence of immobilised ClpS (Figure 2B, lane 2) or from the other control column in which the ClpS-binding mutant ClpSDD/AA was immobilised (Figure 2B, lane 6 and Figure 2D).
Figure 2.
Selected E. coli proteins are specifically eluted from immobilised ClpS using FR dipeptide. (A) Schematic diagram illustrating the procedure for isolation of specific ClpS-interacting proteins. (B) E. coli proteins were separated by Tricine SDS–PAGE after FR dipeptide elution from Ni-NTA agarose beads (lane 2), beads with immobilised ClpS (lane 4) or beads with immobilised ClpSDD/AA (lane 6). These profiles were compared with the control elution profiles, in the absence of extract, from immobilised ClpS (lane 3) or immobilised ClpSDD/AA (lane 5). (C, D) The 2D gel analysis of FR dipeptide-eluted E. coli proteins from Ni-NTA agarose beads containing (C) immobilised ClpS or (D) immobilised ClpSDD/AA.
To determine the identity of the specific ClpS-interacting proteins, Coomassie-stained proteins from two-dimensional (2D) SDS–PAGE were excised and subjected to in-gel tryptic digestion followed by MALDI-TOF mass spectrometry (MS). The peptide mass fingerprints were searched against the SwissProt database using the Mascot search engine and revealed significant matches for 23 cytosolic E. coli proteins (see Supplementary Table S1). The identified proteins are associated with a variety of cellular roles, but fall into two main functional classes, namely metabolic enzymes and stress-related proteins. Interestingly, the majority of the ClpS-interacting proteins isolated are oligomeric, suggesting that ClpS may play a role in the remodelling of (or quality control of damaged) protein complexes.
Dps is truncated to form Dps18 (Dps6−167), an N-end rule ligand from E. coli
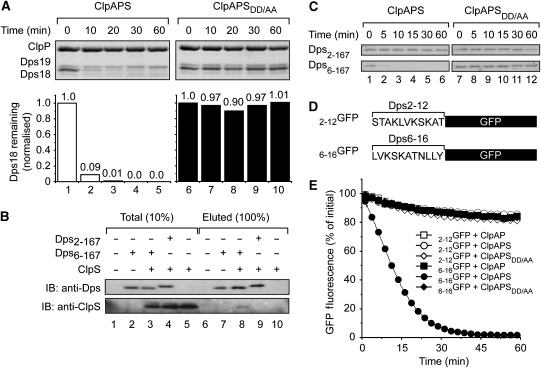
Next, we asked the question, which of the ClpS-interacting proteins, isolated through pull down, are susceptible to degradation by the ClpAPS machinery and, therefore, putative N-end rule substrates. To examine this, we subjected the dipeptide-eluted proteins from the pull down, to a ClpAP degradation assay either in the presence of wild-type ClpS (Figure 3A, lanes 1–5) and as a control in the presence of ClpSDD/AA (Figure 3A, lanes 6–10) or in the absence of ClpS (Supplementary Figure S1). This analysis revealed that two of the three most abundant proteins were degraded: an ∼18-kDa protein (shown in Figure 3A) and an ∼50-kDa protein (Figure 4A). Proteins of low abundance were not visible in the degradation assay. The two most abundant proteins (with a molecular mass of ∼18 and ∼19 kDa, see Figure 2B, lane 4), identified by MS as Dps, are referred to as Dps18 and Dps19, respectively. Interestingly, Dps19 was stable throughout the course of the experiment, suggesting that Dps19 was neither a ClpA- nor a ClpS-dependent substrate of ClpP. In contrast, the ClpAP-mediated degradation of Dps18 showed a strong dependence on wild-type ClpS (Figure 3A, white bars). To further characterise the identity of these proteins (Dps18 and Dps19), the dipeptide-eluted proteins from the pull down were transferred to a PVDF membrane after SDS–PAGE, and subjected to several rounds of N-terminal sequencing by Edman degradation. These data resulted in the identification of Dps19 as Dps2−167, a processed form of Dps lacking the initiating Met, whereas Dps18 lacked the first five residues of Dps (Dps6−167). Importantly, Dps6−167 contained a primary destabilising amino acid (Leu) at its N-terminus. To ensure that Dps6−167 was not simply an artefact of the cell extraction procedure, cells were lysed under denaturing conditions using 8 M urea revealing two forms of Dps (data not shown), suggesting that Dps6−167 was an authentic substrate of the ClpAS or ClpAPS machinery resulting from a specific endoproteolytic cleavage. Initially, we tested whether Dps6−167 was recognised by the ClpAS machine directly or through a trans-targeting mechanism, involving a hetero-oligomeric complex of Dps, as is the case for UmuD/UmuD' (Gonzalez et al, 2000). To address this possibility, we generated both forms of Dps (Dps2−167 and Dps6−167) in vitro using an ubiquitin (Ub)-fusion system (Catanzariti et al, 2004) and performed in vitro coimmunoprecipitation experiments using anti-Dps antisera (Figure 3B). Importantly, the anti-Dps antisera was able to immunoprecipitate both forms of Dps: Dps2−167 (Figure 3B, lane 9) and Dps6−167 (Figure 3B, lane 7), as determined by immunodecoration of the western blot. However, ClpS was only co-precipitated using the anti-Dps antisera in the presence of Dps6−167 (Figure 3B, lane 8) and not in the presence of Dps2−167 (Figure 3B, lane 9) nor in the absence of Dps (Figure 3B, lane 10). Collectively, these data indicate that ClpS can interact with both homo- and hetero-oligomeric (Figure 2B) complexes of Dps containing Dps6−167, but not with homo-oligomeric complexes of Dps2−167.
Figure 3.
Dps6−167 is a ClpS-dependent substrate of the ClpAP protease. (A) The ClpAP-mediated degradation of ClpS-interacting proteins was monitored in the presence of wild-type (lanes 1–5) or mutant ClpS (lanes 6–10). Proteins were separated by Tricine SDS–PAGE and visualised by Coomassie blue staining. (B) Interaction of ClpS with Dps, examined through coimmunoprecipitation. ClpS in the presence of Dps6−167 (lane 3), Dps2−167 (lane 4) or in the absence of Dps (lane 5) was subjected to coimmunoprecipitation with antibodies against Dps. Protein A Sepharose-eluted proteins separated by Tricine SDS–PAGE were analysed by immunoblotting (IB) as indicated. Dps6−167 only (lane 2) served as an IP control. (C) Time course of ClpAP-mediated degradation of Dps2−167 or Dps6−167 in the presence of wild-type (lanes 1–6) or mutant ClpS (lanes 7–12). Samples were separated by Tricine SDS–PAGE and proteins visualised by Coomassie blue staining. (D) Diagrammatic representation of the GFP fusions used in (E). (E) The ClpAP-dependent degradation of 2−12GFP (open symbols) and 6−16GFP (filled symbols) was monitored by fluorescence in the absence of additional components (squares), in the presence of ClpS (circles) or in the presence of ClpSDD/AA (diamonds).
