Abstract
Heavy metal pumps constitute a large subgroup in P-type ion-transporting ATPases. One of the outstanding features is that the nucleotide binding N-domain lacks residues critical for ATP binding in other well-studied P-type ATPases. Instead, they possess an HP-motif and a Gly-rich sequence in the N-domain, and their mutations impair ATP binding. Here, we describe 1.85 Å resolution crystal structures of the P- and N-domains of CopA, an archaeal Cu+-transporting ATPase, with bound nucleotides. These crystal structures show that CopA recognises the adenine ring completely differently from other P-type ATPases. The crystal structure of the His462Gln mutant, in the HP-motif, a disease-causing mutation in human Cu+-ATPases, shows that the Gln side chain mimics the imidazole ring, but only partially, explaining the reduction in ATPase activity. These crystal structures lead us to propose a role of the His and a mechanism for removing Mg2+ from ATP before phosphoryl transfer.
Keywords: crystallography, ion pump, nucleotide binding mode, phosphoryl transfer, Wilson disease
Introduction
P-type ATPases are ATP-powered ion pumps and have crucial functions in ion homoeostasis from bacteria to human. They are characterised by an Asp that is auto-phoshophorylated and dephosphorylated during the reaction cycle in one of the cytoplasmic domains (P-domain). They form a large family that encompasses several distinct subgroups, of which type II and type IB are the largest (Axelsen and Palmgren, 1998). Type II ATPases include the best-studied members of the P-type ATPases, namely Ca2+-ATPases (SERCA), Na+,K+-ATPase and gastric H+,K+-ATPase. Type IB represents heavy metal transporting ATPases, of which Cu+-ATPases are the most prevalent (Argüello, 2003).
These two groups exhibit similarities as well as distinct differences in architecture. X-ray crystallography of the Ca2+-ATPase from skeletal muscle sarcoplasmic reticulum (SERCA1a) has established that the ATPase consists of 3 cytoplasmic domains (i.e. A, actuator; N, nucleotide binding; P, phosphorylation) and 10 transmembrane helices (Toyoshima et al, 2000). The α-subunit of the Na+,K+-ATPase from pig kidney has the same domain organisation as SERCA1a, although it also possesses a β-subunit and an FXYD protein (Morth et al, 2007; Shinoda et al, 2009). Even plant H+-ATPases that belong to type III have the same topology (Pedersen et al, 2007).
Type IB ATPases also comprise three cytoplasmic domains, but posses a metal-binding domain consisting of one to six repeats of a CXXC-motif at the N-terminus, and only eight transmembrane helices (Kühlbrandt, 2004). Furthermore, they apparently have a mode of binding ATP that is different from other subtypes, as the N-domain lacks residues normally critical for ATP binding, including a Phe that stacks the adenine ring and an Arg involved in cross-linking the N- and P-domains through ATP (Toyoshima and Mizutani, 2004). Instead, different residues in the N-domain are absolutely conserved within the type IB ATPases. They include an HP-motif and a Gly-rich sequence. For the Gly-rich sequence, a role similar to the P-loop observed with many ATPases and GTPases (Walker et al, 1982) has been postulated, although P-type ATPases in general do not have it and the crystal structures of the N-domain located the Gly's in a β-sheet (Sazinsky et al, 2006b). Hence, type IB ATPases seem to recognise ATP entirely different from other ATPases, a distinction that has also been suggested by mutational, nucleotide titration and nuclear magnetic resonance (NMR) studies on the N-domain of ATP7B (Tsivkovskii et al, 2003; Morgan et al, 2004; Dmitriev et al, 2006).
X-ray crystallography and NMR spectroscopy have been successfully used to determine the structures of the three cytoplasmic domains of PIB-ATPases (Dmitriev et al, 2006; Sazinsky et al, 2006a, 2006b; Lübben et al, 2007), but failed to resolve bound nucleotides. They showed that the A- and P-domains are very similar to those in PII type ATPases. Even the core architecture of the N-domain is quite similar to that of the PII type, but the critical Phe and Arg are certainly absent. Of particular interest is the functional role of the absolutely conserved His in the HP-motif (Tsivkovskii et al, 2003), as mutations here are frequently identified in patients with Wilson disease and a malfunctioning Cu+-ATPase (ATP7B) (Thomas et al, 1995). A recent molecular dynamics study proposed that the His stacks with the adenine ring (Rodriguez-Granillo et al, 2008).
CopA, a bacterial (Hatori et al, 2007, 2008) or archaeal (González-Guerrero and Argüello, 2008) Cu+-ATPase, is the best-studied member of the PIB-ATPases, as it is much simpler than human Cu+-ATPases ATP7A and 7B. However, it is difficult to say, at present, how different they are, as even kinetic studies on the same archaeal ATPase did not provide consistent results (Mandal and Argüello, 2003; Rice et al, 2006). Systematic studies on domain movements and kinetic intermediates were carried out with bacterial CopA (Hatori et al, 2007, 2008), but are still left to be done for human Cu+-ATPases.
