Abstract
Influenza virus RNA-dependent RNA polymerase is a multi-functional heterotrimer, which uses a ‘cap-snatching' mechanism to produce viral mRNA. Host cell mRNA is cleaved to yield a cap-bearing oligonucleotide, which can be extended using viral genomic RNA as a template. The cap-binding and endonuclease activities are only activated once viral genomic RNA is bound. This requires signalling from the RNA-binding PB1 subunit to the cap-binding PB2 subunit, and the interface between these two subunits is essential for the polymerase activity. We have defined this interaction surface by protein crystallography and tested the effects of mutating contact residues on the function of the holo-enzyme. This novel interface is surprisingly small, yet, it has a crucial function in regulating the 250 kDa polymerase complex and is completely conserved among avian and human influenza viruses.
Keywords: influenza virus, interface, mutant, RNA polymerase, structure
Introduction
Influenza kills >50 000 people in the United States every year on an average (WHO, 2003), and estimates of the death toll in the 1918 pandemic range up to 50 million people worldwide (Taubenberger et al, 2005). Recent outbreaks of highly pathogenic avian influenza viruses in Asia have rapidly spread across continents, and present vaccines and medication seem unlikely to greatly alleviate any epidemic or pandemic, should these viral strains adapt to human hosts (Peiris et al, 2007). The viral RNA (vRNA) polymerase is not yet a target of any approved pharmaceutics, but has recently become a focus for the development of new anti-influenza drugs, as it is highly conserved in strains of influenza virus, which infects both birds and human beings. The vRNA polymerase carries out a number of essential processes in the viral life cycle, but many of these and their regulation remain poorly understood (Elton et al, 2005). The three subunits, PB1, PB2, and PA, play distinct roles within the polymerase, and are all essential for viral replication; but despite considerable functional analyses, relatively little is known about their structure (Tarendeau et al, 2007, 2008; Guilligay et al, 2008; He et al, 2008; Obayashi et al, 2008; Dias et al, 2009; Yuan et al, 2009). Here, we have solved the crystal structure of a complex formed by fragments of PB1 and PB2. This subunit interface is essential for transcription initiation. Similar to the PA–PB1 interface (He et al, 2008; Obayashi et al, 2008), this interaction depends on a short N-terminal fragment of one protein, which raises the possibility that a suitable small molecule may be able to disrupt the interaction in vivo and significantly restrict viral replication.
The RNA polymerase of influenza A virus forms an RNP complex with each of eight negative-strand RNA genome segments and nucleoprotein packaged within the mature virion (Portela and Digard, 2002). Once an RNP complex is released into the host cell cytoplasm, it uses nuclear import machinery to move into the nucleus (Whittaker and Digard, 2005), where it initiates viral mRNA transcription by the process of ‘cap snatching' (Plotch et al, 1981). This process involves cutting an mRNA cap-containing oligonucleotide from host cell pre-mRNA to extend into viral mRNA, which then polyadenylates at the 3′ end (Poon et al, 1999; Zheng et al, 1999). The polymerase synthesizes viral genomic RNA (vRNA) and complementary RNA (cRNA) in appropriate proportions, each with the correct ends and with no cap. The regulation of these processes is not well understood, though some details are known. Cap binding to PB2, for example, requires vRNA binding (Cianci et al, 1995; Li et al, 1998). This may reflect interactions between the three subunits, all of which are essential for both RNA transcription and replication (Huang et al, 1990; Nagata et al, 2008). The nature of the PA–PB1 contact has been determined by functional studies and characterized crystallographically (He et al, 2008; Obayashi et al, 2008). PB2 can also interact with PB1, but there is no direct interaction between PA and PB2 (St Angelo et al, 1987; Digard et al, 1989). Although additional regions of contact are reported between these subunits (Biswas and Nayak, 1996), mutational analyses indicate that the C-terminus of PB1 (residues 712–746) forms the core interaction with the N-terminus of PB2 (Gonzalez et al, 1996; Poole et al, 2007). Toyoda et al (1996) used an immunoprecipitation assay and deletion mutants to show that the N-terminal 249 amino acid residues of PB2 can bind to PB1. However, a subsequent study from the same laboratory detected PB1 by co-precipitation with PB2 minus the N-terminus, suggesting the possibility of another region of interaction with PB1 (Ohtsu et al, 2002). This was supported by Poole et al (2004), who also identified a second PB1-binding site in the C-terminal half of PB2. To clarify the interaction between the two subunits, we have independently determined the binding domains located at the extreme C-terminus of PB1 and the N-terminus of PB2. The crystal structure of the complex formed by these regions has been determined and refined to 1.7 Å resolution, and transcriptional activity assays used to observe the effects of mutating contact residues at this interface.
