Abstract
Small interfering RNAs (siRNAs) directed to gene promoters can silence genes at the transcriptional level. siRNA-directed transcriptional silencing (RdTS) was first described in plants and yeasts and more recently in mammalian cells. RdTS has been associated with the induction of epigenetic changes and the formation of complexes containing RNA interference and chromatin-remodelling factors. Here, we show that a promoter-targeted siRNA inhibits transcription of the c-myc gene. Transcriptional silencing of c-myc did not involve changes of known epigenetic marks. Instead, the c-myc promoter-targeted siRNA interfered with transcription initiation blocking the assembly of the pre-initiation complex. Transcriptional interference depended on Argonaute 2 and a noncoding promoter-associated RNA initiated upstream and overlapping the transcription start site. Silencing of c-myc led to growth arrest, reduced clonogenic potential and senescence of c-myc over-expressing prostate cancer cells with minimal effect on normal cells. RNA-directed transcriptional interference may be a natural mechanism of transcriptional control and siRNAs targeting noncoding RNAs participating in this regulatory pathway could be valuable tools to control expression of deregulated genes in human diseases.
Keywords: c-myc, promoter-associated RNA, RNA interference, transcriptional gene silencing, transcriptional interference
Introduction
Noncoding small RNAs, like small interfering RNAs (siRNAs) and microRNAs (miRNAs), are emerging as important regulators of cellular functions controlling gene expression at multiple levels (Kim and Rossi, 2007; Filipowicz et al, 2008). Recent studies have shown that small double-stranded RNAs—here called promoter-targeted siRNAs to distinguish them from siRNAs complementary to mRNA—can silence genes at the transcriptional level when directed to gene regulatory regions. The phenomenon of siRNA-directed transcriptional silencing (RdTS) was first described in yeasts and plants, where it was shown to involve DNA and chromatin modifications in the targeted genomic region (Matzke and Birchler, 2005; Buhler and Moazed, 2007; Grewal and Elgin, 2007). More recently, RdTS was shown to occur in mammalian cells and, like in yeasts and plants, was associated with the induction of epigenetic events (Morris et al, 2004; Ting et al, 2005; Weinberg et al, 2006; Pulukuri and Rao, 2007). However, the underlying mechanisms are still poorly understood. In human cells, RdTS requires the formation of multi-protein complexes containing elements of the RNA interference (RNAi) machinery, like Argonaute (Ago) proteins (Janowski et al, 2006; Kim et al, 2006). Ago proteins have been shown to localize to siRNA-targeted promoters and their knock-down abolishes RdTS (Janowski et al, 2006; Kim et al, 2006). DNA and chromatin modifying activities, like DNA methyltransferases (DNMTs) and histone methyltransferases (HMTs), are also recruited to the targeted promoter and participate in RdTS (Kim et al, 2006; Weinberg et al, 2006). However, transcriptional silencing by siRNAs can occur without DNA or histone modifications, suggesting that the process can be mediated by multiple, partially distinct mechanisms (Ting et al, 2005; Janowski et al, 2005a, 2006; Schwartz et al, 2008). Unclear is also the target of the siRNAs. It was proposed that siRNAs might bind to single-stranded DNA regions, such as those present at TSSs, forming RNA:DNA hybrids and physically block transcription (Janowski et al, 2005a). Recently, siRNAs have been shown to bind to promoter-associated RNAs (pRNAs) forming RNA:RNA hybrids and create docking sites for the recruitment of gene silencing complexes (Han et al, 2007; Schwartz et al, 2008).
Although still not extensively investigated, RdTS could be a very effective strategy to knock-down expression of genes involved in pathological processes. siRNAs could be designed to act as gene-specific transcriptional repressors and knock-down selectively expression of cancer promoting genes. This approach might be particularly useful for targets, like transcription factors, frequently over-active in cancer cells but difficult to address with conventional small-molecule drugs (Darnell, 2002). In this study, we explored the possibility to use siRNAs to repress transcription of the c-myc gene. c-Myc is a transcription factor and a key regulator of cell proliferation and death. It is one of the most frequently affected oncogenes in human cancers, contributing to deregulated cell proliferation, differentiation, survival and angiogenesis (Pelengaris et al, 2002; Adhikary and Eilers, 2005). c-Myc over-expression is due to gene amplification, chromosomal translocation or activation by other oncogenic pathways. In prostate cancer, c-myc is amplified or over-expressed in 30–80% of cases and is a major player in prostate cancer progression and acquisition of the androgen-independent phenotype (Pelengaris et al, 2002; Adhikary and Eilers, 2005).
We designed siRNAs directed to sequences overlapping the major TSS in the c-myc gene following a strategy first described by Corey's group (Janowski et al, 2005a, 2005b). Our study had a twofold objective. We wanted to assess the ability of the siRNAs to block c-myc transcription and define the underlying mechanism, and second to determine whether silencing of c-myc by this approach resulted in the induction of relevant and stable changes in the phenotype of cancer cells over-expressing the gene. Our study showed that promoter-targeted siRNAs could effectively inhibit c-myc transcription. Analysis of the mechanism revealed that transcriptional silencing did not involve modifications of epigenetic marks earlier associated with RdTS. Instead, the siRNA acted by preventing transcription initiation by a process that could be defined as promoter-specific RNA-directed transcriptional interference (RdTI). RdTI depended on Ago2 and relied on the binding of the siRNA to a pRNA transcribed upstream of the TSS. Silencing of c-myc by the siRNA led to inhibition of proliferation and clonogenic potential of c-myc over-expressing prostate cancer cells with minimal effect on low c-myc expressing normal cells, underscoring the therapeutic potential of this strategy. RdTI may be part of natural RNA-based transcriptional control mechanisms and siRNAs targeting noncoding pRNAs participating in these regulatory pathways could be valuable tools to control the expression of deregulated genes involved in human diseases.