Here, we describe crystal structures at 1.85 Å resolution of CopA-PN, the P- and N-domains of CopA from a hyperthermophile archaea Archaeoglobus fulgidus, with bound adenosine 5′-[β, γ- methylene]triphosphate (AMPPCP) or ADP and Mg2+. Indeed, these structures show that CopA recognises the adenine ring entirely different from the other P-type ATPases and explain the roles of the critical amino-acid residues identified by mutagenesis studies (Tsivkovskii et al, 2003; Morgan et al, 2004; Okkeri et al, 2004). We also describe a crystal structure of the His462Gln mutant with bound Mg2+-AMPPCP at 1.95 Å resolution. The corresponding mutation in ATP7B is the most frequent one found in Caucasian patients with Wilson disease (Thomas et al, 1995). Besides that this residue might be on a genetic hotspot prone to base replacements, the mutant structure provides a plausible explanation. Furthermore, nucleotides bound to protomers related by non-crystallographic symmetry lead us to propose a scenario for removal of Mg2+ from ATP, which is prerequisite to transfer of the γ-phosphate from ATP to the phosphorylated Asp.
Results and discussion
Structure of CopA-PN with bound nucleotides
CopA-PN studied here consists of residues 398–673, which includes the cytoplasmic ends of the two transmembrane helices, corresponding to M4 and M5 in Ca2+-ATPase, and is slightly larger than that studied earlier (Sazinsky et al, 2006b). The structure of CopA-PN with bound AMPPCP and Mg2+ was determined by molecular replacement starting from the model without a bound nucleotide (Sazinsky et al, 2006b) (PDB ID: 2B8E) and refined at 1.85 Å resolution to an Rfree of 22.5% (Table I). The asymmetric unit contained two molecules (MolA and MolB) related by non-crystallographic symmetry, with an r.m.s.d. of 0.76 Å for 267 Cα atoms, showing that the two protomers were similar. An anomalous Fourier difference map for selenomethionine (SeMet)-substituted CopA-PN showed peaks at the sulphur atoms in Met residues in the refined model (Supplementary Figure 1 and Supplementary Table I), corroborating the accuracy of the atomic model. Other crystal structures listed in Table I were determined starting from this atomic model.
Table 1.
Diffraction data and refinement statistics
| Crystal | AMPPCP-Mga | ADP-Mg | His462Glna |
|---|---|---|---|
| Resolution range (Å) | 100.0–1.85 (1.90–1.85) | 100.0–1.85 (1.90–1.85) | 100.0–1.95 (2.01–1.95) |
| Space group | P4322 | P4322 | P4322 |
| Cell a=b (Å) | 90.787 | 89.956 | 90.518 |
| c (Å) | 191.788 | 190.375 | 191.218 |
| Rmerge (%) | 8.1 (29.1) | 12.9 (48.3) | 5.8 (23.2) |
| I/σ (I) | 33.7 (2.8) | 30.6 (5.8) | 32.3 (3.1) |
| Completeness (%) | 99.6 (99.1) | 99.9 (100) | 99.9 (99.8) |
| Redundancy | 6.2 (6.2) | 3.6 (3.4) | 6.5 (5.9) |
| AMPPCP-Mg | ADP-Mg | His462Gln | |
| Resolution range (Å) | 20.0–1.85 | 20.0–1.85 | 20.0–1.95 |
| No. of reflections | 65461 | 63866 | 55707 |
| Rwork/Rfree (%) | 19.5/22.5 | 19.2/22.6 | 20.8/24.5 |
| Number of atoms | 4573 | 4644 | 4411 |
| Overall B-factor (Å2) | 44.9 | 41.0 | 49.7 |
| r.m.s.d. bond (Å) | 0.011 | 0.011 | 0.012 |
| r.m.s.d. angles (deg) | 1.3 | 1.3 | 1.3 |
| Ramachandranb (%) | 93.3/6.5/0.2/0 | 94.6/5.4/0/0 | 93.9/5.4/0.6/0 |
Parentheses denote statistics in the highest-resolution shells. All data were collected at the wavelength of 0.9 Å.
aData from two crystals were merged in AMPPCP-Mg and His462Gln data sets.
bFractions of residues in the most favoured/additionally allowed/generously allowed/disallowed regions of Ramachandran plot according to ProCheck (Collaborative Computational project (1994)). No residue was found in the disallowed region.
Compared with the earlier model without a bound nucleotide, the angle between the P- and N-domains was smaller by ∼6° (Figure 1A). But the difference within the individual domains was very small (r.m.s.d. of 0.47 Å (P) and 0.59 Å (N)), showing that AMPPCP binding hardly changes the conformation. Nevertheless, because a larger polypeptide was used, the turn structure that corresponds to the P7 helix in Ca2+-ATPase, a key element for transmitting structural changes that occur in the P-domain to the A-domain (Toyoshima and Mizutani, 2004), is clearly defined. The P0 strand, which integrates the M4 helix into the P-domain in Ca2+-ATPase, is also clear. Altogether, the present structure reinforces and extends the similarity in the P-domain between the PIB- and PII-ATPases.