Figure 4.
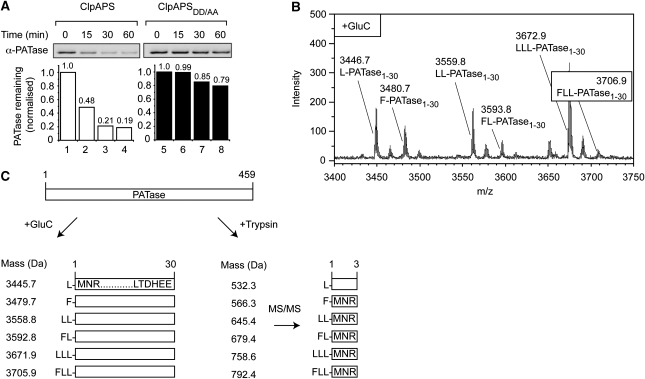
Modified PATase is an N-end rule substrate. (A) The ClpAP-mediated degradation of ClpS-interacting proteins was monitored with time in the presence of wild-type (lanes 1–4) or mutant ClpS (lanes 5–8). Proteins were separated by SDS–PAGE, transferred to PVDF membrane and visualised by immunodecoration with anti-PATase antisera. (B) ClpS-interacting proteins were separated by 2D SDS–PAGE, PATase was excised from the gel and subjected to a GluC in-gel digest. The N-terminal peptides were identified by MALDI-TOF MS. (C) Schematic diagram showing the calculated mass of different N-terminal peptides of PATase (after digestion with GluC or trypsin). The observed amino-acid sequences of the tryptic peptides were determined by MS/MS fragmentation of the peptides (far right).
Although Dps6−167 (in the absence of Dps2−167) did indeed interact with ClpS in vitro (Figure 3B), it remained unclear whether the presentation of Dps6−167 (i.e. in a hetero-oligomeric complex) was required for its ClpS-mediated processing. To test this, we monitored the in vitro degradation of Dps6−167 and Dps2−167 by ClpAP in the presence of wild-type or mutant ClpS (Figure 3C). Consistent with our earlier findings, only Dps6−167 was degraded by the ClpAPS machine (Figure 3C, lanes 1–6). In contrast, Dps6−167 was stable in the presence of ClpSDD/AA (Figure 3C, lanes 7–12). These data suggest that ClpS is essential for the delivery of Dps6−167 to ClpA for degradation by ClpP. To confirm the importance of the N-terminal residues of Dps for degradation, we fused the first 11 amino acids of Dps2−167 or Dps6−167 to GFP to generate 2−12GFP and 6−16GFP, respectively (Figure 3D). Attachment of the first 11 amino acids of Dps2−167 to GFP resulted in a modest amount of ‘untargeted' degradation of GFP (∼15%) over the time course of the experiment (Figure 3E, open squares). This ‘untargeted' ClpAP-mediated degradation of 2−12GFP also occurred in the presence of wild-type (Figure 3E, open circles) or mutant ClpS (Figure 3E, open diamonds). The same amount of ‘untargeted' degradation was also observed for 6−16GFP (containing an N-degron) either in the absence of ClpS (Figure 3E, filled squares) or in the presence of ClpSDD/AA (Figure 3E, filled diamonds). Only in the presence of wild-type ClpS, did the rate of ClpAP-mediated degradation of 6−16GFP increase dramatically (Figure 3E, filled circles; Supplementary Figure S2). Collectively, these data indicate that Dps6−167 is an N-end rule ligand and validates the affinity purification approach taken as a means to isolate further N-end rule substrates from E. coli.
Post-translational modification of PATase generates an authentic N-degron
Initial in vitro degradation results, of the dipeptide-eluted material from the ClpS pull down, indicated that an abundant protein of ∼50 kDa identified by MS as PATase was degraded by ClpAP in a ClpS-dependent fashion (Supplementary Figure S1). To confirm that PATase was indeed a target of the ClpAPS machine, we generated a PATase-specific rabbit polyclonal antisera (Supplementary Figure S3A) and subjected the dipeptide-eluted ClpS-interacting proteins to an in vitro degradation assay. Samples from the degradation assay were separated by SDS–PAGE and analysed by immunoblotting with anti-PATase antisera (Figure 4A). This analysis confirmed that PATase was rapidly degraded by the ClpAP machine in the presence of wild-type ClpS (Figure 4A, lanes 1–4), but was largely stable in the presence of the ClpSDD/AA (Figure 4A, lanes 5–8), indicating that PATase was a potential N-end rule substrate. To ensure that PATase was indeed a cytosolic protein, and not a periplasmic protein as annotated in a few databases, we fractionated wild-type E. coli cells (Supplementary Figure S3B). As expected for a substrate of the ClpAPS machinery, these data indicated that PATase co-localised with DnaK, a cytosolic marker protein and not with Maltose-Binding Protein, a periplasmic marker protein.
To determine the N-terminal sequence of PATase, we took two complementary approaches. Initially, affinity-isolated PATase was subjected to six cycles of Edman degradation (data not shown). These data indicated that the first amino acid of PATase was Leu or Phe and not Met, suggesting that the N-terminus of PATase was modified. In the second approach, an in-gel digest (using either GluC or trypsin) was performed and N-terminal peptides were examined by MALDI-TOF MS. After digestion of PATase with GluC, a family of six different N-terminal peptides were identified with masses that were consistent with the addition of various combinations of Leu and Phe to the N-terminal 30 residues of PATase (Figure 4B and left panel of Figure 4C). To confirm the identity of the different N-terminal sequences of PATase, we performed a tryptic digest and analysed the samples by MS. Consistent with the GluC digest, six different tryptic peptides were identified by LC-MS with masses that were larger than expected for the N-terminal tryptic peptide (MNR) of unmodified PATase (Figure 4C, right panel). Importantly, fragmentation of these tryptic peptides by MS/MS (with the exception of the 532.3-Da peptide) enabled the amino-acid sequence determination of each peptide (Figure 4C, far right panel). For example, a double-charged peptide was fragmented by MS/MS, revealing the sequence FLLMNR (Supplementary Figure S4). Collectively, these data indicate that endogenous PATase, isolated by pull down, contains various post-translational modifications, all of which result from the addition of primary destabilising amino-acids Leu or Phe or combinations of both to the N-terminus of the protein.