Results
Inhibition of c-myc expression by promoter-targeted siRNAs
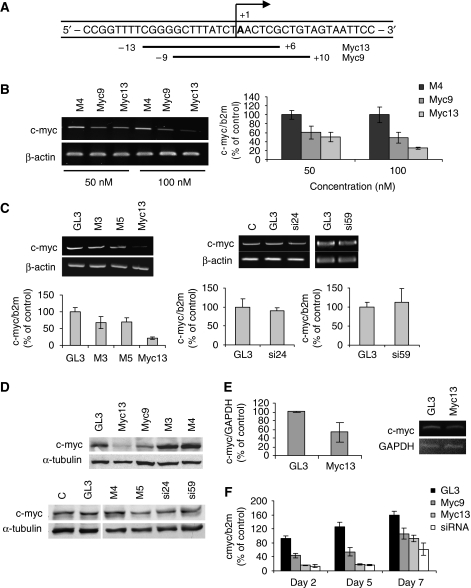
c-Myc is one of the most frequently over-expressed genes in human cancers (Pelengaris et al, 2002; Adhikary and Eilers, 2005). Like other oncogenic transcription factors, c-Myc is an ideal but difficult target for conventional drugs. To assess the ability of siRNAs to block c-myc transcription, we designed siRNAs directed to sequences overlapping the c-myc TSS (Figure 1A). The sequences targeted by myc9 and myc13 started at nucleotide-9 and -13 relative to the TSS, respectively, and encompassed the 11-nucleotide region from −9 to +2 that is unwound in the initiation complex and found to be important for transcriptional silencing (Holstege et al, 1997; Janowski et al, 2005a, 2005b). PC3 prostate cancer cells were transfected with siRNAs and harvested 72 h later to measure c-myc mRNA and protein level. c-myc mRNA was reduced in a dose-dependent manner in cells transfected with myc9 and myc13 compared with control siRNA-treated cells, with myc13 been more effective than myc9 as shown by RT–PCR and quantitative RT–PCR (qRT–PCR) (Figure 1B). Mismatched and scrambled control siRNAs (i.e. M3, M4, M5 and GL3) as well as siRNAs directed to sequences near the TSS (−24 to −59 from the TSS) but not overlapping the critical 11-nucleotide sequence did not have any effect on c-myc expression as shown by RT–PCR and qRT–PCR (Figure 1C). Consistently, c-Myc protein level was reduced in cells treated with active siRNAs (myc9 and myc13) and not affected in cells treated with control siRNAs as shown by immunoblotting (Figure 1C). c-myc expression was also reduced by the siRNAs in DU145 and LNCaP prostate cancer cells expressing high and intermediate levels of the gene (Supplementary Figure S1). As further demonstration of sequence and target specificity, transfection of active and control siRNAs in PC3 cells did not induce detectable levels of INF-α (Supplementary Figure S2), which could be a potential source of off-target effects (Hornung et al, 2005; Judge et al, 2005). Nuclear run-on assays were performed to determine whether reduced c-myc expression by myc13 was due to transcriptional inhibition. A decrease of nascent c-myc transcripts was seen in cells treated with the siRNA compared with control-treated cells, confirming reduced transcription of the gene (Figure 1E). siRNA generally induce a transient reduction of transcript level, which is reversed within few days (Kim and Rossi, 2007). To determine whether the promoter-targeted siRNAs had a similar behaviour, c-myc mRNA was measured by qRT–PCR at 2, 5 and 7 days after transfection. The effect of myc 9 and myc13 on c-myc RNA was similar at days 2 and 5, whereas it reversed partially at day 7 (Figure 1F), consistent with earlier data obtained using a similar approach (Janowski et al, 2005a). Under similar conditions, an siRNA targeting the mRNA reduced c-myc RNA to the same extent as myc13 and exhibited a similar kinetics of recovery, as shown earlier with a reporter gene system (Bartlett and Davis, 2006).
Figure 1.
Inhibition of c-myc gene expression by promoter-targeted siRNAs. (A) Position of the siRNA target sequences relative to the major transcription start site (+1) in the c-myc promoter. Myc9 and myc13 siRNAs were directed to sequences starting 9 and 13 nucleotide upstream to the TSS, respectively. (B) PC3 prostate cancer cells were transfected with 50 or 100 nM of siRNAs. Total RNA was extracted after 3 days and analysed by RT–PCR (left panel) or qRT–PCR (right panel). M4 was used as mismatched control siRNA. P<0.05 for my9 and myc13 compared with control-treated cells. (C) PC3 cells were transfected with 100 nM of myc13 along mismatched and control siRNAs (M3, M5 and GL3) or with siRNAs (si24 and si59) directed to sequences in the c-myc promoter not overlapping the TSS. Cells were harvested after 3 days and mRNA analysed by RT–PCR (top panels) or qRT–PCR (bottom panels). (C) Mock-transfected control cells. P<0.05 for myc13 compared with control-treated cells. (D) PC3 cells were transfected with 100 nM of the indicated siRNAs and harvested after 3 days to assess c-Myc protein by western blotting. (E) PC3 cells were transfected with 100 nM of siRNAs and harvested after 3 days. Nuclei were incubated in the presence of biotin-UTP. Nascent RNA was purified using streptavidin-agarose beads and analysed by RT–PCR with primers specific for c-myc and GAPDH. Right panel, gel densitometry data (mean±s.d.) from three independent experiments. P<0.05 compared with control-treated cells. Left panel, gel scan of a representative experiment. (F) PC3 cells were transfected with 100 nM of Gl3, myc9, myc13 and a mRNA targeting siRNA and harvested after 2, 5 and 7 days. mRNA level was measured by qRT–PCR. Values are normalized for the level in mock-transfected cells at day 2. P<0.05 for myc9, myc13 and siRNA compared with mock-and GL3-transfected cells at days 2 and 5.