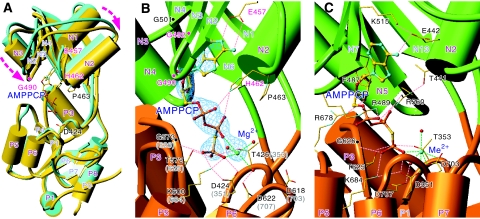
Figure 1.
Overall structures of CopA-PN with bound nucleotides. (A) Superimposition of models for AMPPCP-Mg2+-bound form (yellow) and -unbound form (Sazinsky et al, 2006b) (cyan) (PDB entry 2B8E) of CopA-PN aligned with the P-domain. AMPPCP is shown in ball-and-stick representation. Purple dashed arrows show the movements of the N-domain induced by the binding of AMPPCP. (B, C) Details of the nucleotide-binding sites of the AMPPCP-Mg2+-bound form of CopA-PN (B) and the E1-AMPPCP-Me2+ form of Ca2+-ATPase (Toyoshima and Mizutani, 2004) (PDB entry 1VFP) (C). Small spheres represent Mg2+ (green in B) and Me2+ (physiologically Mg2+ but most likely Ca2+ in the crystal (Picard et al, 2007); cyan in C), and coordinating water molecules (red). The pink broken lines show likely hydrogen bonds, as defined with HBPLUS (McDonald and Thornton, 1994). The cyan net in B represents a simulated annealed omit map contoured at 4.5σ of AMPPCP-Mg2+ and its coordinating water molecules. The residues associated with Wilson disease are labelled in red. Corresponding residue numbers in the Ca2+-ATPase are shown in parentheses.
A Fourier difference (∣Fobs∣−∣Fcalc∣) map before introducing AMPPCP and Mg2+ into the atomic model (Figure 1B) clearly located the bound nucleotide near the hinge between the N- and P-domains. Yet, electron density for Mg2+ and that for the γ-phosphate were nearly equal in strength. Therefore, a Mn2+ derivative was prepared and the position of the Mn2+ was identified by anomalous scattering (Supplementary Figure 2). This position was also consistent with the one obtained using a Mg2+-free derivative, which was made by dialysing Mg2+ away from preformed crystals. These maps indicated that one Mg2+ is bound to AMPPCP in MolA, but none in MolB. No hint of Mg2+ was found in the P-domain in either protomers.
The Fourier difference map showed that the adenine ring of AMPPCP is inserted into a hydrophobic cleft between an α-helix (N2) and the central β-sheet in the N-domain, and that the triphosphate moiety extends towards the phosphorylation residue Asp424 in the P-domain (Figures 1 and 2). The γ-phosphate of AMPPCP forms hydrogen bonds with residues in the P-domain conserved over the entire P-type ATPase family (Figures 1 and 3). Thus, the bound nucleotide bridges the P- and N-domains as in Ca2+-ATPase (Toyoshima and Mizutani, 2004).
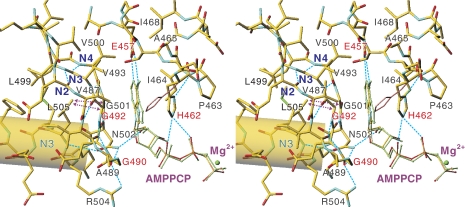
Figure 2.
A stereo view of the adenine-binding site of CopA-PN. The atomic model represents MolA, with AMPPCP in MolB (light green) and His462 in the nucleotide-unbound form (pink) superimposed. Dotted arrows indicate van der Waals contacts between Leu505 and the Cα atoms of Gly492 and Gly501. Broken cyan lines show likely hydrogen bonds. The cylinder represents the Nα3 helix.
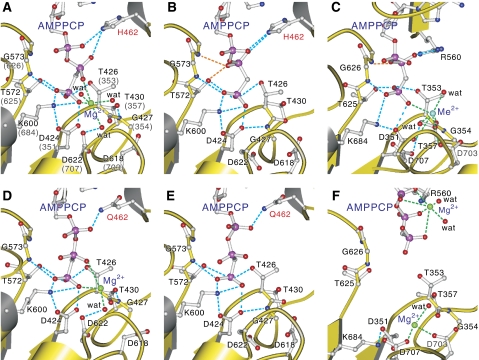
Figure 3.
Details of the triphosphate-binding site in the P-domain. MolA (A) and MolB (B)of CopA-PN wild type, those of His462Q mutant (D, E) and Ca2+-ATPase (C: E1·AMPPCP form (Toyoshima and Mizutani, 2004), F: E2·ATP(TG) form: PDB entry 2DQS). Side chains of important residues and AMPPCP are shown in ball-and-stick. Broken lines in light blue show likely hydrogen bonds, and those in green show coordination of the divalent cation. Broken lines in orange indicate hydrogen bonds specific to MolB (B) and Ca2+-ATPase (C). Small spheres represent Mg2+ (green), Me2+ (most likely Ca2+ in the crystal structure (Picard et al, 2007); cyan), and coordinating water molecules (red). Prepared with Molscript (Kraulis, 1991).