Results
PB1–PB2 interaction domain
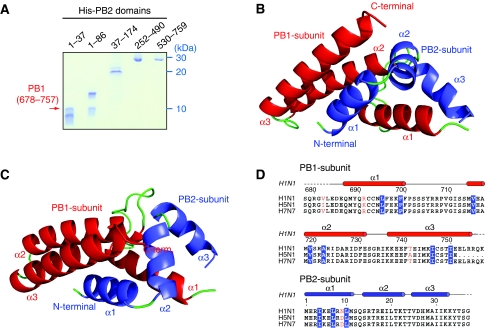
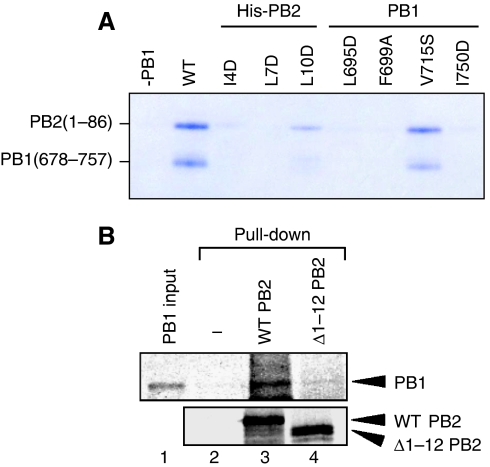
To characterize the interaction between PB1 and PB2 in more detail, we used a co-precipitation assay to observe binding between fragments of the C-terminus of PB1 with the N-terminus of PB2. It was found that only a short region, residues 678–757, of PB1 was required for tight binding, in agreement with the earlier results (Poole et al, 2007). This fragment (called PB1-C) was tested with residues 1–37, 1–86, 37–174, 252–490, and 530–759 of PB2, but only the 1–37 and 1–86 fragments of PB2 showed binding (Figure 1A). Residues 37–177 of PB2 did not bind to the C-terminus of PB1, in agreement with Perales et al (1996), who showed that deletion of 27 amino acids from the N-terminus of PB2 dramatically diminished viral RNA polymerase activity. They further showed that the N-terminal 124 residues of PB2 behave as a dominant-negative inhibitor of virus transcription. Furthermore, a PB2-specific monoclonal antibody raised against the N-terminus of the protein is able to inhibit the initiation step of transcription in vitro, presumably by interfering with binding to PB1 (Barcena et al, 1994; Ochoa et al, 1995). In the co-precipitation assay, PB2 residues 530–759, including the proposed second PB1-binding site, were not found to interact with the C-terminus of PB1. These results clearly show that the C-terminus of PB1 and the N-terminus of PB2 form the principal, if not only, subunit interface. The interacting fragments are notably short sequences from each subunit, only 80 and 37 residues of PB1 and PB2, respectively. Together, these fragments comprise around only 6% of the total molecular weight of the holo-enzyme, yet are responsible for crucial communication between subunits.
Figure 1.
The nature of the PB1–PB2 interface. (A) Co-precipitation experiments of the C-terminus of PB1 (residues 678–757) co-expressed with different fragments of PB2 carrying a hexa-histidine tag at the N-terminus. The red arrow indicates the PB1-C peptide, which is retained on a Ni-NTA column only with the constructs in lanes 1 (residues 1–37) and 2 (residues 1–86). (B) An overall ribbon diagram showing the structure of the complex, with helices from PB1 coloured red, and helices from PB2 coloured blue. Coil regions are coloured green. (C) The same model as (B), but rotated 90° around a horizontal axis to show the separation between the three helices of the N-terminal peptide of PB2. The principal contacts all involve helix 1 of PB2 (residues 1–12). (D) The sequences of the complexed fragments with a sequence alignment of human (H1N1) influenza, an avian strain (A/Duck/Hong Kong/2000, H5N1), and H7N7 (A/Equine/London/1416/1973). Secondary structure is indicated with red or blue bars showing helices in PB1 and PB2, respectively, and broken lines showing disordered regions. Amino acid residues shown in white on blue form hydrophobic contacts across the PB1–PB2 interface; residues shown in red are not conserved between different viral strains, and, therefore, unlikely to have an essential function. Overall, the interface region between PB1 and PB2 is very highly conserved.