LFTR is required for the post-translational modification and turnover of PATase in vivo
Next, we considered which enzyme might be responsible for the modification of PATase in vivo. As LFTR, encoded by aat, is responsible for the addition of Leu and Phe to an acceptor protein, and the post-translational modification of PATase involved the addition of Leu and Phe, we asked the question, is LFTR required for the modification of PATase in vivo and if so what is the consequence of this modification. Initially, we repeated the ClpS pull-down experiment using lysates from various E. coli strains (wild-type, ΔclpA and Δaat) and monitored the amount of PATase recovered using the anti-PATase antisera (Figure 5A). These data revealed that the steady state levels of PATase, during stationary phase, were higher in both Δaat cells (Figure 5A, lane 2) and ΔclpA (Figure 5A, lane 3) when compared with wild-type E. coli cells (Figure 5A, lane 1). As expected, the stabilisation of PATase in ΔclpA cells leads to an improved recovery of PATase, relative to wild-type cells, in the dipeptide-eluted fraction (Figure 5A, compare lanes 4 and 6). In contrast, PATase was not recovered from the pull down using a Δaat cell lysate (Figure 5A, lane 5), despite the increased levels of PATase in the cell (Figure 5A, lane 2). These data are consistent with our MS/MS analysis of PATase isolated from ΔclpA cells, which suggest that PATase is translated with an N-terminal Met, and hence, lacks an N-degron in Δaat E. coli. To gain direct evidence for this, we isolated PATase from Δaat cells by immunoaffinity chromatography using an anti-PATase antisera and then performed a GluC digest of the PATase spot, excised from the gel after separation by 2D-PAGE. From this analysis, we identified a peptide of mass 3334.6 Da (Supplementary Figure S5A), which is indicative of the N-terminal peptide of PATase (PATase1−30 containing an N-terminal Met). Importantly, this peptide fragment was absent from the GluC profile of PATase isolated from ΔclpA cells (Supplementary Figure S5B). Consistently, all of the modified N-terminal peptides (of PATase isolated from ΔclpA cells) were absent from GluC profile of PATase isolated from Δaat cells.
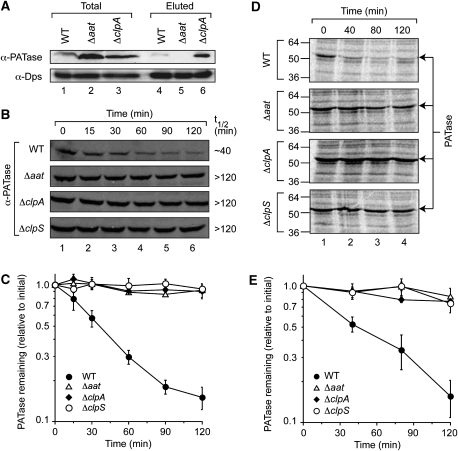
Figure 5.
The degradation of PATase in vivo is dependent on the ClpAPS protease and modification by LFTR. (A) Extracts from E. coli strains indicated (lanes 1–3) were applied to Ni-NTA agarose beads containing immobilised ClpS. The ClpS-interacting proteins eluted using FR dipeptide are shown (lanes 4–6). After elution, proteins were separated by glycine SDS–PAGE and transferred to a PVDF membrane for immunodecoration with specific antisera as indicated. (B) The steady state turnover of PATase, in various E. coli strains, was analysed by immunoblotting after the addition of spectinomycin. (C) The relative amount of PATase was determined by quantification of the immunoreactive band from four independent experiments (using the GelEval software). Error bars represent the standard error of the mean determined from four independent experiments. (D) The half-life of newly synthesised PATase, in various E. coli strains, was analysed by pulse-chase IP of radiolabelled proteins using anti-PATase antisera. (E) The relative amount of newly synthesised PATase was determined by quantification of radiolabelled PATase from three independent experiments (using the GelEval software). Error bars represent the standard error of the mean determined from three independent experiments.
To determine whether modification of PATase by LFTR was responsible for the turnover of PATase in vivo, we added spectinomycin to various E. coli strains (wild-type, Δaat, ΔclpA and ΔclpS), during logarithmic growth phase to inhibit protein synthesis and then monitored the levels of PATase in these cells. In wild-type cells, PATase was degraded with a half-life of ∼40 min (Figure 5B and C), whereas a control protein (ClpP) was stable throughout the time course of the experiment (Supplementary Figure S6). In contrast to wild-type cells, deletion of aat, clpA or clpS, leads to a stabilisation of PATase (half-life >120 min) during logarithmic growth phase (Figure 5B and C), suggesting that modification of PATase by LFTR is required for ClpS interaction and its subsequent ClpAP-mediated turnover, but did not affect the stability of ClpP (Supplementary Figure S6). To examine whether the ClpAPS-mediated turnover of newly synthesised PATase was dependent on modification by LFTR, we performed pulse-chase immunoprecipitation (IP) experiments using the same E. coli strains as above (i.e. wild-type, Δaat, ΔclpA and ΔclpS). Consistent with the above translation arrest experiments, we observed a half-life of ∼30 min for newly synthesised PATase in wild-type cells (Figure 5D and E), whereas its half-life was significantly increased in each of the deletion strains tested (Figure 5D and E). Taken together, these data indicate that LFTR plays a critical role in the ClpAPS-mediated turnover of endogenous PATase in vivo.
Only modified PATase is a ClpS-dependent ClpAP substrate
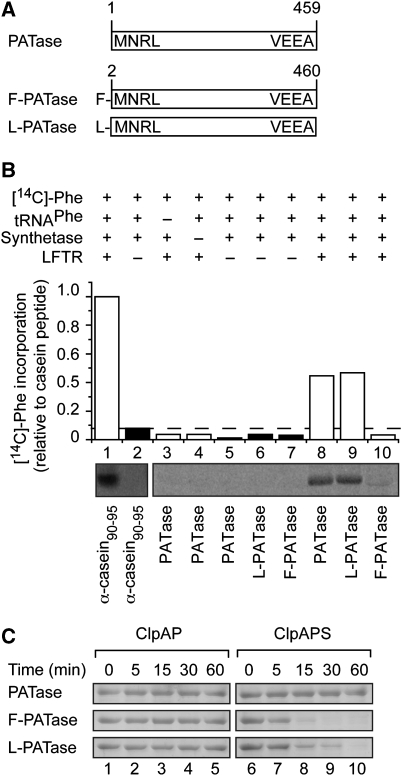
Although it was evident that aat was required for the turnover of PATase in vivo, a direct role for LFTR in the post-translational modification of PATase was unexpected given the proposed specificity of LFTR. Therefore, to determine whether LFTR was directly responsible for the conjugation of Leu and/or Phe to PATase, we reconstituted the conjugation reaction in vitro. First, using the Ub-fusion system, we generated various recombinant forms of PATase, including PATase, which represents the unmodified cellular form of the protein and contains an N-terminal Met (Figure 6A), and F-PATase or L-PATase, which represent post-translationally modified forms of the protein and containing either an N-terminal Phe or Leu, respectively (Figure 6A). As a control, we tested the ability of LFTR to catalyse the attachment of [14C]-Phe to a known substrate (casein fragment 90–95) in the presence (Figure 6B, lane 1) or absence (Figure 6B, lane 2) of LFTR. Next, we tested whether recombinant PATase could act as a substrate for LFTR in the conjugation reaction. Consistent with our earlier in vivo findings (Figure 5) and mass spectroscopy analysis (Figure 4), PATase (Figure 6B, lane 8) and L-PATase (Figure 6B, lane 9), but not F-PATase (Figure 6B, lane 10) were radiolabelled in the presence of [14C]-Phe-tRNAPhe and LFTR (Figure 6B, white bars). To ensure that this radiolabelling of PATase and L-PATase was specific, several control experiments were performed. In the absence of LFTR (Figure 6B, black bars), neither PATase (Figure 6B, lane 5) nor L-PATase (Figure 6B, lane 6) was radiolabelled. Likewise, the absence of either tRNAPhe or aminoacyl-tRNA synthetase prevented radiolabelling of PATase (Figure 6B, lanes 3 and 4, respectively).