However, the recognition of the adenine ring is entirely different between CopA and Ca2+-ATPase, even though the dispositions of α-helices and β-sheets in the N-domain are rather similar (Figure 1B and C). In Ca2+-ATPase, the adenine ring stacks with Phe487 (Toyoshima and Mizutani, 2004), a very well-conserved residue in PIA (Haupt et al, 2006)-, PII (Morth et al, 2007)- and PIII (Pedersen et al, 2007)-type ATPases, sticking out from the β-sheet. In CopA, the adenine ring is flanked by the β-sheet itself on one side and, on the other, by the side chains of Ile464 and His462 at the end of the N2 helix. A part of the β-sheet is extraordinarily flat because two Gly's (Gly492 and 501) in the β-sheet take unfavourable dihedral angles, so that all the Gly atoms lie in the plane of the β-sheet and make contact with the adenine ring. Hence, any substitution of Gly492 will disallow such a configuration of the β-strand and the side chain will protrude on the opposite side of the β-sheet to the adenine ring. Therefore, in many cases, mutations of this residue could well be tolerated (Okkeri et al, 2004). The β-sheet has a distinct ridge consisting of three Val's (Val487, 493 and 500) at the middle, which lines an upper corner of the binding cleft. At the very top is located Glu457, a critical residue, the carboxyl group of which forms hydrogen bonds with the N1 and N6 atoms of the adenine ring (Figure 2). The orientation of the Glu457 carboxyl is fixed by four hydrophobic residues surrounding it. The structural role of this invariant Glu, substitutions of which abolish ATPase activity (Okkeri et al, 2004) and adenine binding (Morgan et al, 2004), is now clarified.
At one end of the β-sheet is located another critical Gly (Gly490), the carbonyl of which forms hydrogen bonds with Arg504 and Asn502 side chains. As the side chain amide of Asn502 in turn forms a hydrogen bond with an OH-group of ribose, its positioning seems critically important. Any substitution of Gly490 will disallow such hydrogen bonds, because the Cβ atom of the substituted residue will collide with the Ala489 carbonyl, consistent with the involvement of the corresponding Gly1099 in Wilson disease (Cox and Moore, 2002).
Such unfavourable main chain conformations of Gly492 and Gly501 seem to be imposed by van der Waals contacts with Leu505 located at the end of the N3 helix (Figure 2). In the NMR structure of ATP7B (Dmitriev et al, 2006), the Wilson disease protein, Trp1153 side chain fulfils this role. The main chain amide of Leu505 forms a hydrogen bond with Asn502, which is made possible by the unfavourable main chain conformation of Gly501; the orientation of the side chain of Leu505 is determined by the surrounding hydrophobic residues. Rather tight packing of the N3 helix against the β-sheet seems to be stabilised by the clustering of hydrophobic residues at the interface. Thus, the functional roles of these conserved Gly's are evident and distinctly different from those in the P-loop (Walker et al, 1982). Their conformations are not affected by the binding of nucleotide.
On the other side of the adenine ring is located the invariant 462HP. Here, the role of His462 is obvious (Figure 2). A nitrogen atom (Nɛ1) of His forms hydrogen bonds with the α- and β-phosphates, and thereby orients the γ-phosphate towards the phosphorylation residue (Asp424). The other nitrogen atom (Nɛ2) on the imidazole ring is used for making hydrogen bond with the Ile464 amide to orient the imidazole ring correctly. Its positioning is evidently aided by van der Waals contacts with Pro463 and the adenine ring. This conformation of the His462 side chain seems to be a less favourable one when nucleotide is absent (Sazinsky et al, 2006b) (pink stick in Figure 2). In the AMPPCP-bound form, Pro463 is in van der Waals contacts with the side chain of Thr426 in the P-domain (3.95 Å), suggesting that it might work as a stopper or a reporter of the N-domain inclination.
Structure of the His462Gln mutant
In Wilson disease, which is a genetic disorder associated with mutations in human Cu+-ATPase ATP7B causing Cu+ accumulation in the liver, brain and other tissues (Das and Ray, 2006), a Gln-substitution of the invariant His1069 in the HP-motif is by far the most frequent mutation in Caucasian patients (Thomas et al, 1995). To more deeply understand the role of His462 and the effects of the mutation, we determined the crystal structure of His462Gln mutant of CopA-PN with bound AMPPCP and Mg2+.