The crystal structure of the complex form
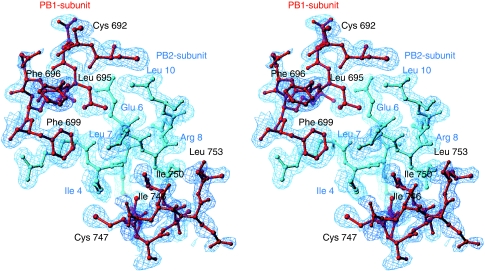
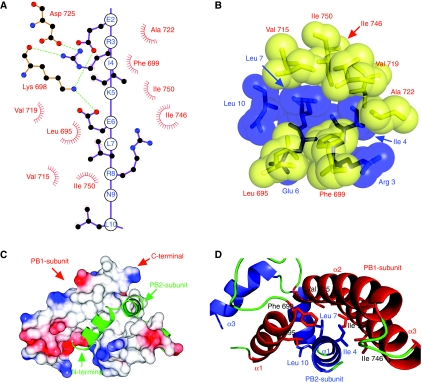
Co-expression in Escherichia coli of PB1-C (residues 678–757 of PB1) with PB2-N (residues 1–37 of PB2) yielded a stable complex that we could purify and crystallize with two copies in the asymmetric unit. The X-ray crystal structure was initially refined to 2.1 Å resolution in space-group P21, revealing the complex to be a single compact domain (Figure 1B and C), which is very highly conserved among all influenza virus strains (Figure 1D). Nearly all the residues of the two polypeptide chains are visible in the electron density, with only a few residues at the chain termini being disordered. The final electron density map at 1.7 Å resolution (in space-group C2) covering key interface residues is shown in Figure 2. Both PB1-C and PB2-N consist of three α-helices, but neither polypeptides alone adopt a stable tertiary structure. Helix 1 of PB2-N lies against helices 2 and 3 of PB1-C, and helix 1 of PB1-C is held between all three helices of PB2-N. PB2-N has an extended shape with almost no intermolecular contacts between its three helices. N-terminal fragments of PB2 could be readily expressed and purified with an N-terminal GST tag, but these fusion proteins showed no binding to PB1 in vitro, suggesting that they are not properly folded. The complex could only be produced by co-expression of the PB1 and PB2 domains. The interface buries over 1400 Å2 of surface area, consistent with tight binding, and includes four salt bridges: between Glu 2 and Lys 698, Arg 3 and Asp 725, Arg 3 and Lys 698, and Glu 6 and Lys 698 (Figure 3A). All the other eight hydrogen bonds between the polypeptides involve main-chain atoms. Analysis of the model by PISA (Krissinel and Henrick, 2007) suggested that a similar interface is present in the KIX domain of mouse CREB-binding protein (PDB 1kdx), but direct superposition of the model shows a rather different interaction between polypeptide chains. No subunit interface in PDB was found to share the same ‘3 plus 3' helix structure, and the most similar ones, including 1kdx, have a buried surface area less than half that of PB1–PB2. Unlike the interaction between the C-terminus of PA and the N-terminus of PB1 (He et al, 2008; Obayashi et al, 2008), which has a predominantly hydrophobic character, the PB1–PB2 interface shows more polar interactions and is more extensive in sequence length and buried surface area (Figure 3B and C). However, the majority of the interaction energy appears to be contributed by helix 1 of PB2-N, which involves not only the four salt bridges to PB1-C, but also the key apolar contacts, such as Ile 4 and Leu 7 (Figure 3B and D). These two residues are completely buried in the protein interface.
Figure 2.
Electron density map. A stereo view of the final electron density map (2mFo-DFc) covering the key residues of the complex. PB1 is shown in red and PB2 in blue. The map has a resolution of 1.7 Å and is contoured at 1.3 σ.
Figure 3.
Interactions between PB1 and PB2. (A) A schematic diagram showing the interactions between the two polypeptides. Helix 1 of PB2-N is drawn as a linear model, with the side chains touching PB1 shown in a two-dimensional ball and stick form. Lys 698 and Asp 725 of PB1 form the only salt bridges across the interface, shown as green dotted lines. These salt bridges are not found in every copy of the complex, and mutation studies (replacing Glu 2 or Arg 3 with alanine) show that they have little effect on PB1–PB2 binding (data not shown). Apolar residues of PB1 are shown in red as simple dashed arcs to indicate hydrophobic contacts between 3.4 and3.9 Å in length. This figure was prepared using LIGPLOT (Wallace et al, 1995). (B) A space-filling representation, with PB1 residues shown in yellow and labelled in red. PB2 residues are shown and labelled in blue. The van der Waals surface of each atom is shown semi-transparent. (C) Schematic diagram showing the molecular surface of PB1, coloured by charge (blue positive, red negative). The potential scale ranges from −1 kT/e (red) to 1 kT/e (blue). PB2 is shown as a green ribbon to reveal the PB1-binding surface beneath it is largely apolar. (D) A ribbon diagram showing the helices of PB1-C and PB2-N in red and blue, respectively, with coil regions in green. Side chains selected for mutagenesis are shown as stick models.
The RNA polymerase activity of PB1 or PB2 double mutants
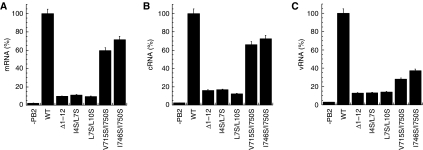
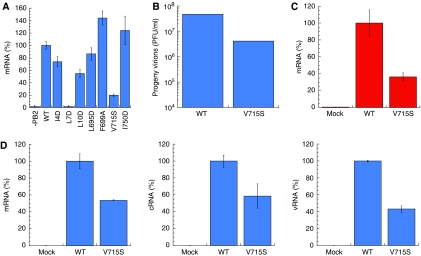
To test the model functionally, we examined the effects of different PB2 mutations on the level of viral RNA synthesis in vivo (Figure 4). Without PB2, no product RNA is detectable in the assay. Deleting helix 1 (residues 1–12) of PB2 reduces the RNA polymerase activity by 90%. Mutant PB2-N in which Ile 4 and Leu 7 were replaced with serine residues also showed a dramatic reduction in product RNA yield. Simultaneously replacing Leu 7 and Leu 10 with serine produced a similar effect. Two more double mutants were prepared, in which PB1 residues Val 715 and Ile 750, or Ile 746 and Ile 750 were replaced with serine. Both these PB1 mutants showed a significantly reduced yield of vRNA and a substantial but smaller drop in the yield of cRNA and mRNA (Figure 4). These results are compatible with the structural model, in which Leu 7 is buried within the hydrophobic core of the structure. The side chain of Val 715 is buried close to that of Leu 7, but nearby polar residues at the protein surface, including Ser 713 and Arg 754, should be able to accommodate a serine side chain comfortably. Ile 750 lies close to the protein surface in the model, which presumably allows a polar residue to occupy this position without preventing PB1–PB2 binding.