Figure 6.
The N-terminal conjugation of PATase in vitro is dependent on LFTR and generates a ClpS-dependent N-end rule substrate of the ClpAP machine. (A) Diagrammatic representation of the various recombinant forms of PATase used in (B and C). (B) N-terminal conjugation of [14C]-Phe to various acceptor proteins (α-casein fragment (lanes 1 and 2), PATase (lanes 3–5 and 8) L-PATase (lanes 6 and 9) and F-PATase (lanes 7 and 10)) was performed in the presence (white bars) or absence (black bars) of LFTR. The level of PATase conjugation was determined by quantitation of the radiolabelled band (using the GelEval software), relative to conjugation of the control α-casein peptide (lane 1). (C) The ClpAP-mediated degradation of various recombinant forms of PATase was monitored in the absence (lanes 1–5) or presence of ClpS (lane 6–10).
To confirm the importance of the N-terminal modification of PATase with respect to its turnover, we compared the ability of each PATase variant to interact with ClpS using a pull-down assay (Supplementary Figure S7A). Only wild-type ClpS and not ClpSDD/AA bound the modified forms of PATase (F-PATase and L-PATase), whereas the control protein (PATase) failed to interact with either ClpS proteins (Supplementary Figure S7A). Next, we monitored the ClpAP-mediated degradation of modified or unmodified PATase in the absence (Figure 6C, lanes 1–5) or presence (Figure 6C, lanes 6–10) of ClpS. Unmodified PATase was stable both in the absence and the presence of ClpS with a half-life of >60 min (Figure 6C). In contrast, both modified forms of PATase (F-PATase and L-PATase) were rapidly degraded by ClpAP, but, importantly, only in the presence of ClpS (Figure 6C, lanes 6–10).
Bacterial N-end rule substrates contain a hydrophobic element required for ClpAP-mediated degradation
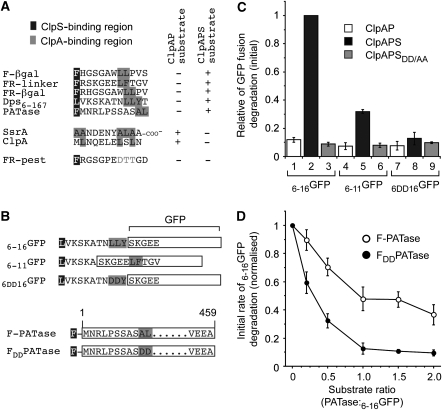
Armed with a collection of N-end rule substrates (both model and natural), we aligned their N-terminal amino-acid sequences and searched for a common feature. From this analysis, we identified a conserved motif composed of two to three hydrophobic amino acids within the first 12 amino acids of the sequence (Figure 7A). This hydrophobic motif was generally located at least six residues C-terminal of the N-degron. Notably, this motif was absent from FR-PEST-GFP, a ClpS-binding protein (Figure 1D), which is not degraded by the ClpAPS machine (data not shown), suggesting that this region may be required for transfer to ClpA. Interestingly, the ClpA-binding site of a well-characterised ClpAP substrate, the SsrA tag, is composed of two hydrophobic patches separated by a similar distance as observed for the proposed ClpS and ClpA interaction sites (Figure 7A). Therefore, we speculated that by combining FR (for ClpS recognition) and the SsrA tag (for ClpA binding) to create FRssrA, we could generate a robust ClpAPS substrate. As proposed, the addition of FR to the N-terminus of ssrA-GFP was sufficient to transform ssrA-GFP into a robust ClpS-mediated substrate of the ClpAP machine (Supplementary Figure S8). Next, we generated several mutant proteins (within the Dps–GFP fusion and PATase) to further examine the role of this hydrophobic element in N-end rule substrates (Figure 7B). Initially, we tested its role within the Dps degradation signal tagged to GFP (e.g. 6−16GFP), either through deletion (6−11GFP) or mutation (6DD16GFP) of the Dps tag (Figure 7B). In the absence of this motif, a small reduction in the ClpAP-mediated degradation of these substrates was observed either in the absence of ClpS (Figure 7C, compare lane 1 with lanes 4 and 7) or in the presence of the ClpS mutant (Figure 7C, compare lane 3 with lanes 6 and 9). The ClpAPS-mediated degradation of 6−16GFP (Figure 7C, lane 2) was dramatically reduced by deletion (Figure 7C, lanes 5) or mutation (Figure 7C, lanes 8) of the hydrophobic element within the substrate. Importantly, neither deletion nor mutation of this element affected ClpS binding (Supplementary Figure S7B). Taken together, these data suggest that modification of the hydrophobic element effects substrate processing by the ClpAP protease, and we speculate that this reflects an inability of ClpA to accept the substrate from ClpS. A similar mutation within the hydrophobic element of PATase (FDDPATase) also reduced the ClpAPS-mediated degradation of this substrate significantly (Supplementary Figure S9). These data indicate that the hydrophobic element may be an important feature common to all bacterial N-end rule substrates. Next, to further analyse the role of the hydrophobic element in substrate delivery, we compared the ClpAPS-mediated degradation of 6−16GFP in the presence of either F-PATase (Figure 7D, open circles) or FDDPATase (Figure 7D, filled circles). As expected, both F-PATase and FDDPATase inhibited the degradation of 6−16GFP; however, the concentration of substrate required for maximal inhibition varied considerably. In the case of F-PATase, the rate of 6−16GFP degradation was reduced by half at equimolar substrate levels (Figure 7D) consistent with that expected for reversible substrate binding. In contrast, sub-stoichiometric levels of FDDPATase (i.e. PATase: GFP ratio of 0.3) were already sufficient to inhibit the rate of 6−16GFP degradation by ∼50% (Figure 7D). Hence, these data suggest that FDDPATase functions as an irreversible inhibitor of the ClpAPS-mediated degradation reaction.