The crystal was generated similarly, although higher concentrations of AMPPCP (14 mM versus 2 mM) and Mg2+ (20 mM versus 4 mM) were required. The atomic model for the mutant was almost the same as that of the wild type (r.m.s.d. of 0.28 Å) except around AMPPCP. The mutant binds AMPPCP similarly, as the Gln side chain partially mimics the imidazole ring of His (Figure 4). The carbonyl group of Gln side chain forms a hydrogen bond with Ile464 amide, as in the wild type (Figure 4), and thereby fixes the Gln side chain. At this position, however, the amide group donates its hydrogen to the α-phosphate only, leaving the β-phosphate free. It is impossible for AMPPCP to take the same zigzag conformation as in the wild type, because the oxygen atom in the α-phosphate resides too far away from the side chain amide of Gln462 (Supplementary Table II). As a result, the conformations of the phosphate groups in MolA and MolB are different from those in the wild type (Figure 3).
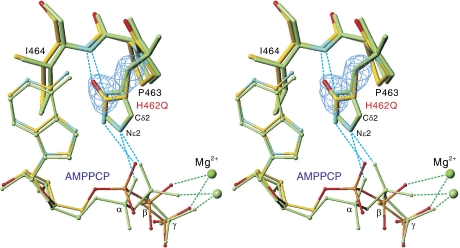
Figure 4.
Adenine recognition by His462Gln mutant. Superimposition of the atomic models for His462Gln mutant (atom colour) and wild type (green) of CopA-PN. His/Gln462 -Pro463-Ile464 are shown in stereo. The cyan net represents an ∣Fobs∣−∣Fcalc∣ electron density map contoured at 4σ when Gln462 was refined as Gly using the mutant data.
Conversely, the conformation of AMPPCP in the mutant crystal cannot be realised in the wild type, because the α-phosphate comes too close to His462 and there is no room for β-phosphate to rotate, so that it can form a hydrogen bond with His462 (Supplementary Figure 3 and Table II). This is because the angle between the ribose and the oxygen atom bridging to the α-phosphate (O5*) is larger in the wild type (Supplementary Figure 3), suggesting that the α-phosphate is pushed away from His462 by van der Waals contacts (Supplementary Table II). This in turn makes enough space for the β-phosphate to rotate. These observations highlight the importance of the α-phosphate in correctly positioning the β-phosphate in the zigzag conformation of the wild-type structure.
Isothermal titration calorimetry (ITC) showed that the His462Gln mutation reduces the affinity for AMPPCP ∼20-fold (Kd=∼0.1 mM for the wild type and ∼2.2 mM for the mutant; Supplementary Figure 4). Indeed, at least 8 mM of AMPPCP was necessary for generating mutant crystals, whereas 2 mM was sufficient for the wild type. In Enterococcus hirae CopB (Bissig et al, 2001) and Escherichia coli ZntA (Okkeri et al, 2004), a zinc-transporting ATPase, the ATPase activity and phosphoenzyme formation from ATP were reduced by the corresponding mutations, but only to 40–20% of the wild type at high ATP concentrations. In contrast, other mutations of the same residue abolished ATPase activity in ZntA (Okkeri et al, 2004). Thus, in archaeal and bacterial PIB-ATPases, the Gln-substitution of the invariant His decreases, but retains some ability for phosphoryl transfer, consistent with the crystal structures.
In human ATP7B, however, phosphoryl transfer was not detected with the His1069Gln mutant even at 1 mM ATP (Tsivkovskii et al, 2003). With the isolated N-domain of ATP7B, ITC measurements showed that the mutation reduces the affinity for nucleotides ∼16-fold (Kd of ∼0.075 to ∼1.2 mM) (Morgan et al, 2004) similarly to CopA-PN. Thus, the consequence of the His to Gln mutation seems to be different in human and archaeal/bacterial PIB-ATPases. One possible explanation is that the HP-motif is not solely involved in ATP binding. As the HP-motif is located on the outermost surface of the N-domain and proximal to the A-domain, it might be involved in interaction with other domains. In fact, from a low-resolution 3D electron microscopy of the full length CopA, Wu et al (2008) suggested that the N-terminal metal-binding domain (NMBD) may interact with the HP-motif and the A-domain. The loop (Ser424-Val437) in the N-domain of Ca2+-ATPase, one of the two contact sites with the A-domain in the E1·AMPPCP form (Toyoshima and Mizutani, 2004), is certainly absent in CopA or ATP7B. It has also been reported that the NMBD interacts with the N-domain in both human ATP7B (Tsivkovskii et al, 2001) and bacterial CopA (Lübben et al, 2009). As the number of the CxxC repeat in the NMBD is variable (one or two in bacterial and archaeal PIB-ATPases to six in human ATP7B), and because its correct interaction with the main body of the ATPase is critical in ATP hydrolysis (Hatori et al, 2007, 2008), it is conceivable that the His to Gln mutation causes different effects depending on the variations in the interaction with the NMBD.