Promoter-targeted siRNAs block assembly of the transcription pre-initiation complex
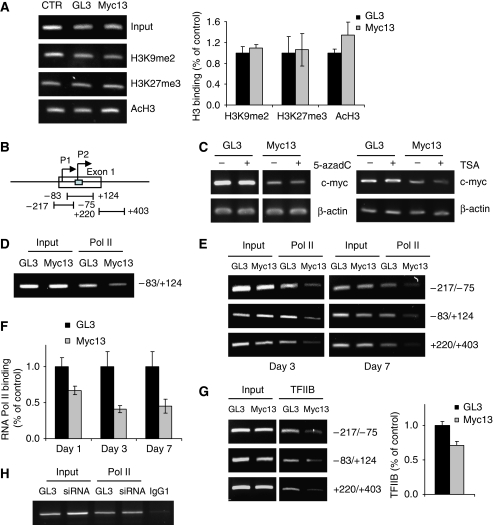
The myc13 siRNA induced an efficient and stable block of c-myc transcription. To investigate the mechanism by which the siRNA acted on the c-myc promoter, we examined the effects on epigenetic marks earlier reported to be involved in RdTS in mammalian cells. Using chromatin immunoprecipitation (ChIP), we assessed the level of histone H3K9 dimethylation (H3K9me2) and H3K27 trimethylation (H3K27me3), two repressive epigenetic marks (Li et al, 2007) shown to increase as a result of RdTS (Ting et al, 2005; Kim et al, 2006). The level of both H3K9me2 and H3K27me3 in the c-myc promoter did not change on transfection of myc13 (Figure 2A). H3K9me2 and H3K27me3 were enriched, however, in the promoter of the transcriptionally silenced genes p16 and RARB2 in PC3 cells (Kondo et al, 2008), confirming the adequacy of the assay conditions (Supplementary Figure S3). Adjacent regions of the c-myc promoter region (Figure 2B) were also examined with no evidence of increased H3K9me2 or H3K27me3 up to 7 days after transfection (data not shown). The level of histone H3 acetylation, a marker of transcriptionally active chromatin (Li et al, 2007), was also unaffected by treatment with the siRNA (Figure 2A). Furthermore, treatment of PC3 cells with the DNMT inhibitor 5-azadeoxycytidine (5-azadC) or the pan HDAC inhibitor trichostatin (TSA) did not affect c-myc silencing, indicating that DNMTs and HDACs were unlikely to be involved in the process (Figure 2C). These results argued against the involvement of these epigenetic events in silencing of c-myc by the siRNA targeting the TSS and were in agreement with earlier reports indicating that RdTS could be independent of histone and DNA modifications (Ting et al, 2005; Janowski et al, 2005a, 2006).
Figure 2.
siRNA-directed inhibition of pre-initiation complex formation on the c-myc promoter. (A) PC3 cells were transfected with 100 nM of myc13 and GL3 and harvested after 3 days. Chromatin immunoprecipitation (ChIP) was performed with antibodies against dimethylated histone H3K9 (H3K9me2), trimethylated histone H3K27 (H3K27me3) and acetylated histone H3 (AcH3). Input and immunoprecipitated DNA were measured by PCR with primers amplifying the −83/+124 region of the c-myc promoter (left panel) and SYBR Green qPCR (right panel). (B) Map of the c-myc promoter showing the regions probed in ChIP experiments. (C) PC3 cells untreated or treated with 5-azadC (left panel) or TSA (right panel) were transfected with siRNAs. Total RNA was extracted after 3 days and c-myc mRNA was measured by RT–PCR. (D) PC3 cells were transfected with siRNAs and ChIP was performed after 24 h with an anti-RNA polymerase II (Pol II) antibody. PCR was performed with primer sets amplifying the −84/+124 region. (E) PC3 cells were transfected with siRNAs and ChIP was performed after 3 (left panel) and 7 (right panel) days using an antibody for RNA polymerase II. PCR was performed with primer sets amplifying the indicated regions of the c-myc promoter. (F) ChIP assays were performed after 1, 3 and 7 days from transfection of siRNAs and RNA Pol II binding assessed by SYBR Green qPCR. P<0.01 compared with control-treated cells. (G) ChIP was performed with an antibody directed to TFIIB and the amount of input and immunoprecipitated DNA determined by PCR with primers amplifying the indicated regions of the c-myc promoter (left panel) and SYBR Green qPCR (right panel). P<0.05 compared with control-treated cells. Representative data at day 3 are shown. (H) PC3 cells were transfected with 100 nM of an siRNA targeting c-myc mRNA and harvested after 3 days to assess RNA Pol II binding to c-myc promoter by ChIP. PCR was performed with primers amplifying the −84/+124 region.
To address the mechanism of transcriptional repression by the siRNA, we considered the possibility that it could interfere directly with PIC assembly at the TSS. To test this hypothesis, PC3 cells were transfected with siRNAs, and ChIP was performed to assess the binding of components of the PIC to the c-myc promoter. Binding of RNA Pol II was reduced in myc13-transfected cells as early as 24 h after transfection, indicating that it was an early event induced by the siRNA (Figure 2D). Decreased RNA Pol II binding was seen reproducibly with distinct primer sets and up to 7 days after transfection (Figure 2E). ChIP data on RNA Pol II binding were confirmed by qPCR with primers spanning the TSS (Figure 2F). Assembly of the PIC requires the binding of general transcription factors (GTFs) that direct RNA Pol II to the core promoter, stabilize the complex at the TSS and catalyse the steps necessary to initiate transcription (Hahn, 2004). To further determine the effects of the siRNA on PIC formation, we assessed binding of TFIIB. This GTF is a central component of PIC (Hahn, 2004; Deng and Roberts, 2007). TFIIB makes sequence-specific contact with the core promoter DNA and is absolutely required for RNA Pol II recruitment to the TSS (Hahn, 2004; Deng and Roberts, 2007). Binding of TFIIB to the c-myc promoter was reduced in myc13-transfected cells parallel to the decrease of RNA Pol II (Figure 2G). Thus, promoter occupancy by essential PIC components was reduced by the siRNA targeting the TSS according to a mechanism closely resembling transcriptional interference (Goodrich and Kugel, 2006; Martianov et al, 2007; Mazo et al, 2007). Unlike the promoter-targeted siRNA, the siRNA targeting c-myc mRNA did not affect RNA Pol II binding to the promoter, indicating that the two siRNAs acted by distinct mechanisms and reduced promoter occupancy by myc13 was not a consequence of reduced c-myc level (Figure 2H).