Figure 7.
N-end rule substrates contain a putative ClpA recognition motif downstream of the N-degron. (A) Sequence alignment of known ClpA and ClpS recognition motifs. The proposed ClpS-binding region is highlighted in black, known and proposed ClpA-binding regions are highlighted in grey. (B) Diagrammatic representation of the various GFP fusions and PATase mutants used in (C) or (D). (C) The ClpAPS-mediated degradation of GFP-fusion proteins 6−16GFP (lanes 1–3), 6−11GFP (lanes 4–6) and 6DD16GFP (lanes 7–9) was monitored in the absence of additional components (white bars), the presence of ClpS (black bars) or in the presence of ClpSDD/AA (grey bars). The rate of degradation of all GFP-fusion proteins was determined relative to ClpAP-mediated degradation of 6−16GFP in the presence of ClpS (lane 2). Error bars represent the standard error of the mean determined from quantification of three independent experiments. (D) The ClpAPS-mediated degradation of 6−16GFP was monitored in the presence of increasing concentrations of F-PATase (open circles) or FDD-PATase (filled circles). The rate of 6−16GFP degradation is relative to ClpAPS degradation in the absence of additional substrate. Error bars represent the standard error of the mean determined from quantification of three independent experiments.
Discussion
Although many components of the N-end rule pathway in E. coli have been defined, so far no bacterial substrates have been identified. Here, we report the identification of 23 ClpS-interacting proteins from E. coli (Figure 2; Supplementary Table S1), two of which—Dps and PATase—undergo some form of post-translational modification to generate an N-degron. Both modified proteins fulfil each of the requirements of a model N-end rule substrate—not only do they contain an N-terminal destabilising residue, but they also contain a downstream hydrophobic element to facilitate transfer of the substrate to ClpAP for degradation (Figure 7). Although the method of generation of the Dps N-degron and its physiological significance is unclear, the N-degron of PATase is generated from the enzymatic attachment of a primary destabilising residue (Leu or Phe) to its N-terminus. Significantly, modification of PATase in vivo implicates Met as a newly identified secondary destabilising residue, which may be conditional in nature, and extends the complexity of the N-end rule pathway in E. coli. Surprisingly, the modification of PATase is performed by LFTR both in vitro and in vivo and as such identifies an undescribed specificity of this enzyme (Figures 4, 5 and 6). This modification of PATase is not only essential for its ClpS-mediated recognition, but also for its subsequent ClpAP-mediated degradation both in vitro and in vitro (Figures 5 and 6). These and other data lead us to the following model for the N-end rule pathway in E. coli (Figure 8).
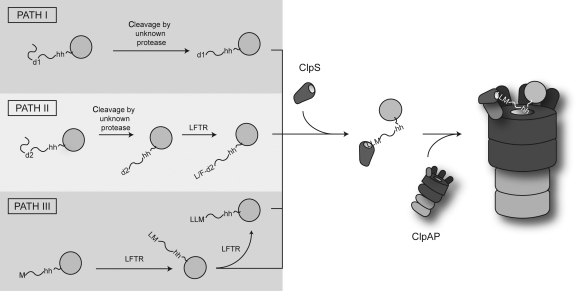
Figure 8.
Model for the generation of N-end rule substrates in E. coli. N-end rule proteins in E. coli contain an internal destabilising residue (d1 or d2), which after cleavage reveals a primary (e.g. Leu, Phe, Tyr, Trp; Path I) or secondary (e.g. Arg, Lys and possibly Met; Path II) destabilising residue at the N-terminus. Secondary destabilising residues are converted into primary destabilising residues by LFTR. In a newly proposed pathway (Path III), the initiating Met functions as a conditional secondary destabilising residue, which is converted by LFTR, into a primary destabilising residue. The addition of multiple primary destabilising residues to an N-terminal Leu (as observed for PATase) may only occur in the artificial setting of a cell lacking ClpA. It is nevertheless tempting to speculate that multiple modifications to the substrate N-terminus increase the linker length and may be required to optimise substrate delivery. All primary N-degrons (generated by Path I, II or III) bind to ClpS and are delivered to the ClpAP machine possibly through an interaction between the hydrophobic element (hh) within the substrate and the pore of ClpA.
Naturally occurring E. coli N-end rule substrates reveal a strict requirement for ClpS and a hydrophobic element for targeted degradation by ClpAP
Until now, the description of the N-end rule pathway in E. coli has been limited to model N-end rule substrates. Our isolation of several ClpS-interacting proteins from E. coli provides the first example of a bacterial N-end rule substrate. Importantly, two of these proteins were extensively characterised and contain all of the proposed features identified in model N-end rule substrates (Tobias et al, 1991; Erbse et al, 2006). Not only do they contain a destabilising residue at the N-terminus, but they also possess an overall positive charge near the unstructured N-terminus of the protein, as was proposed for ClpS binding (Erbse et al, 2006). Further to these properties, we have identified a new element within these N-end rule substrates, a hydrophobic motif located approximately 7 to 10 residues C-terminal of the N-degron, which appears to play a critical role in substrate processing by ClpAP. We speculate that docking of ClpS to the N-domain of ClpA, positions the hydrophobic element of the substrate in close proximity to the central ring of ClpA pore residues, which is necessary for ClpA to disengage the ClpS–substrate complex (Figure 8). Hence, not only the length of the linker sequence, but also the composition of this region, are important determinants for ClpA-mediated degradation of N-end rule substrates. In fact, it is plausible that a strong hydrophobic motif, near the N-terminus of a protein lacking an N-degron, may also be degraded by ClpAP in the absence of ClpS. Coincidently, this element is present in the linker region of various model N-end rule substrates used in the earlier studies (Tobias et al, 1991; Erbse et al, 2006; Wang et al, 2007). In essence, this hydrophobic element serves an analogous function to the internal Lys residue, within eukaryotic N-end rule substrates, required for Ub attachment and subsequent proteosomal degradation (Ravid and Hochstrasser, 2008).
Cleavage of Dps generates an N-degron
In bacteria, Dps is an important multifunctional protein that accumulates in response to a variety of cellular stresses, including nutrient or oxidative stress (Almiron et al, 1992). It binds to and protects DNA against various chemical challenges, including oxidative stress (Martinez and Kolter, 1997) and UV damage (Nair and Finkel, 2004). Generally, it is believed that oligomeric complexes of Dps bind to and protect DNA by forming a physical barrier to the stress. In the case of oxidative damage, Dps is also believed to protect the cell by reducing the accumulation of hydroxyl radicals, resulting from the Fenton reaction, through its Fe-chelating activity.