Binding of the triphosphate
We have so far focused on the recognition of the adenine ring in which there is virtually no difference between the two protomers. However, the structures around the β- and γ-phosphates of AMPPCP are distinctly different and also differ from those in Ca2+-ATPase (Figure 3). First, Mg2+ is found only with AMPPCP bound to MolA. It is coordinated by the β- and γ-phosphates of AMPPCP and three water molecules. The geometry of coordination is far from ideal: distances from the coordinating oxygen atoms are too long and the sixth coordination by a water molecule is sterically hindered by Thr426 (Figure 3A). Hence, the affinity of this Mg2+ seems rather low. Second, the β-phosphate has different configurations between MolA and MolB and the one in MolB is stabilised by two extra hydrogen bonds (orange dotted lines in Figure 3B) with Thr572 and Asp574 in the conserved 572TGD-motif. Third, the main chain of Gly427 in the π-helix that connects the phosphorylation site to the N-domain is flipped in MolB to form a normal α-helix (Figure 3B). This π-helix is one of the structural motifs that characterises the haloacid dehalogenase superfamily to which P-type ATPase belongs and is thought to have an important function in changing the water accessibility of the active site (Burroughs et al, 2006). As a result of this flipping, the Gly427 amide is hydrogen bonded to the Asp424 carboxyl, instead of the Thr430 hydroxyl in MolA (Figure 3).
These structural features around the phosphate chain are also different from those in the crystals of Ca2+-ATPase. AMPPCP in the E2·ATP(TG) form has a distinctly different conformation (Figure 3F). AMPPCP in the E1·AMPPCP form (Figure 3C) exhibits a conformation of triphosphate moiety similar to that in MolB. The hydrogen bonding pattern is also closer to that in MolB and the AMPPCP is devoid of bound Mg2+ (Figure 3). Small differences in hydrogen bonding pattern (e.g. lack of the hydrogen bond between the β-phosphate and the Thr625 hydroxyl in Ca2+-ATPase) may be attributed to the difference in hydrogen bond donor, which are His462 in CopA and Arg560 in Ca2+-ATPase. However, in Ca2+-ATPase, the peptide bond between Thr353 and Gly354 in the π-helix is not flipped and a Me2+ (physiologically Mg2+, but most likely to be Ca2+ in the crystal (Picard et al, 2007)) is coordinated by the γ-phosphate of AMPPCP, Asp351 and Asp703. Thus, the conformation of AMPPCP and the ATPase structure around the γ-phosphate are distinctly different between CopA-PN and Ca2+-ATPase.
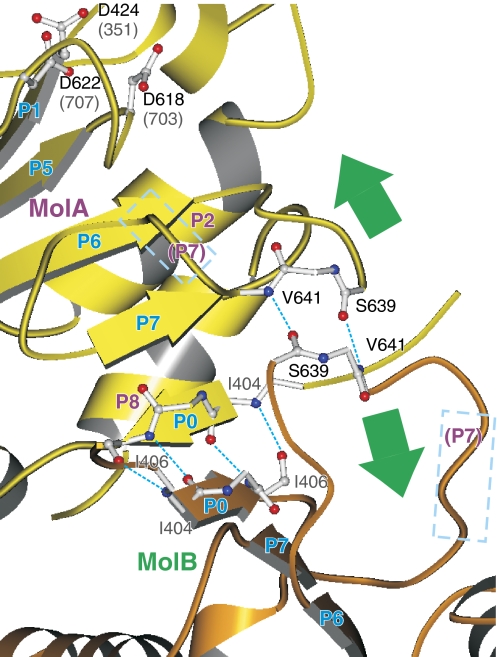
Yet, in all three structures, that is, MolA and MolB of CopA-PN and E1·AMPPCP form of Ca2+-ATPase, the γ-phosphate is stabilised by hydrogen bonds with Lys600/684, Thr572/625, Thr426/353 and possibly with Asp424/351 (if protonated), suggesting that these three different arrangements of the γ-phosphate, Mg2+ and the π-helix are all related and physiologically relevant. Particularly important here is that Mg2+ would be able to bind to AMPPCP as in MolA of CopA-PN in the E1·AMPPCP form of Ca2+-ATPase, if Mg2+ did not bind to the P-domain to bring Asp703 towards the γ-phosphate. In CopA-PN, Mg2+ probably could not bind to the P-domain because crystal packing suppressed the movements of the P-domain (arrows in Figure 5) required for Mg2+ binding. The P-domain of either protomers is very tightly packed by extending the central β-sheet over adjacent protomers (Figure 5).
Figure 5.
Crystal contact between the P-domains of adjacent protomers. Dotted lines in cyan show hydrogen bonds between the β-strands in neighbouring protomers (MolA, yellow; MolB, orange). Rectangles in broken lines show the P7 helices, which has an important role in transmitting the binding signal of Mg2+ to the transmembrane domain in Ca2+-ATPase (Toyoshima and Mizutani, 2004). Green arrows indicate the movements expected for the part of the P-domain induced by the binding of Mg2+.
An interesting possibility then is that the two structures around the γ-phosphate in CopA-PN crystal may represent transient states before the final, productive structure realised in the Ca2+-ATPase crystal, and may actually exhibit a mechanism for removing bound Mg2+ from ATP in the normal reaction cycle. This is an important step, as the removal of Mg2+ from the γ-phosphate is a prerequisite to its binding to the P-domain, which pulls electrons from the carboxyl group of Asp, and thereby makes the phosphoryl transfer to the Asp feasible. We must remember that the free energy liberated by hydrolysis of aspartylphosphate is larger (11.7 kcal/mol) than that of ATP (7.3 kcal/mol) under the standard conditions, implying that the contribution of Mg2+ is essential.