Figure 4.
RNA synthesis activity of PB1 or PB2 double mutants in HeLa cells. Bar charts showing the level of viral mRNA (A), cRNA (B), or vRNA (C) synthesis by different RNA polymerase double mutants, compared with that of the wild-type (WT) polymerase or with PB2 absent (−PB2). RNA was isolated from HeLa cells transfected with plasmids for expression of viral RNP components. Using primers specific for viral mRNA, cRNA, or vRNA, the production of each RNA type could be separately assessed by quantitative PCR (see Materials and methods). In the absence of the PB2 subunit, enzyme activity is negligible. The results are mean and s.d. for three independent experiments.
The RNA polymerase activity of PB1 or PB2 single mutants
Further experiments were conducted with mutant PB2-N, in which a single residue was changed at a time. The RNA synthesizing activity significantly dropped in the case of the I4D mutation, but a much more dramatic reduction in mRNA yield was found when Leu 7 was replaced with aspartate (Figure 5A). Similar experiments were performed, in which PB1 residues Leu 695 and Ile 750 were individually replaced with aspartate, Phe 699 with alanine, and Val 715 with serine. None of these mutants showed a significantly reduced yield of mRNA except V715S, for which the mRNA yield dropped by about 80% (Figure 5A). Leu 695 and Ile 750 are accessible to solvent water, which presumably allows an aspartate residue to replace either of them without preventing PB1–PB2 binding. The nearby Arg 8 on PB2 may even allow a novel interaction to form with the carboxylate group of Asp 750 in the mutant. The side chains of both Val 715 and Phe 699 are buried close to that of Leu 7. Replacing Phe 699 of PB2 with alanine is expected to introduce a substantial cavity within the interface, and extra flexibility may account for the significantly enhanced activity in the functional assay with this mutant. The very strong depression of the enzyme activity in the V715S mutant was not predicted from the crystal structure, as mentioned above. In particular, the model gives no reason to believe that the valine to serine mutation will prevent or greatly weaken the PB1–PB2 interaction, which prompted us to investigate this mutation further.
Figure 5.
RNA synthesis activity of PB1 or PB2 single mutants. (A) Bar chart showing the level of viral mRNA synthesis of different RNA polymerase single mutants compared with that of the wild-type polymerase (WT). The mRNA production in HeLa cells was assayed as in Figure 4A. The L7D mutation effectively abolishes polymerase activity. (B) The yield of progeny virus. MDCK cells were infected with either wild-type or PB1-V715S virus at an MOI=1. After 24 h post infection, the supernatants were collected, and the plaque titer was determined using MDCK cells. The wild-type virus showed roughly 10-fold greater yield than the PB1-V715S mutant. (C) The level of vRNA synthesis in MDCK cells infected with wild-type virus or PB1-V715S virus. MDCK cells were infected with either wild-type or PB1-V715S virus in the presence of 100 μg/ml cycloheximide to block protein synthesis. The real-time quantitative PCR assays were carried out with a primer set specific for NP mRNA, showing that mRNA synthesis is severely curtailed by the single mutation. (D) An identical experiment to (B), but without the addition of cyclohexmide. Production of mRNA (left-hand panel), cRNA (centre) and segment 5 vRNA (right-hand panel) were assayed separately. The mutant polymerase shows significantly reduced activity for each product. The β-actin mRNA was used as an internal control for the whole procedure (see Materials and methods).
Analysis of the Val715 mutation in PB1
By reverse genetics, a recombinant virus carrying a PB1 genome segment possessing the V715S mutation was created. The seven other genome segments are all wild type. This allowed us to analyse the effect of the single-site mutation at the level of primary transcription from infecting vRNP. We succeeded in recovery of V715S virus, although the virus titer was less than that of wild type (Figure 5B). As RNA polymerase is a structural part of the vRNP, the isolation of virus indicates that the PB1–PB2 interaction is not prevented completely. We examined the level of viral primary transcription from infecting vRNP in the presence of cycloheximide (CHX), a potent protein synthesis inhibitor. It is known that inhibition of viral protein synthesis represses new vRNP formation, thereby resulting in degradation of replicated viral genomic RNA, but not of viral mRNA (Vreede et al, 2004). In this way, we could evaluate the viral transcription activity independent of viral genome replication or the efficiency of RNA polymerase complex formation. The level of primary transcription from infecting V715S vRNP was found to be decreased significantly compared with that from wild type (Figure 5C). As expected from the lower level of primary transcription, the synthesis of vRNA, cRNA, and viral mRNA in cells infected with V715S virus was also reduced in the absence of CHX (Figure 5D).