In our analysis, we have found that Dps is susceptible to proteolytic cleavage, by an as yet unidentified protease, converting a pre-N-degron into an N-degron (Dps6−167). Although technical challenges have currently limited our ability to clearly define a difference in steady state levels of Dps6−167 when comparing wild-type and ΔclpA cells, truncated Dps was present in urea-treated whole cells, suggesting that this fragment is not the result of post-lysis proteolysis. Interestingly, the N-terminal region of Dps consists of several basic residues and appears to have two important functions: it has been implicated in DNA condensation (Ceci et al, 2004) and is required for recognition by the ClpXP protease (Flynn et al, 2003). Although much is known about the role of Dps in DNA condensation, little is known about the disassembly of these Dps–DNA complexes. We speculate that the N-terminal cleavage of Dps, which prevents its ClpXP-mediated turnover, may play a role in the disassembly of hetero-oligomeric Dps–DNA complexes, exiting stationary phase—and these complexes are then primed for disassembly by ClpA in a ClpS-dependent manner. Currently, we are unable to determine whether truncated Dps is disassembled and degraded by the ClpAPS protease or merely disassembled by the ClpAS unfoldase, in which it may be subsequently removed from the cell by other cellular proteases.
Nevertheless, this type of regulated degradation controls a range of other cellular processes in bacteria and often the ultimate removal of these fragments is mediated by adaptor proteins. For example, in E. coli after DNA damage, ClpX uses UmuD as an adaptor protein to deliver UmuD' to ClpXP for degradation (Gonzalez et al, 2000). Similarly, an N-terminal fragment of RseA, which is generated as part of the periplasmic stress response, is delivered to ClpXP by the adaptor protein SspB (Flynn et al, 2004). Endoproteolytic processing also has an important function in Saccharomyces cerevisiae chromosome stability. In fact, cleavage of the cohesion protein SCC1 generates an unstable C-terminal fragment bearing and N-terminal Arg, which is recognised by UBR1 and degraded by the N-end rule pathway (Rao et al, 2001).
PATase and the role of L/F-transferase in the N-end rule
To maintain optimal cell growth, bacteria need to balance the level of various cellular components. For example, polyamines are not only critical for a variety of cellular functions, but their accumulation can also be detrimental to the cell by inhibiting protein synthesis (Fukuchi et al, 1995). In E. coli, PATase, as part of the L-arginine pathway, catalyses the aminotransferase reaction from putrescine to 2-oxoglutarate to generate L-glutamate and 4-aminobutanal (Samsonova et al, 2003), and hence, contributes to the level of polyamines in the cell.
Here, we show that both the steady state levels and the half-life of PATase are dependent on the activity of LFTR in vivo. This LFTR-dependent modification of PATase is consistent with earlier findings, in which a protein of similar molecular mass (∼50 kDa) was identified as a target of LFTR (Soffer and Savage, 1974). Therefore, we propose that the N-end rule pathway contributes to putrescine homeostasis by modulating PATase levels through the combined action of LFTR and the ClpAPS machinery.
Remarkably, the N-terminal residue of PATase, before its conjugation by LFTR, is neither Arg nor Lys, but Met, which suggests that LFTR exhibits a broader specificity than previously proposed. Consistent with this notion, our in vitro conjugation reaction showed not only that PATase is the first natural substrate of LFTR, but also confirmed that the specificity of LFTR recognition includes both N-terminal Met and Leu (in addition to Arg and Lys). As a consequence of these findings, the potential pool of LFTR substrates within the cell could be dramatically increased given that ∼30% of E. coli proteins retain their initiating Met (Frottin et al, 2006). Despite this, we speculate that Met is unlikely to act as an unrestricted secondary destabilising residue, rather we propose that the specificity of LFTR may be influenced by the identity of the penultimate residue in the acceptor, and hence, only a handful of these potential substrates (containing an N-terminal Met) will be recognised by LFTR. Consistently, a recent structural study showed that the penultimate residue of the acceptor makes direct contact with the enzyme (Watanabe et al, 2007). However, a systematic study examining a role for the penultimate residue has not yet been performed, and so this role remains speculative. Interestingly, from MS sequencing of the tryptic PATase fragments, we observed (1) multiple attachments of Leu (up to three) to the N-terminus of PATase and (2) the attachment of Phe was exclusively N-terminal. These observations have a number of implications; first, Leu (in addition to Met, Arg and Lys) can act as an acceptor for LFTR, and hence, is a secondary destabilising residue. Although this finding may merely be an artefact of the ΔclpA system used in these experiments, it does have a potentially interesting biological implication. Multiple attachments (of Leu) to the N-terminus of a substrate provides the cell with a mechanism to increase the length between the primary destabilising residue (required for ClpS binding) and the hydrophobic element (for ClpA binding) to mediate efficient delivery to ClpA for ClpP-dependent degradation. Second, in contrast to Leu, Phe is not an acceptor for LFTR, and hence, the reaction can be terminated by the attachment of Phe to the N-terminus of the substrate.
The characterisation of two N-end rule substrates from E. coli shows the utility of our approach. Using this method, we have compiled a list of putative N-end rule substrates. Intriguingly, several of these putative substrates are hetero-oligomers (in which both components of the complex were isolated). Hence, it is likely that some of these interacting proteins merely co-purify with authentic substrates. Similarly, it is likely that the molecular chaperones DnaK and ClpB have been co-purified through a common substrate. Future experiments to determine protein turnover in various strains will be crucial to determine the physiological relevance of the ClpS-interacting proteins identified in this study, and hence, it remains a challenge to elucidate the full biological significance of the N-end pathway in E. coli.
Materials and methods
Strains and culture conditions
E. coli K12 strain BW25113 (rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1) and single gene knockout mutants, ΔclpA (JW0866), ΔclpS (JW0865), ΔclpP (JW0427), Δaat (JW0868), Δdps (JW0797) and ΔygjG (JW5510), in BW25113 background were obtained from the Keio collection (Baba et al, 2006). Stationary phase cultures of E. coli were grown at 37°C for 26 h in LB media supplemented with 50 μg/ml kanamycin, when appropriate. For half-life experiments, the different E. coli strains were grown to mid-logarithmic phase before translation was stopped by the addition of spectinomycin as described (Dougan et al, 2002b).