Conformation of the triphosphate and flipping of a peptide bond in the π-helix
At this stage, we can say that the flipping of the peptide bond between Thr426 and Gly427 in the π-helix is not the result of unbinding of Mg2+ from AMPPCP or extra hydrogen bonds at the β-phosphate. This is because we observed the same difference between MolA and MolB in the ADP derivative, in which the bound AMPPCP was replaced with ADP by soaking the preformed crystals (Supplementary Figure 5). In the ADP derivative, the conformation of ADP was indistinguishable between MolA and MolB. Hence, the flipping seems to be a result of crystal packing. In fact, though not described earlier, two conformations of Thr426-Gly427 are found in different protomers in the nucleotide-free form of CopA-PN (PDB ID: 2B8E).
Furthermore, the crystal structure of the His462Gln mutant indicates that the presence or absence of Mg2+ is unrelated to the configuration of the β-phosphate. The β-phosphate in MolB is free to take the same configuration as that in MolA, even though Mg2+ is absent (Figure 3D, E). This means that whether Mg2+ can bind to AMPPCP or not is determined by the protein structure around the γ-phosphate, and the only difference is the flipping of Thr426-Gly427 peptide bond. Then, the question to be addressed is how the flipping affects the coordination of the Mg2+. Superimposition of the atomic models for MolA and MolB shows that the flipping does not sterically interfere with either Mg2+ or water molecules directly coordinating with Mg2+.
From the crystal structure of the Asp351Ala mutant of Ca2+-ATPase (Marchand et al, 2008), we know that the contribution from the Asp351 carboxyl is not essential for Mg2+ binding to the P-domain between Asp351 and Asp703, and the affinity of ATP is much higher in the mutant (McIntosh et al, 2004). Then a likely cause for the dissociation of Mg2+ is the introduction of the amide group of Gly427 near Mg2+ and resultant decrease of electro-negativity there. This flipped form is evidently the less stable one, as all the other crystal structures of the P-type ATPases (Toyoshima, 2008) do not show it. Nonetheless, a modelling experiment with Ca2+-ATPase shows that the flipped form is quite possible.
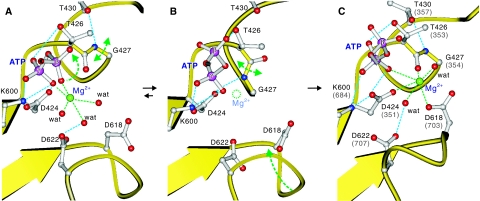
These considerations lead us to propose a possible scenario for the dissociation of Mg2+ from ATP and binding to the P-domain (Figure 6). In this model, ATP binds to the ATPase in an extended conformation with Mg2+ between the β- and γ-phosphates (Figure 6A). This is the conformation of Mg2+·ATP most frequently observed in many crystal structures of ATPases and kinases. In this state, the Mg2+ is not coordinated by any protein atom (Figures 3A and 6A), far from Asp424, the phosphorylation residue, and from Asp618, the Mg2+-coordintating residue, absolutely conserved in all P-type ATPases. The Mg2+ bound to ATP dissociates when the Thr-Gly peptide bond flips, possibly by thermal movement of the π-helix or possibly by inclination of the N-domain, and the main chain amide of Gly comes close to the Mg2+ (Figure 6B). This dissociation and thermal movement of the P-domain allow a change in ATP conformation to a zigzag form and the binding of Mg2+ (Figure 6C), so that it bridges the γ-phosphate, Asp424 and Asp618, as realised in the E1·AMPPCP form of Ca2+-ATPase (Figure 3C). In this zigzag form, the β-phosphate of ATP is firmly fixed by three to four hydrogen bonds, presumably to make the γ-phosphate approach the phosphorylation residue (Asp424) with the correct geometry. Then the Mg2+ will pull electrons from the γ-phosphate and the Asp424 carboxyl to facilitate a covalent bond formation between them.
Figure 6.
A scenario for the removal of Mg2+ from ATP and re-binding to the P-domain before phosphoryl transfer. Models were built from the crystal structures of MolA (A) and MolB (B) of CopA-PN and the E1·AMPPCP form of Ca2+-ATPase (C), and the residue numbers refer to those of CopA; those of Ca2+-ATPase are shown in parentheses (C). Only the β- and γ-phosphates of ATP are shown. Lys425 to Leu429 form a π-helix (A, C), which is converted to a normal α-helix when Thr426-Gly427 peptide bond is flipped (B). Mg2+ bound to ATP (A) dissociates by the flipping (B) and binds to the P-domain bridging the γ-phosphate of ATP, the phosphorylation residue (D424) and a Mg2+-coordinating residue (D618). Broken lines show likely hydrogen bonds (cyan) and coordination of Mg2+ (green). Small spheres represent Mg2+ (green) and coordinating water molecules (red). Arrows indicate expected movements during the Mg2+ relocation.