RdTI and promoter-associated noncoding RNAs
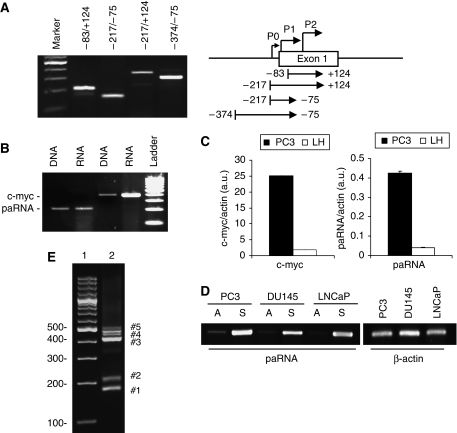
Noncoding RNAs have a prominent role in RdTS in plants and yeasts (Buhler and Moazed, 2007; Grewal and Elgin, 2007). Noncoding pRNAs have been shown recently to be the target of RdTS in mammalian cells (Han et al, 2007; Schwartz et al, 2008). Thus, we examined whether transcription occurred upstream and overlapping the c-myc TSS. RT–PCR with different primer sets showed the presence of low copy transcripts in the region −400 to +120 relative to the TSS (Figure 3A). Promoter-associated transcripts were much less abundant than c-myc mRNA as determined by semiquantitative RT–PCR (Figure 3B) and qPCR [∼50-fold lower than mRNA] (Figure 3C). The level of pRNAs correlated positively with c-myc mRNA expression, with higher levels in c-myc over-expressing PC3 cells than immortalized prostate epithelial LH cells (Figure 3C). Strand-specific RT–PCR was used to determine strand orientation of the promoter-associated transcripts. Sense transcripts were predominant over antisense transcripts in PC3, DU145 and LNCaP prostate cancer cells (Figure 3D) and no products were seen in no-RT reactions excluding contamination with genomic DNA (Supplementary Figure S4). 5′ rapid amplification of cDNA ends (5′RACE) confirmed the presence of multiple transcripts initiating upstream of the major TSS (Figure 3E). Cloning and sequencing of the 5′RACE products showed that the promoter-associated transcripts initiated within 300 and 800 bp from the major TSS (Supplementary Figure S5).
Figure 3.
Detection of promoter-associated transcripts in the c-myc gene. (A) Total RNA was isolated from PC3 cells and amplified using distinct primer sets shown in the left panel to identify promoter-associated transcripts in the region surrounding the major c-myc transcription start site (P2). (B) Total RNA and genomic DNA from PC3 cells were amplified with primers specific for c-myc mRNA and pRNA (F−83/R+124). Genomic DNA was amplified in parallel to control for amplification efficiency. (C) RNA isolated from PC3 and normal prostate epithelial (LH) cells was examined by real time RT–PCR to assess c-myc mRNA and pRNA levels. (D) Total RNA was extracted from PC3, DU145 and LNCaP cells and analysed by strand-specific RT–PCR with F−83/R+124 primers to identify sense (S) and antisense (A) promoter-associated transcripts. (E) 5′RACE products from PC3 cells were amplified with a nested gene-specific primer and the Abridged Universal Amplification Primer (Invitrogen). The marked PCR products were cloned and sequenced to confirm their identity.
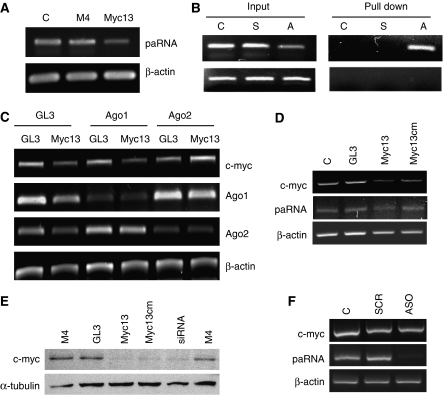
As proposed for other noncoding RNAs (Mattick and Makunin, 2006; Prasanth and Spector, 2007) pRNAs in the c-myc promoter could have a regulatory function, perhaps maintaining promoter accessibility to transcription factors and assisting in transcription initiation. The presence of pRNAs also argued in favour of a model in which siRNAs bound to the noncoding transcript and formed a complex that switched off transcription as recently proposed (Han et al, 2007; Schwartz et al, 2008). In favour of this hypothesis, the level of the c-myc pRNA was reduced, although only partially, in myc13-treated cells indicating an interaction between the two RNA species (Figure 4A). The physical interaction was confirmed by biotin-linked siRNA pull-down assays in which cells were transfected with an siRNA having either the sense or antisense strand labelled with biotin. The biotin-labelled antisense strand of myc13 was found to bind to the pRNA, whereas no signals were detected with the biotin-labelled sense strand or unlabelled siRNA (Figure 4B). Furthermore, no signal was detected in biotin pull-down samples by PCR after RNase treatment, indicating the absence of contaminating genomic DNA (Figure 4B).
Figure 4.
Interaction of promoter-directed siRNA with promoter-associated RNA. (A) PC3 cells were transfected with siRNAs and total RNA extracted after 3 days. RT–PCR was performed with primers specific for pRNA (F−217/R+124) and β-actin. (B) PC3 cells were transfected with the siRNA myc13 with biotin-labelled sense (S) or antisense (A) strand and control (C) siRNA. Cells were harvested after 24 h and RNA bound to the biotin-labelled siRNA was isolated with streptavidin-agarose beads and examined by RT–PCR with primers specific for the c-myc pRNA (top panel). Input and pull-down samples were subjected to PCR after RNAse treatment to exclude the presence of contaminating DNA in pull-down samples (bottom panel). (C) PC3 cells were transfected with myc13 along with GL3 and siRNAs targeting Ago1 or Ago2. Cells were harvested after 3 days to measure c-myc, Ago1, Ago2 and β-actin mRNA by RT–PCR. (D) PC3 cells were transfected with the fully complementary siRNA (myc13) or the siRNA with central mismatches relative to the target sequence (myc13cm). Cells were harvested after 3 days to measure c-myc and β-actin mRNA by RT–PCR. (E) PC3 cells were transfected with myc13, myc13 cm and the indicated positive and negative control siRNAs and harvested after 3 days to assess of c-Myc protein level by western blot. (F) PC3 cells were transfected with 100 nM of a scrambled (SCR) or antisense (ASO) oligonucleotide directed to the pRNA. c-myc mRNA and pRNA level was assessed by RT–PCR.
Ago 2 mediates RdTI in the c-myc gene
To establish whether the RNAi machinery was involved in RdTI, we knocked-down Ago1 and Ago2 and examined the effect on siRNA-directed c-myc silencing. Ago proteins are integral components of the RNAi pathway (Kim and Rossi, 2007; Tolia and Joshua-Tor, 2007) and are involved in RdTS in mammalian cells (Janowski et al, 2006; Kim et al, 2006). Efficient silencing of both Ago1 and Ago2 was achieved using specific siRNAs both in single- (Supplementary Figure S6) and double-transfection experiments (Figure 4C). In the latter, knock-down of Ago2 prevented c-myc silencing by myc13, whereas knock-down of Ago1 and a control siRNA did not have any effect (Figure 4C). This indicated that c-myc silencing relied mainly on Ago2, whereas previously both Ago1 and Ago2 had been shown to mediate RdTS (Janowski et al, 2006; Kim et al, 2006). As Ago2 has slicer activity (Kim and Rossi, 2007), we examined whether cleavage of the pRNA was required for RdTI. For this purpose, we used an siRNA (myc13cm) with three central mismatched bases relative to the target sequence, which would abrogate target RNA cleavage (Filipowicz et al, 2008). Silencing of c-myc was not significantly affected by the presence of central mismatches as myc13cm was as effective as myc13 and the mRNA targeting siRNA in decreasing c-myc mRNA and protein (Figure 4D and E), indicating that RdTI did not depend on Ago2 slicer activity. Thus, binding of the siRNA to the pRNA rather than its elimination was required for silencing of c-myc. Consistent with this hypothesis, an antisense oligonucleotide that almost completely knocked-down the pRNA did not affect c-myc transcription (Figure 4F). Taken together, these data indicated that formation of the siRNA:pRNA complex with the contribution of Ago2 was responsible for reduced assembly of the functional PIC and blockade of transcription initiation in the c-myc promoter.