Proteins and peptides
ClpA, ClpP, ClpS (wild-type and mutants) and GFP-SsrA were over-expressed in E. coli and purified as described earlier (Dougan et al, 2002b). For the generation of ClpS-His6 and mutants, PCR-amplified clpS and mutant clpS encoding ClpSDD/AA were cloned into pUHIS and expressed in E. coli strain XL1-blue and purified under native conditions using Ni-NTA agarose (Qiagen). Recombinant model (FR-linker-GFP, FR-PEST-GFP, 2−12GFP, 6−16GFP, 6−11GFP, 6DD16GFP), authentic N-end rule substrates (L-PATase, F-PATase, FDDPATase and Dps6−167), control proteins (M-PEST-GFP, PATase and Dps2−167) and LFTR were generated using the Ub-fusion system (Catanzariti et al, 2004). Briefly, genes encoding proteins of interest were amplified by PCR either from E. coli genomic DNA or pDD173 encoding GFP (Dougan et al, 2002b) and cloned into pHUE (Catanzariti et al, 2004). Proteins were expressed as linear His6-Ub-fusion proteins in E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene) and purified using Ni-NTA agarose (Qiagen). The His6-Ub was cleaved with His6-Usp2cc (Catanzariti et al, 2004) and the untagged proteins isolated through a reverse affinity chromatography step. Untagged LFTR was purified to homogeneity essentially as described above, except that the extraction buffer was 50 mM Tris–HCl pH 6.8, 500 mM KCl, 6 mM 2β-ME, 10 mM imidazole and 5% (v/v) glycerol as described (Suto et al, 2006). All proteins were >95% pure as judged by Coomassie-stained SDS–PAGE. Protein concentrations refer to the protomer and were determined using Bio-Rad Protein Assay using BSA (Pierce) as a standard. Dipeptides were synthesised by GenScript Corporation (New Jersey, USA) to >98% purity.
In vitro binding assays and affinity isolation of N-end rule proteins
For binding assays using purified proteins, N-end rule substrates were added to Ni-NTA agarose (10 μl settled beads) without and with immobilised ClpS-His6 or ClpSDD/AA-His6 (∼10 μg) in 500 μl of buffer B (20 mM Hepes-KOH pH 7.5, 50 mM KOAc, 10 mM MgOAc, 10% glycerol), including 0.5% (v/v) Triton X-100 and 10 mM imidazole, and incubated, end-over-end, for 30 min at 4°C. The beads were washed with 500 μl of buffer B, including 0.25% (v/v) Triton X-100 and 20 mM imidazole, and N-end rule proteins were eluted with FR dipeptide (1 mg/ml) in wash buffer. For preparation of E. coli lysates (wild-type, ΔclpA and Δaat), cell pellets were resuspended in ice-cold-binding buffer (3 ml/g of wet cells) supplemented with EDTA-free protease cocktail inhibitor (Roche), lysozyme (0.2 mg/ml) and DNase I (10 μg/ml) and mixed (30 min) before clarification by centrifugation. E. coli lysates (100 mg/100 μl of Ni-NTA agarose beads) were mixed end-over-end for 30 min at 4°C with pre-equilibrated Ni-NTA agarose either lacking or containing immobilised ClpS-His6 or ClpSDD/AA-His6 (∼100 μg). The beads were collected by centrifugation, the unbound fraction removed and the bead slurry transferred to 1 ml Mobicol columns (MoBiTec) and washed with 8 ml of binding buffer and then 4 ml of wash buffer. Proteins specifically bound to ClpS were eluted with 100 μg FR or Met-Ser dipeptide in buffer B, including 20 mM imidazole.
Substrate degradation assays
Degradation of all GFP-fusion proteins was monitored by changes in fluorescence (excitation 410 nm and emission 500 nm) on a Molecular Devices Spectra Max M2. The reactions were carried out as described (Dougan et al, 2002b). Non-fluorescent degradation assays were preformed as described earlier (Erbse et al, 2008). Unless otherwise stated, 1.2 μM ClpA, 2.8 μM ClpP and 1.2 μM ClpS (wild-type or mutant) were used. Samples were removed from the reactions at the indicated time points and stopped by the addition of sample buffer. After separation by SDS–PAGE, proteins were detected by Coomassie blue staining and the bands were quantified using the GelEval1.21 software (Frogdance).
Coimmunoprecipitation of ClpS with anti-Dps antiserum
Recombinant purified Dps2−167 or Dps6−167 (1 μM) or buffer alone were pre-mixed in ice with and without ClpS (1 μM) in IP buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 40 mM MgOAc, 10% (v/v) glycerol, 0.25% (v/v) Triton X-100). The protein samples were then added to Protein A Sepharose containing pre-bound antibodies directed against Dps and incubated end-over-end for 2 h at 4°C. The unbound fraction was removed, the beads washed with ice-cold-IP buffer followed by elution of bound proteins with 50 mM glycine–HCl, pH 2.5.
2D gel electrophoresis
ClpS-interacting proteins (90–120 μg) were acetone precipitated and then solubilised in strip rehydration solution (8 M Urea, 2% (w/v) CHAPS, 0.5% (v/v) IPG buffer, 20 mM DTT, 0.002% (w/v) Bromophenol blue). Using an Ettan IPGphor II Manifold with cup loading, protein samples were applied to the anodic end of rehydrated (>12 h in rehydration solution) Immobiline DryStrip gels (13 cm, linear pH 4–7 gradient strips), and isoelectric focussing performed at 100 V for 0.5 h, 100–500 V for 2 h, 500–1000 V for 1 h, 1000–8000 V for 3.5 h followed by 8000 V for 1 h using an Ettan IPGphor II unit at 20°C. Gel strips were incubated (15 min) in equilibrated buffer (50 mM Tris–HCl pH 8.8, 6 M Urea, 30% (v/v) glycerol, 2% (w/v) SDS), containing 1% (w/v) DTT followed by incubation (15 min) in equilibration buffer containing 135 mM iodoacetamide. The second-dimension protein separation was performed using Tricine SDS–PAGE with a 4–16% linear polyacrylamide gradient run at 30 mA. Proteins were stained with Coomassie blue.
Edman degradation and mass spectrometry
Protein on PVDF membrane was subjected to 5–6 cycles of automated Edman degradation using an Applied Biosystems 494 Procise Protein sequencing system. Commassie-stained proteins were excised from 1D or 2D-SDS–PAGE gels and in-gel proteolytic digestion performed with trypsin or GluC. The peptides from the GluC digest were spotted on an Anchorchip target using DHB as matrix and analysed by MALDI-TOF MS (Bruker-Daltonics). The tryptic peptides were injected onto a reversed phase column (LC-Packings pepmap100, C18 300 μm) under the control of an Ultimate3000 HPLC (Dionex) and eluted into an HCTultra ion trap (Bruker Daltonics) for MS and MS/MS analysis.