The results of the in vitro and in vivo functional assays strongly suggest that the Val 715 residue in PB1 is involved in one or more steps in RNA synthesis. To exclude the possibility that the V715S mutation simply blocks PB1–PB2 binding, co-precipitation experiments were carried out, in which the co-expressed PB1–PB2 complex was attached to a Ni-NTA column using a histidine tag fused to PB2-N. The column was washed before eluting with imidazole, and the loss or retention of PB1 was determined by gel electrophoresis. Free PB2-N was easily degraded and not detected in this assay. The L695D, F699A, and I750D mutants all showed no binding to PB2-N, whereas the V715S mutation allowed PB1 to remain bound to PB2, as expected from the crystal structure (Figure 6A). Clearly, the co-precipitation assay is not a test of equilibrium binding, but also depends on the dissociation rate of the partner proteins. Nevertheless, the results irrefutably show that the V715S mutation does not block PB1–PB2 binding. A weakened interaction between PB1 and PB2 is not apparently incompatible with the enzyme activity under the assay conditions used, although full-length PB1 and PB2 were used in the polymerase activity assays. The V715S mutant, however, shows both significant PB2 binding and greatly reduced enzyme activity, which suggests that a slightly altered mode of interaction may have an effect on the efficiency of the polymerase activity. The enzyme activity is not lost in this case by failure of PB1 and PB2 to bind to each other.
Figure 6.
Binding assay of PB1 and PB2 mutants. (A) Co-precipitation experiments using PB1-C (residues 678–757) co-expressed with the N-terminus of PB2 (residues 1–86) carrying a hexa-histidine tag at the N-terminus. The complex was loaded on a nickel affinity column and washed before eluting with imidazole and assaying by SDS–PAGE gel. The Coomassie blue stained gel shows that the PB2 fragment is degraded in the absence of PB1 (lane: −PB1). The wild-type PB1 sequence and the V715S mutant both bound strongly to wild-type PB2-N, giving a band for each denatured polypeptide. All the other single mutations tested show significant or complete loss of binding, and consequent degradation of PB2-N. (B) Immunoprecipitation assay of PB1–PB2 interaction. Full-length wild-type PB2, PB2(Δ1–12), and full-length wild-type PB1 were separately expressed in an in vitro (rabbit reticulocyte lysate) translation system (Promega). 35S-methionine labelling was carried out according to the manufacturer's protocol. The recombinant PB1 was incubated with wild-type PB2 (lane 3) or PB2(Δ1–12) (lane 4) at room temperature for 1 h, and then immunoprecipitated using anti-PB2 antibody and protein A-sepharose beads. Protein eluted from the beads was analysed by 7.5% acrylamide SDS–PAGE and visualized by autoradiography. Lane 1 shows PB1 alone, and lane 2 represents a mock experiment. Lane 4 shows a faint band corresponding to PB1, indicating a much weaker interaction with the mutant PB2 than with wild type (lane 3).
The crystal structure presented here shows that the PB1-C and PB2-N peptides form a novel fold depending on the presence of both partner chains. This structure clearly represents the principal contact between the subunits, but does not exclude the possibility that there is another contact surface in the holo-complex. In fact, we found by pull-down assay that full-length PB2 lacking helix 1 does apparently interact with full-length PB1, albeit weakly (Figure 6B), in agreement with Poole et al who showed that PB2 interacts with PB1 through its N- and C-termini (Poole et al, 2004). The weak band (Figure 6B, lane 4) in pull-down assays is, however, far from definitive evidence of a specific, biologically relevant interaction. In contrast, subunit interaction through the PB1-N- and PB2-C-termini is readily demonstrable without resorting to autoradiography.
Discussion
Earlier reports have shown that a mutation in one of the polymerase subunits may affect the function of other subunits and be suppressed by a compensating mutation in another subunit (Treanor et al, 1994; Fodor et al, 2002). This suggests that there are regulatory mechanisms of the different polymerase functions involving communication between subunits, and that Val 715 in PB1 may assist the transcription of virus genes by signalling between PB1 and PB2. In this scenario, the V715S mutation allows PB1 and PB2 to bind, but interferes with communication between them. A major reorganization of T7 RNA polymerase occurs during RNA synthesis (Yin and Steitz, 2002). If the influenza RNA polymerase undergoes a similar, drastic conformational change, then inhibition of the switch, or destabilization of either state, could also explain the reduced polymerase activity of the V715S mutant. The crystal structure presented here offers no obvious explanation for the loss of activity, as a serine side chain is not much smaller than valine, and the hydroxyl group could hydrogen bond to nearby solvent water. The tight PB1–PB2 binding observed with the V715S mutant is, therefore, completely in agreement with the molecular model. As the V715S mutant does not weaken the subunit interaction, its effects apparently occur through structural or dynamic changes in the complex during RNA synthesis. The strong sequence conservation of the PB1–PB2 interface also argues against a simple, rigid contact, which simply serves to hold the two subunits together.