c-myc promoter-targeted siRNAs are effective inhibitors of cancer cell proliferation
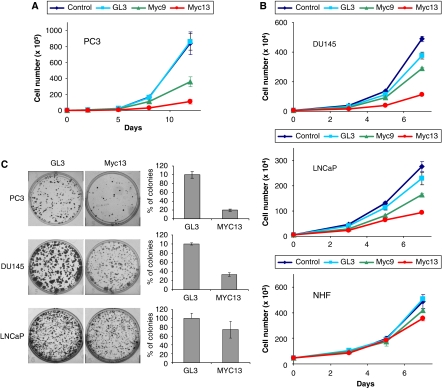
Myc is a key regulator of cell proliferation and death (Pelengaris et al, 2002; Adhikary and Eilers, 2005). Cells that over-express c-myc may depend on the continuous production of the protein and be highly sensitive to its down-regulation. To determine whether cell proliferation was affected as a consequence of c-myc knock-down induced by promoter-targeted siRNAs, we monitored growth of siRNA-transfected PC3 cells over a 12-day period. Untreated and control-treated cells behaved identically, whereas proliferation of cells treated with myc9 and myc13 was markedly reduced (Figure 5A). In fact, growth of myc13-treated cells was almost completely arrested. At day 8, growth was reduced by 40 and 80% and after 12 days by 60 and 90% for myc9- and myc13-treated cells, respectively, relative to control cells. Overall, the extent of cell growth inhibition by the two siRNAs correlated well with their relative potency as c-myc transcriptional repressors. Similar results were seen in clonogenic assays with myc13 (see below), consistent with persistent silencing of the c-myc gene.
Figure 5.
Reduced proliferation and clonogenic potential of siRNA-transfected prostate cancer cells. (A) PC3 cells were transfected with 100 nM of siRNAs in 12-well plates and counted with an automated cell counter over a 12-day period. P<0.005 compared with control cells at days 5, 8 and 12 with myc13, and days 8 and 12 with myc9. (B) DU145 (top panel) and LNCaP (middle panel) prostate cancer cells and normal human fibroblasts (bottom panel) were transfected with 100 nM of siRNAs and counted over a 7-day period as indicated above. P<0.005 compared with control cells at days 5 and 7 for DU145 and LNCaP cells treated with myc13. (C) PC3, DU145 and LNCaP cells were transfected with 100 nM of siRNAs and plated at clonal density in six-well plates. Colonies were stained with crystal violet (left panels) and counted after 12 days using an automated colony counter (right panels). P<0.005 for PC3 and DU145, and P<0.05 for LNCaP cells.
Variations in the transcriptional activity of a gene may affect the ability of siRNAs to bind to the promoter and inhibit its transcription. The combination of high targeting efficiency and cell addiction to the target may make this approach highly selective towards cancer cells with deregulated expression of the target, while sparing normal cells with a physiological expression level. We explored this concept by examining the effects of siRNAs in two other prostate cancer cell lines (DU145 and LNCaP) and normal human fibroblasts (NHFs). Prostate cancer cells with high and intermediate c-myc levels were sensitive to the siRNA (Figure 5B). In contrast, NHF with low c-myc expression were minimally affected. Among prostate cancer cells, PC3 cells with the highest c-myc level were the most sensitive both in cell proliferation (Figure 5B) and clonogenic assays (Figure 5C), as further indication that higher transcriptional activity increases targeting efficiency.
c-myc gene knock-down by promoter-targeted siRNAs results in growth arrest and cell senescence
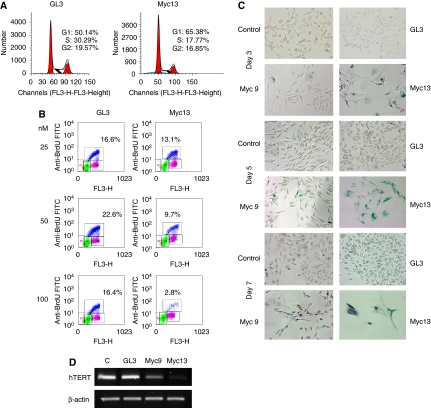
Given the role of c-myc in cell-cycle regulation (Pelengaris et al, 2002; Adhikary and Eilers, 2005), we investigated whether its down-regulation by promoter-targeted siRNAs resulted in cell-cycle perturbations consistent with the effects seen on cell proliferation. PC3 cells were transfected with siRNAs and harvested after 3 days. FACS analysis after PI staining revealed a marked increase in G1 (50 versus 65%) and decrease in S-phase (30 versus 18%) in myc13-treated cells compared with control cells (Figure 6A). Similar but smaller effects on the percentages of G1 (58%) and S-phase (26%) cells were observed in myc9-transfected cells (data not shown). The effect on cell proliferation was also examined by BrdU pulse labelling. Cells transfected with myc13 showed a dose-dependent decrease of the percentage of proliferating cells compared with control-treated cells (Figure 6B). The fraction of BrdU-positive cells decreased on transfection with 25, 50 and 100 nM of myc13 by 21, 57 and 80%, respectively, compared with control cells. Unlike the effect on cell proliferation, c-myc down-regulation had only a small effect on the survival of PC3 cells as judged by the absence of a prominent subG1 peak in siRNA-treated cells (Figure 6A). To confirm this finding, the level of active caspase-3 was determined by FACS along with c-Myc protein level in myc13- and control-treated cells. Despite the reduced c-Myc level, only a small fraction of PC3 cells (∼3%) showed signs of caspase-3 activation (Supplementary Figure S7).
Figure 6.