In vitro aminoacyl-transferase assay
E. coli tRNAPheV was amplified by PCR from genomic DNA using KNT50 (5′-tgtaatacgactcactatagcccggatagctcagtcg-3′), which includes a T7 promoter sequence and KNT51 (5′-tggtgcccggactcggaatcgaac-3′). Transcription of tRNAPheV was performed using T7 RNA polymerase (Riboprobe in vitro transcription system, Promega) essentially as described by the manufacturer with the following exceptions: 50–100 ng of PCR-amplified DNA template was used in a 100-μl reaction. The aminoacylation of tRNAPhe and N-terminal aminoacylation (conjugation) of substrates were performed essentially as described (Suto et al, 2006). Specifically, 5 μM α-casein fragment 90–95 (RYLGYL, Sigma) or 5 μM PATase (PATase, L-PATase and F-PATase) was incubated at 37°C for 8 min in reaction buffer (50 mM Tris–HCl pH 8.0, 100 mM KCl, 10 mM MgOAc, 1 mM DTT containing 2 mM ATP, 150 nM tRNAPhe, 35 μM [14C]-Phe (18.3 GBq/mmol, PerkinElmer), 500 Units E. coli aminoacyl-tRNA synthetase (Sigma) and 0.12 μM LFTR). Samples were analysed by 12.5% glycine SDS–PAGE (for PATase) or 16.5% Tricine SDS–PAGE (for α-casein fragment 90–95). After separation, polypeptides were fixed, gels dried and radioactivity detected by phosphoimage analysis, after parallel exposure of the gels, using a Typhoon Trio Variable Mode Imager (GE Healthcare). Radiolabelled substrates were quantified using the GelEval software.
Pulse-chase IP experiments
Selected E. coli strains (wild-type, Δaat, ΔclpA, ΔclpS and ΔygjG) were grown overnight at 37°C in 2xYT media. The overnight culture was washed twice in minimal media (1 × M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% (w/v) glucose) before the cells were finally resuspended in minimal media to a starting OD600 of ∼0.1. Cells were then grown at 37°C, with shaking, to an OD600 of 2.5–3.0, labelled with [35S]-Met/Cys (50 μCi/ml) for 1 h and then the labelled cellular proteins were chased by the addition of 0.1% (w/v) Met. After the addition of unlabelled Met, cells were collected for analysis at various times (as indicated). Each cell pellet was resuspended in lysis buffer (50 mM Tris–HCl pH 8, 5 mM EDTA, 2 mg/ml lysozyme, 1 mM PMSF), then supplemented with 1.5% (w/v) SDS and heat treated for 5 min at 95°C. Cell lysates (46 μl) were then diluted 12-fold into pulse-chase (PC) buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 0.5% (v/v) Triton X-100, 1 mM EDTA, 1 mM PMSF) and after centrifugation (10 min at 13 000 r.p.m.), the supernatants were subjected to IP. Briefly, anti-PATase antiserum covalently cross-linked to Protein A Sepharose (25 μl settled beads) was added to 500 μl of each cell lysate and incubated at 4°C for 1 h with end-over-end mixing. The Sepharose beads were washed three times with 500 μl of PC buffer and then treated with SDS loading buffer and heat treated for 5 min at 95°C. Radiolabelled proteins were then separated by 12.5% glycine SDS–PAGE, fixed in the gel (30% (v/v) methanol and 10% (v/v) acetic acid) and dried before phosphoimage analysis.
Miscellaneous
Glycine and Tricine SDS–PAGE was performed essentially as described (Laemmli, 1970; Schagger and von Jagow, 1987). Polyclonal antibodies against ClpS, Dps and PATase were raised in rabbits using untagged purified recombinant proteins. Western transfer was performed using a semi-dry transfer system and proteins detected using horseradish peroxidase-coupled secondary antibodies together with enhanced chemiluminescence detection reagents (GE Healthcare) and ChemiGenius2 Bio-Imaging System (SynGene). Digital images were captured using GeneSnap software (SynGene).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Information
Acknowledgments
We thank Rohan Baker for providing plasmids pHUE and pUsp2cc, Fiona Durand for technical assistance and APAF for performing Edman degradation. E. coli strains used in this study were provided by the National BioResource Project (NIG, Japan). DAD and KNT are supported by QEII Fellowships from the Australian Research Council (ARC). RLN is the recipient of an Australian Postgraduate Award. This work was funded by an ARC Discovery Project DP0450051 to DAD and KNT.
References
- Almiron M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6: 2646–2654 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT (2004) An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci 13: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E (2004) DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res 32: 5935–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Bukau B (2002a) Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol Life Sci 59: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B (2002b) ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683 [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Erbse AH, Wagner JN, Truscott KN, Spall SK, Kirstein J, Zeth K, Turgay K, Mogk A, Bukau B, Dougan DA (2008) Conserved residues in the N-domain of the AAA+ chaperone ClpA regulate substrate recognition and unfolding. FEBS J 275: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Sauer RT, Baker TA (2004) Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev 18: 2292–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11: 671–683 [DOI] [PubMed] [Google Scholar]
- Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T (2006) The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics 5: 2336–2349 [DOI] [PubMed] [Google Scholar]
- Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A, Igarashi K (1995) Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J Biol Chem 270: 18831–18835 [DOI] [PubMed] [Google Scholar]
- Gonzalez M, Rasulova F, Maurizi MR, Woodgate R (2000) Subunit-specific degradation of the UmuD/D' heterodimer by the ClpXP protease: the role of trans recognition in UmuD' stability. EMBO J 19: 5251–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Hengge-Aronis R (2003) Regulation by proteolysis in bacterial cells. Curr Opin Microbiol 6: 163–172 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lupas AN, Koretke KK (2003) Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol 141: 77–83 [DOI] [PubMed] [Google Scholar]
- Martinez A, Kolter R (1997) Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179: 5188–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B (2007) The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Nair S, Finkel SE (2004) Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186: 4192–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Uhlmann F, Nasmyth K, Varshavsky A (2001) Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410: 955–959 [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonova NN, Smirnov SV, Altman IB, Ptitsyn LR (2003) Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol 3: 2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Shrader TE, Tobias JW, Varshavsky A (1993) The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol 175: 4364–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer RL (1974) Aminoacyl-tRNA transferases. Adv Enzymol Relat Areas Mol Biol 40: 91–139 [DOI] [PubMed] [Google Scholar]
- Soffer RL, Savage M (1974) A mutant of Escherichia coli defective in leucyl, phenylalanyl-tRNA-protein transferase. Proc Natl Acad Sci USA 71: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K (2006) Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J 25: 5942–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Kwon YT (2007) The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci 32: 520–528 [DOI] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A (1991) The N-end rule in bacteria. Science 254: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Varshavsky A (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2005) Regulated protein degradation. Trends Biochem Sci 30: 283–286 [DOI] [PubMed] [Google Scholar]
- Wang KH, Sauer RT, Baker TA (2007) ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev 21: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K (2007) Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449: 867–871 [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401: 90–93 [DOI] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286: 1888–1893 [DOI] [PubMed] [Google Scholar]
- Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA (2002) Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol 9: 906–911 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Information