Functional studies confirm the importance of helix 1 of PB2-N to viral RNA synthesis. As shown in Figure 4, deleting this helix (residues 1–12) greatly reduces the RNA polymerase activity. Additional experiments conducted with mutant PB2 also showed a dramatic reduction in mRNA synthesis with various interface mutants, such as L7D, which blocks PB1 binding to PB2-N. In contrast, some of the PB1 mutants show very different effects on subunit binding and polymerase activity. For example, the F699A and I750D mutants show weak PB2 binding, but increased enzyme activity. These results show that the PB1–PB2 interface is not merely a passive attachment surface by which the partner proteins come together, but that it has an important function in regulating the overall enzyme activity. This interface is surprisingly small, yet it has a crucial function in regulating the 250 kDa polymerase complex. It is completely conserved among avian and human influenza viruses, notably including strains associated with high mortality, but unlike any other structure in the Protein DataBank. Given its importance to viral replication and strict conservation, the PB1–PB2 interface appears as a promising target for novel anti-influenza drugs of use against all strains of influenza A virus. It is hoped that the structure presented here will assist the search for such compounds.
Materials and methods
Cloning, expression, and purification of the PB1–PB2 complex
Cloning and purification were essentially carried out as for the PA–PB1 complex described earlier (Obayashi et al, 2008). The sequences used are from influenza A/Puerto Rico/8/1934. Fragments of the PB2 gene encoding residues 1–37, 1–86, 37–174, 252–490, and 530–759 were cloned into pET28b with a hexa-histidine tag and TEV cleavage site at the N-terminus. The PB1-C coding region was cloned downstream of the PB2 gene with a Shine–Dalgarno sequence. The resulting co-expression plasmid was transformed into E. coli BL21(DE3)RILP codon-plus strain, and cells were cultured at 15°C overnight after induction with 0.5 mM IPTG. The PB1–PB2 complex was purified by chromatography using Ni-NTA agarose (Qiagen), followed by SP and Q (GE Healthcare) sepharose. The histidine tag was removed by TEV protease digestion after Ni-NTA chromatography, and the purified complex was then concentrated to 5 mg/ml by centricon YM-3 (Millipore) for crystallization. The co-precipitation assay was carried out basically by the same method as for purification of above PA–PB1 complex. After purification by Ni-NTA, proteins were analysed by SDS–polyacrylamide gel electrophoresis (15%) and staining with Coomassie blue.
Crystallization and data collection
Crystals of the PB1–PB2 complex were originally grown by the hanging drop vapour diffusion method against a crystallization buffer containing 0.1 M potassium phosphate (pH 5.8) and 15% PEG 4000 at 20°C. Diffraction data were collected from a crystal cooled to −180°C. Crystallization buffer containing 25% glycerol was used to prevent icing. X-ray diffraction data were collected at beam-line 17A at the Photon Factory in Japan. One selenomethionyl-substituted crystal was used to collect datasets at three different X-ray energies around the Se-K absorption edge. Data were measured using an ADSC Quantum 270 CCD detector. The crystals formed in space-group P21 with a=44.27 Å, b=61.48 Å, c=45.47 Å, β=103.4°, and contained two copies of the complex in the asymmetric unit. Subsequently, higher quality crystals were grown using buffer containing 0.8 M sodium citrate and 20% PEG 4000 at 20°C. The crystals formed in space-group C2 with a=60.70 Å, b=69.99 Å, c=61.35 Å, β=97.9°, and contained two molecules in the asymmetric unit. All diffraction data integration, scaling, and merging were performed using HKL2000 and SCALEPACK (Otwinowski and Minor, 1997).
Structure determination and refinement
The positions of 12 out of 14 possible selenium sites were found by analysing the P21 multi-wavelength datasets with SHELXC and SHELXD (Sheldrick, 1986). Phase determination was carried out with SOLVE (Terwilliger and Berendzen, 1999). After solvent flattening with RESOLVE (Terwilliger, 2003), the electron density was interpreted and traced using COOT (Emsley and Cowtan, 2004). The model was refined with REFMAC (Murshudov et al, 1997). Solvent molecules were placed at positions where spherical electron density peaks were found above 1.3 σ in the ∣2Fo−Fc∣ map and above 3.0 σ in the ∣Fo−Fc∣ map, and where stereochemically reasonable hydrogen bonds were allowed. Structural evaluation of the final models of the PB1–PB2 complex using PROCHECK (Laskowski et al, 1993) indicated that 95% of the residues are in the most favourable regions of the Ramachandran plot, with no residues in ‘disallowed' regions. The final model contains 109 of the 117 residues in the sequence, with residues 678–684 of PB1 and 36–37 of PB2 unobserved. A summary of the data collection and refinement statistics is given in Table I. The structure in space-group C2 was solved by molecular replacement, and refinement carried out as for the first structure. Atomic coordinates and structure factors of the complex have been deposited in the Protein Data Bank under accession code 2ZTT (P21) and 3A1G (C2).
Table 1.