Induction of growth arrest and senescence by c-myc promoter-directed siRNAs in prostate cancer cells. (A) PC3 cells transfected with siRNAs were harvested after 3 days, stained with propidium iodide and analysed by FACS to determine cell-cycle distribution. (B) siRNA-transfected PC3 cells were pulse-labelled with BrdU 3 days after transfection. Fixed cells were stained with FITC-conjugated anti-BrdU antibody and 7-AAD and analysed by FACS to determine the percentage of BrdU-positive proliferating cells. (C) siRNA-transfected PC3 cells were stained to detect senescence-associated β-galactosidase (SA-β-gal) activity after 3 (top panel), 5 (middle panel) and 7 (bottom panel) days. Images were collected with an inverted microscope equipped with a digital camera. Dark blue cells are positive for SA-β-gal activity. (D) PC3 cells were transfected with 100 nM of siRNAs and after 3 days hTERT expression was determined by RT–PCR.
Microscopy examination of siRNA-treated cells confirmed the absence of signs of apoptotic cell death but revealed morphological changes compatible with the induction of a senescence-like process. PC3 cells became progressively enlarged, flattened and more dispersed on treatment with siRNAs. To confirm this observation, cells were stained to detect senescence-associated β-galactosidase (SA-β-gal) activity, an established marker of cell senescence. Most cells exhibiting cell enlargement and flattening were SA-β-gal positive. Increasing numbers of SA-β-gal positive cells were detected in cultures of myc13 and, to a lesser extent, myc9-transfected cells from 3 to 7 days after transfection, parallel to the overall decrease in cell number (Figure 6C). Thus, growth arrest was associated with the induction of a senescence-like phenotype in siRNA-treated PC3 cells. As an additional marker of reduced c-myc function and induction of cell senescence, we examined the expression of the telomerase subunit hTERT. hTERT encodes the catalytic subunit of human telomerase and is transcriptionally regulated by c-myc (Horikawa and Barrett, 2003). Telomerase is required for cell immortalization and its down-regulation results in reduced cell proliferation and senescence (Shay and Wright, 2005). hTERT expression was reduced in cells incubated with myc9 and myc13 compared with control cells, concomitant to the reduced c-myc level (Figure 6D). Thus, c-myc promoter-targeted siRNAs reduced the expression of a known c-myc target (Horikawa and Barrett, 2003) and induced cell senescence, a phenotype earlier associated with c-myc down-regulation (Guney et al, 2006) and consistent with the known functions of this oncogene.
Discussion
In this study, we show that siRNAs targeting the TSS in the c-myc gene interfere with transcription initiation. The transcriptional silencing effect was associated with reduced assembly of the PIC at the TSS as demonstrated by the decreased binding of both RNA Pol II and TFIIB to the c-myc promoter. TFIIB binds the core promoter DNA and directs RNA Pol II to the TSS promoting assembly of the functional PIC (Hahn, 2004; Deng and Roberts, 2007). Thus, the concomitant loss of TFIIB and RNA Pol II suggests that the siRNA prevents early steps in PIC formation. Transcriptional interference in the c-myc gene relied on the presence of a sense pRNA overlapping the TSS. RdTI depended also on Ago2, suggesting that Ago2 was guided by the siRNA to bind the c-myc pRNA. Thus, the complex formed by the siRNA and pRNA with the assistance of Ago2 could physically impair PIC assembly in the c-myc promoter. In this case, the interaction of the siRNA with pRNA might be analogous to the miRNA:mRNA interaction, which results in block of translation and does not lead necessarily to mRNA degradation (Filipowicz et al, 2008). Alternatively, binding of the siRNA may result in cleavage and degradation of the pRNA, which might have a positive role in transcription perhaps assisting in the loading of RNA Pol II and the assembly of the PIC. Degradation of the pRNA could prevent a critical step in transcription initiation. However, we found that Ago2 slicer activity was not essential for RdTI and pRNA knock-down by an antisense oligonucleotide did not affect c-myc transcription. Recent data from other groups also support the idea that degradation of the pRNA by siRNAs is not required for transcriptional silencing (Han et al, 2007; Schwartz et al, 2008).
RNA-directed transcriptional control is likely a complex process involving different players and mechanisms (Han et al, 2007; Martianov et al, 2007; Kim et al, 2008; Place et al, 2008; Schwartz et al, 2008; Wang et al, 2008; Yu et al, 2008). Here, we show that transcriptional interference by siRNAs occurs without changes of epigenetic marks earlier associated with RdTS. This finding suggests that siRNAs can silence genes by at least two partially different mechanisms: one involving recruitment of epigenetic factors, like HMTs and DNMTs, and the other involving direct interference with transcription initiation. The concomitant reduction of RNA Pol II and TFIIB shown here is evidence of reduced PIC formation by the siRNA. On the other hand, epigenetic silencing by recruitment of Polycomb group proteins does not affect binding of RNA Pol II and GTFs to the targeted promoter (Dellino et al, 2004) and RdTS was shown to require RNA Pol II activity (Weinberg et al, 2006). The induction of different mechanisms of gene silencing by siRNAs may depend on the position of the target sequence relative to the TSS or be an intrinsic property of the target gene and its regulatory set up. In both cases, gene silencing is initiated by Ago proteins and likely depends on recruitment of additional factors to the promoter (Janowski et al, 2006; Kim et al, 2006). The ability of Ago proteins to elicit distinct and even opposite effects is not surprising. miRNA-loaded Ago can alternatively promote translation repression or activation depending on the cell conditions and the protein partners recruited to the complex (Filipowicz et al, 2008).
Transcription of noncoding pRNAs is a widespread phenomenon in the human genome (Mattick and Makunin, 2006; Prasanth and Spector, 2007; Kapranov et al, 2007a). The role of this class of noncoding RNAs in transcriptional regulation is largely unknown and just begins to be investigated (Mattick and Makunin, 2006; Prasanth and Spector, 2007). Early work on RdTS in mammalian cells did not take in consideration the presence of promoter-associated transcripts and their possible involvement in this process (Morris et al, 2004; Ting et al, 2005; Janowski et al, 2005a). Only recently, the presence of pRNAs and the consequences of their targeting by siRNAs in human cells have been investigated (Han et al, 2007; Schwartz et al, 2008). In this study, we identified promoter-associated transcripts upstream and overlapping the TSS in the c-myc gene. Consistent with our findings, recent tiling array and high-throughput studies have shown the presence of low abundance transcripts initiated near the TSSs of many genes and transcribed both in sense and antisense direction relative to the protein coding sequences (Kapranov et al, 2007a; Core et al, 2008; Preker et al, 2008; Seila et al, 2008). The level of the promoter-associated transcripts correlated with that of the adjacent protein coding sequences with high level in highly transcribed genes, as we have seen here for the c-myc gene, suggesting that pRNAs might have a role in transcriptional regulation and chromatin remodelling (Kapranov et al, 2007a; Core et al, 2008; Preker et al, 2008; Seila et al, 2008). Our study indicates that pRNAs might have a role in transcription initiation and reveals a yet unrecognized level of RNA-based transcriptional control in mammalian cells.