Data collection and refinement statistics
| Data sets | Se-Met | Remote | Inflection | Peak |
|---|---|---|---|---|
| Space group | C2 | P21 | ||
| Unit cell (Å) | a=60.70, b=69.99, c=61.35, β=97.9° | a=44.27, b=61.48, c=45.47, β=103.4° | ||
| Resolution range (Å) | 20.0–1.70 | 20.0–2.1 | 20.0–2.1 | 20.0–2.1 |
| Reflections (measured/unique) | 130 675/25 865 | 72 079/13 052 | 72 082/12 849 | 73 974/12 930 |
| Completeness (overall/outer shell, %)a | 92.2/73.0 | 92.9/85.6 | 93.0/81.5 | 94.3/83.7 |
| Rmergeb (overall/outer shell, %) | 4.4/12.4 | 4.9/13.1 | 8.4/15.8 | 9.5/16.1 |
| Redundancy (overall) | 5.1 | 5.6 | 5.7 | 5.8 |
| Mean 〈I/σ (I)〉(overall) | 25.0 | 20.5 | 21.1 | 22.1 |
| Phasing (20.0–2.1 Å) | ||||
| Mean FOMc after RESOLVE phasing | 0.70 | |||
| Refinement statistics | ||||
| R-factor/free R-factor (%)d | 23.9/29.3 | 23.2/27.2 | ||
| RMSD bond lengths (Å)/bond angles (deg) | 0.023/1.8 | 0.022/2.1 | ||
| Number of water molecules | 64 | 33 | ||
| Average B-factor (PB1/PB2/water, Å2) | 35/30/39 | 52/47/45 | ||
| Ramachandran plot | ||||
| Residues in most favourable regions (%) | 95.6 | 94.6 | ||
| Residues in additional allowed regions (%) | 4.4 | 5.4 | ||
aCompleteness and Rmerge are given for overall data and for the highest resolution shell. The highest resolution shells for the datasets are 2.18–2.10 and 1.76–1.7 Å, respectively.
bRmerge=Σ∣Ii−〈I〉∣/Σ∣Ii∣, where Ii is intensity of an observation and 〈I〉 in the mean value for that reflection and the summations are over all equivalents.
cFigure of merit (FOM)=∣Fbest∣−∣F∣.
dR-factor=Σh∣Fo(h)−Fc(h)∣/Σ∣hFo(h)∣, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. The free R-factor was calculated with 5% of the data excluded from the refinement.
Reconstitution of viral RNP in transfected cells
The in vivo viral RNA polymerase assay using reconstituted viral RNP was performed as described earlier (Hara et al, 2006). HeLa cells were transfected with viral protein expression plasmids encoding PA, PB1 (either wild-type or mutant), PB2 (either wild-type or mutant), NP, and pHH21-vNS-Luc reporter plasmid. This reporter plasmid carries the luciferase gene in reverse orientation sandwiched between 23 nucleotide-long 5′- and 26 nucleotide-long 3′-terminal promoter sequences of the influenza virus segment 8, which is placed under the control of the human Pol I promoter. After incubation for 16 h, total RNA purified from the cells was subjected to reverse transcription with different primers to assess the level of vRNA, cRNA, and mRNA. The 5′-TATGAACATTTCGCAGCCTACCGTAGTGTT-3′, which corresponds to the luciferase coding region between nucleotide sequence positions 351 and 380, was used to measure the vRNA level. The 5′-AGTAGAAACAAGGGTGTTTTTTAGTA-3′, which is complementary to the 3′ portion of the segment 8 cRNA, was used to measure cRNA synthesis, and oligo(dT)20 was used to measure mRNA. The synthesized single-stranded cDNAs were subjected to real-time quantitative PCR analysis (Thermal Cycler Dice Real Time System TP800; TaKaRa) with two specific primers, 5′-TATGAACATTTCGCAGCCTACCGTAGTGTT-3′ corresponding to the luciferase coding region between nucleotide sequence positions 351 and 380, and 5′-CCGGAATGATTTGATTGCCA-3′ complementary to the luciferase coding region between nucleotide sequence positions 681 and 700. NP mRNA transcribed from the expression plasmid was used as an internal control.
Generation of recombinant virus
A recombinant virus carrying the PB1-V715S mutation was generated by the plasmid-based transfection method (Neumann et al, 1999). The PB1-V715S genome segment and seven other wild-type genome segments were generated by cellular RNA polymerase I and wild-type PB1, PB2, PA, and NP were produced from plasmids encoding these proteins by cellular RNA polymerase II. After incubation for 48 h post transfection, an aliquot of the cell culture supernatant was used for virus amplification in MDCK cells. At 48 h post infection, the culture fluid was collected and stored at −80°C until use.
Assay of RNA polymerase activity in infected-MDCK cells
MDCK cells were infected with either wild-type or V715S virus for 3 h. Total RNA was subsequently isolated and reverse transcribed with either 5′-AGTAGAAACAAGGGTATTTTTCTTTA-3′, which is complementary to the 3′ portion of the segment 5 cRNA, oligo(dT)20, or 5′-GACGATGCAACGGCTGGTCTG-3′ corresponding to the viral NP gene between nucleotide positions 424 and 444. These different primers allowed cDNA templates to be produced, which in turn could be assayed to find the level of cRNA, mRNA, and vRNA, respectively. These single-stranded cDNAs were subjected to real-time quantitative PCR analysis with two specific primers, 5′-GACGATGCAACGGCTGGTCTG-3′ and 5′-AGCATTGTTCCAACTCCTTT-3′, which are complementary to NP mRNA between bases 424 and 444, and 595 and 614. As a standard, β-actin mRNA was also amplified with two specific primers, 5′-ATGGGTCAGAAGGATTCCTATGT-3′ and 5′-GGTCATCTTCTCGCGGTT-3′, which are complementary to β-actin mRNA between bases 139 and 161, and 343 and 360, respectively. The relative amounts of vRNA, cRNA, and mRNAs were calculated by using the second derivative maximum method.
Acknowledgments
We thank staff at beam-line Photon Factory BL17A for assistance in data collection. This work was supported in part by the ISS applied research partnership program to S-YP.
References
- Barcena J, Ochoa M, de la Luna S, Melero JA, Nieto A, Ortin J, Portela A (1994) Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol 68: 6900–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Nayak DP (1996) Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J Virol 70: 6716–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci C, Tiley L, Krystal M (1995) Differential activation of the influenza virus polymerase via template RNA binding. J Virol 69: 3995–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW (2009) The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914–918 [DOI] [PubMed] [Google Scholar]
- Digard P, Blok VC, Inglis SC (1989) Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171: 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D, Digard P, Tiley L, Ortin J (2005) Structure and function of the influenza virus RNP. In Current Topics in Influenza Virology, Kawaoka Y (ed), pp 1–92. Norfolk: Horizon Scientific Press [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG (2002) A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76: 8989–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Zurcher T, Ortin J (1996) Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res 24: 4456–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S (2008) The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15: 500–506 [DOI] [PubMed] [Google Scholar]
- Hara K, Schmidt FI, Crow M, Brownlee GG (2006) Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol 80: 7789–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y (2008) Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Huang TS, Palese P, Krystal M (1990) Determination of influenza virus proteins required for genome replication. J Virol 64: 5669–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Li ML, Ramirez BC, Krug RM (1998) RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J 17: 5844–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nagata K, Kawaguchi A, Naito T (2008) Host factors for replication and transcription of the influenza virus genome. Rev Med Virol 18: 247–260 [DOI] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y (1999) Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA 96: 9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY (2008) The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454: 1127–1131 [DOI] [PubMed] [Google Scholar]
- Ochoa M, Barcena J, de la Luna S, Melero JA, Douglas AR, Nieto A, Ortin J, Skehel JJ, Portela A (1995) Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res 37: 305–315 [DOI] [PubMed] [Google Scholar]
- Ohtsu Y, Honda Y, Sakata Y, Kato H, Toyoda T (2002) Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol Immunol 46: 167–175 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Peiris JS, de Jong MD, Guan Y (2007) Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 20: 243–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales B, de la Luna S, Palacios I, Ortin J (1996) Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol 70: 1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858 [DOI] [PubMed] [Google Scholar]
- Poole E, Elton D, Medcalf L, Digard P (2004) Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321: 120–133 [DOI] [PubMed] [Google Scholar]
- Poole EL, Medcalf L, Elton D, Digard P (2007) Evidence that the C-terminal PB2-binding region of the influenza A virus PB1 protein is a discrete alpha-helical domain. FEBS Lett 581: 5300–5306 [DOI] [PubMed] [Google Scholar]
- Poon LL, Pritlove DC, Fodor E, Brownlee GG (1999) Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol 73: 3473–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Digard P (2002) The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 83: 723–734 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (1986) SHELXS86-Program for Crystal Structure Solution. Germany: University of Gottingen [Google Scholar]
- St Angelo C, Smith GE, Summers MD, Krug RM (1987) Two of the three influenza viral polymerase proteins expressed by using baculovirus vectors form a complex in insect cells. J Virol 61: 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ (2007) Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14: 229–233 [DOI] [PubMed] [Google Scholar]
- Tarendeau F, Crepin T, Guilligay D, Ruigrok RW, Cusack S, Hart DJ (2008) Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog 4: e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG (2005) Characterization of the 1918 influenza virus polymerase genes. Nature 437: 889–893 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda T, Adyshev DM, Kobayashi M, Iwata A, Ishihama A (1996) Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J Gen Virol 77(Part 9): 2149–2157 [DOI] [PubMed] [Google Scholar]
- Treanor J, Perkins M, Battaglia R, Murphy BR (1994) Evaluation of the genetic stability of the temperature-sensitive PB2 gene mutation of the influenza A/Ann Arbor/6/60 cold-adapted vaccine virus. J Virol 68: 7684–7688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreede FT, Jung TE, Brownlee GG (2004) Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol 78: 9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Whittaker GR, Digard P (2005) Entry and intercellular transport of influenza virus. In Current Topics in Influenza Virology, Kawaoka Y (ed), pp 37–64. Norfolk: Horizon Scientific Press [Google Scholar]
- World Health Organization (WHO) (2003) Influenza fact sheet. 〈http://www.who.int/mediacentre /factsheets/2003/fs211/en/〉
- Yin YW, Steitz TA (2002) Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y (2009) Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458: 909–913 [DOI] [PubMed] [Google Scholar]
- Zheng H, Lee HA, Palese P, Garcia-Sastre A (1999) Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J Virol 73: 5240–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]