Altogether, these findings point to the existence of complex RNA-based transcriptional regulatory networks (Mattick and Makunin, 2006; Prasanth and Spector, 2007; Kapranov et al, 2007b; Core et al, 2008; Preker et al, 2008; Seila et al, 2008). Noncoding RNA species participating in these transcriptional regulatory pathways could represent novel classes of endogenous targets and effector molecules to modulate gene expression for experimental and therapeutic purposes (Han et al, 2007; Martianov et al, 2007; Kim et al, 2008; Place et al, 2008; Schwartz et al, 2008; Wang et al, 2008; Yu et al, 2008). Here, we show that promoter-targeted siRNAs can effectively inhibit c-myc transcription. Silencing of this oncogene by the siRNAs led to profound inhibition of proliferation and clonogenic potential of prostate cancer cells over-expressing the target. On c-myc down-regulation, the majority of prostate cancer cells was growth arrested and acquired a senescent-like phenotype. These effects were consistent with the known c-myc functions and confirmed both the efficacy and specificity of this approach. The potential of promoter-targeted siRNAs for therapeutic applications is currently being tested pre-clinically both in vitro and in vivo. If successful in these settings, promoter-targeted siRNAs may be readily available for clinical testing and represent new tools for treatment of cancer and other diseases.
Materials and methods
Cell lines, siRNAs and cell transfection
Prostate cancer cell lines PC3, DU145 and LNCaP and NHFs were maintained either in RPMI or DMEM under standard cell culture conditions (Cangemi et al, 2008). Immortalized prostate epithelial (LH) cells expressing hTERT and SV40 large T antigen were maintained in PrEGM medium (Cangemi et al, 2008). siRNAs were purchased from Ambion as 21-nucleotide duplexes annealed and HPLC purified (Supplementary Table S1). The position of the TSSs was determined by consulting a public TSS database (Wakaguri et al, 2008). The c-myc mRNA targeting Silencer Select siRNA was purchased from Ambion. Phosphorothioate modified antisense oligonucleotides (Supplementary Table S1) were purchased from Sigma. Cell transfection was done in six-well plates using Lipofectamine2000 according to standard protocols (Cangemi et al, 2008). Transfected cells were either harvested at the required times or re-plated 24 h after transfection when indicated. Treatment of PC3 cells with 5-aza-2′-deoxycytidine (5 μM) and TSA (200 nM) was done for 3 days as described earlier (Cangemi et al, 2008; Pulukuri and Rao, 2007).
RNA and protein analysis
Total RNA was extracted using Trizol (Invitrogen) and RNeasy Mini Kit (Qiagen). All RNA samples were treated with DNase I (Qiagen) to remove any contaminant genomic DNA. RT–PCR was performed using SuperScript One Step RT–PCR system (Invitrogen) with gene-specific primers (Supplementary Table S2). PCR products were analysed by agarose gel electrophoresis, visualized by ethidium bromide and quantified using AlphaImager and AlphaEase software (Innotech). Strand specific RT–PCR was performed by omitting either forward or reverse primer in the RT step. Reactions without template (negative control) or with genomic DNA (positive control) were performed to assess specificity and efficiency of amplification of each primer set. No products were detected in reactions lacking template or without the RT step. For qRT–PCR, 1 μg of RNA was reverse transcribed using Superscript first strand kit (Invitrogen) The cDNA was then diluted and used as template for SYBR Green qPCR on the ABI 7000 (Applied Biosystems) machine (Cangemi et al, 2008). Alternatively, qRT–PCR was performed using Express One-Step SYBR GreenER system (Invitrogen). The amount of c-myc transcript was determined using β-actin or β2-microglobulin as reference and either the ΔCt or standard curve method. Nuclear run-on assays were performed as described (Zhang et al, 2005). Nuclei were isolated from siRNA-transfected cells and incubated in nuclear run-on reaction buffer containing rNTPs and biotin-16-UTP at 30°C for 45 min. This was followed by DNAse I (Sigma) and proteinase K (Roche) treatment and RNA extraction using Trizol (Invitrogen). Biotinylated nascent RNA was purified using streptavidin-agarose beads, eluted from the beads and analysed by RT–PCR with primers for c-myc and GAPDH (Supplementary Table S2). Band intensity was determined using the AlphaEase software and normalized to the reference gene. For immunoblotting, cells were lysed as described earlier (Cangemi et al, 2008). Protein samples were separated on 10% polyacrylamide–SDS gels and transferred to nitrocellulose membranes that were probed with antibodies against c-myc (clone 9E10, Santa Cruz Biotechnology, Santa Cruz, CA) and α-tubulin (Oncogene Research Products). Blots were developed using peroxidase-conjugated secondary antibodies and the enhanced chemiluminescence system (Amersham). All the experiments were repeated at least three times and representative results or mean±s.d. are shown.
Cell proliferation and senescence assays
Cells transfected with siRNAs were seeded in 12-well plates, collected and counted at the indicated times using an automated cell counter (Beckman Coulter Counter). Clonogenic assays were performed by plating transfected cells (1 × 103/well) in six-well plates (Cangemi et al, 2008). After 12 days, colonies were stained with crystal violet and counted using an automated colony counter system (AlphaImager). Detection of senescence-associated β-galactosidase activity was done using the senescent cells staining kit (Sigma). siRNA-transfected cells were plated in 24-well plates and at each time point stained according to the manufacturer's protocol. Cells were then covered with 70% glycerol and examined with an inverted microscope (Zeiss Axiovert 100) equipped with a digital camera. Each experiment was performed in triplicate and repeated at least three times. Either representative results or mean±s.d. are shown. Student's t-test was performed to assess statistical significance of the differences among groups.
Cell cycle, BrdU incorporation and apoptosis assays
siRNA-transfected cells were harvested and fixed with 70% ice-cold ethanol. Fixed cells were stained with propidium iodide (50 mg/ml in 0.1% sodium citrate) and analysed by FACS (FACSCalibur, Becton Dickinson) as described (Carbone et al, 2004). To determine the fraction of proliferating cells, siRNA-transfected cells were pulse-labelled with BrdU (10 μM for 1 h) 3 days after transfection. Cells were collected and stained with FITC-conjugated anti-BrdU antibody and 7-AAD according to the BrdU Flow Staining Kit protocol (BD Pharmingen). The percentage of BrdU-positive cells was determined by FACS. Active caspase-3 was detected using the anti-active caspase 3 staining kit (BD, Biosciences) according to the manufacturer's protocol. To detect c-Myc, cells were incubated in Fixation medium for 15 min at 37°C, recovered by centrifugation and incubated on ice-cold 90% methanol for 30 min. Cells were incubated with anti-c-Myc antibody (9E10) for 45 min at room temperature followed by 30-min incubation with a FITC-labelled anti-mouse secondary antibody (BD Biosciences). Samples were analysed by FACS with 515 nm and 610 bandpass filters for FITC and caspase-3 detection, respectively. All the experiments were repeated at least three times and representative results are shown.
Chromatin immunoprecipitation
siRNA-transfected cells were collected, cross-linked with formaldehyde and processed using the EZ-ChIP kit (Upstate Biotehcnology). After sonication, chromatin was immunoprecipitated with antibodies for dimethylated histone H3 at lysine 9 (H3K9me2) and trimethylated histone H3 at lysine 27 (H3K27me3), acetylated histone H3, RNA polymerase II (Upstate Biotechnology) and TFIIB (Santa Cruz). DNA–protein cross-links were reversed and DNA purified from total cell lysates (input) and immunoprecipitated fractions. PCR was performed with Taq Gold (Roche) and primers amplifying three adjacent regions surrounding the c-myc transcription start site (Supplementary Table S2). PCR products were separated on 2% agarose gels, stained with ethidium bromide and quantified using AlphaEase software. ChIP experiments were repeated at least three times and representative results are shown. Quantitative real time PCR was performed using SYBR Green qPCR as described (Cangemi et al, 2008), and the primer sets indicated in Supplementary Table S2. The amount of immunoprecipitated DNA was calculated in reference to a standard curve and normalized to input DNA.
Biotin-labelled siRNA pull-down assay
PC3 cells were transfected in 10-cm dishes with 100 nM myc13 siRNA in which either the sense or antisense strand were 5′-biotin labelled. Biotin-labelled siRNA pull-down assay was performed as described earlier (Han et al, 2007). Briefly, cells were harvested after 48 h from transfection and exposed to formaldehyde to cross-link nucleic acid-protein complexes. Total nucleic acids were extracted with DNeasy extraction kit (Qiagen) and incubated with streptavidin-agarose beads for 2 h at 4°C. The eluted material was incubated for 1 h at 65°C to reverse cross-links and then treated for 30 min at 37°C with DNase I. RNA was extracted using Trizol and analysed by RT–PCR using primers specific for the c-myc promoter-associated transcript (F−83/R+124) followed by nested PCR (F−44/R+68). In parallel, samples were subjected to RNase treatment and PCR (without RT) followed by nested PCR to exclude the presence of contaminating genomic DNA.
5′ rapid amplification of cDNA ends
5′RACE was performed using the Invitrogen 5′RACE System. First strand cDNA was synthesized from 1 μg of total RNA isolated from PC3 cells using gene-specific reverse primer 1 (GSP1; Supplementary Table S2) followed by RNase treatment. The resulting cDNA was purified using SNAP columns and tailed with dCTP using TdT. dC-tailed cDNA was then amplified using gene-specific primer 2 (GSP2) and the Abridged Anchor Primer (Invitrogen). PCR products were reamplified using gene-specific primer 3 (GSP3) and the Abridged Universal Amplification Primer. The final PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the Swiss National Science Foundation (CVC), the Tessin Foundation for Cancer Research (CVC) and a Mario Luvini Foundation fellowship (CP).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Bartlett DW, Davis ME (2006) Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res 34: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D (2007) Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Cangemi R, Mensah A, Albertini V, Jain A, Mello-Grand M, Chiorino G, Catapano CV, Carbone GM (2008) Reduced expression and tumor suppressor function of the ETS transcription factor ESE-3 in prostate cancer. Oncogene 27: 2877–2885 [DOI] [PubMed] [Google Scholar]
- Carbone GM, Napoli S, Valentini A, Cavalli F, Watson DK, Catapano CV (2004) Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res 32: 4358–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE Jr (2002) Transcription factors as targets for cancer therapy. Nat Rev Cancer 2: 740–749 [DOI] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V (2004) Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893 [DOI] [PubMed] [Google Scholar]
- Deng W, Roberts SG (2007) TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 116: 417–429 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF (2006) Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol 7: 612–616 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney I, Wu S, Sedivy JM (2006) Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a). Proc Natl Acad Sci USA 103: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S (2004) Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol 11: 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV (2007) Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA 104: 12422–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT (1997) Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J 16: 7468–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa I, Barrett JC (2003) Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis 24: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G (2005) Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11: 263–270 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR (2005a) Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol 1: 216–222 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR (2006) Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13: 787–792 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR (2005b) Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat Chem Biol 1: 210–215 [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I (2005) Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 23: 457–462 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V et al. (2007a) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR (2007b) Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ (2007) Strategies for silencing human disease using RNA interference. Nat Rev Genet 8: 173–184 [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ (2008) MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 105: 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ (2006) Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13: 793–797 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP (2008) Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 (Spec No 1): R17–R29 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW (2007) Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci 120(Pt 16): 2755–2761 [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292 [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G (2002) c-MYC: more than just a matter of life and death. Nat Rev Cancer 2: 764–776 [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA 105: 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH (2008) RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Pulukuri SM, Rao JS (2007) Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res 67: 6637–6646 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA (2008) Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15: 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE (2005) Mechanism-based combination telomerase inhibition therapy. Cancer Cell 7: 1–2 [DOI] [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG, Baylin SB (2005) Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet 37: 906–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L (2007) Slicer and the argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K (2008) DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res 36 (Database issue): D97–D101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Rei, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV (2006) The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12: 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H (2008) Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MX, Ou H, Shen YH, Wang J, Wang J, Coselli J, Wang XL (2005) Regulation of endothelial nitric oxide synthase by small RNA. Proc Natl Acad Sci USA 102: 16967–16972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information