Such flipping of the peptide bond has been observed with metal-binding sites in an ion pump and a channel. They include Glu309-Gly310 in Ca2+-ATPase (Obara et al, 2005) and Val76-Gly77 in the selective filter in the KcsA potassium channel (Bèrneche and Roux, 2000; Zhou et al, 2001). In Ca2+-ATPase, Glu309 provides two carboxyl oxygen atoms to Ca2+, and the peptide bond flipping is a part of the conformational changes required for accommodating the movement of the transmembrane helix M4. In the KcsA crystal structure (Zhou et al, 2001), the Val76 carbonyl points to the inside of the pore in the ‘high-K+' structure, but away from the pore in ‘low-K+'. Although the substitution of the Gly by Ala in Ca2+-ATPase did not produce substantial effects (Maruyama et al, 1989), that Gly is an absolutely conserved residue in all P-type ATPases and might have a role as proposed here.
Conclusion
We have described that CopA-PN, the P- and N-domains of an archael Cu+-ATPase, a representative member of heavy metal pumps, recognises the adenine ring entirely different from other P-type ATPases, despite the fact that the arrangements of the secondary structure elements are very similar (Figures 1B and C). Yet, we can perhaps say that the basic mechanism is the same, because what seems to be required for adenine recognition is a flat surface (either the aromatic ring of Phe or an ultra-flat β-sheet) and an electrostatic interaction that stabilises N6 (and N1) of the adenine ring (Figure 1). For this purpose, Glu457 is used in CopA, whereas it is Glu442 in Ca2+-ATPase. P-type ATPases do not use the most common mechanism involving the main chain carbonyl and amide groups (Cappello et al, 2002). Another important device is a guide to orient the γ-phosphate to the phosphorylation residue with the correct geometry—key for efficient phosphoryl transfer. In many ATPases and GTPases, the P-loop does this job. In Ca2+-ATPase, the phosphate chain takes a characteristic zigzag conformation because of Arg489 and Arg560 making hydrogen bonds with the α- and β-phosphates, respectively (Toyoshima and Mizutani, 2004). We observed a similar zigzag conformation of the phosphate chain in CopA, except His462 alone is responsible. The His462Gln-mutant structure informs the importance of the positioning of the α-phosphate. The structure of the imidazole ring and particularly its rigidity, backed by the adjacent Pro residue, seem critical for this purpose. In Ca2+-ATPase, Thr441 takes an equivalent position to His462 and fixes the Arg560 side chain (Figure 1C); Arg489, in turn, fixes the α-phosphate with respect to the ribose, and thereby orients the β-phosphate to interact with the Arg560. Such involvement of multiple residues could be useful for accelerating partial reactions by ATP. In Na+,K+-ATPase and Ca2+-ATPase, residues that stabilise domain interactions are also recruited for ATP binding, thereby facilitating transitions to next steps (Shinoda et al, 2009). Another possibility for acceleration is to destabilise the binding of ADP after phosphoryl transfer. The zigzag conformation of the triphosphate chain might be a manoeuvre for this, by placing strain on the triphosphate chain. In Ca2+-ATPase, a conformational change of the Phe487 side chain (stacked with the adenine moiety) will easily destabilise the binding of the adenine ring. In CopA, ATP and ADP have similar affinities (Morgan et al, 2004) as the adenine ring is firmly held by hydrogen bonds with Glu457 and by unusually flat β-sheet. These structural features may explain, at least partially, a much slower turnover of the ATPase reaction cycle in CopA (3–4 s−1 at 60°C) (Hatori et al, 2008) compared with Ca2+-ATPase (∼30 s−1 at 37°C).
The arrangements of AMPPCP and Mg2+ in the two protomers constituting the CopA-PN crystal are different (Figure 3A, B) and also differ from those in the E2·ATP(TG) and E1·AMPPCP forms of Ca2+-ATPase (Figure 3C, F). We regard that the two different conformations of AMPPCP, only one of which has bound Mg2+, represent two snapshots before phosphoryl transfer, and propose a scenario for dissociation of Mg2+ from ATP and its binding to the P-domain (Figure 6). This Mg2+ binding to the P-domain is one of the key events in active transport by P-type ATPases, because it is a prerequisite to the formation of the covalent bond between the γ-phosphate and the carboxyl group of the aspartate, and to pull up the transmembrane M1 helix towards the cytoplasm to occlude ions in the transmembrane-binding sites (Toyoshima and Mizutani, 2004).
Materials and methods
Cloning and purification of CopA-PN
CopA-PN from A. fulgidus, containing the P- and N-domains of CopA (residues Lys398-Lys673), was polymerase chain reaction (PCR) amplified from genomic DNA. The PCR product, with 5′-XhoI and 3′-HindIII restriction endonuclease sites flanking the coding sequence, was cloned into a modified pBAD vector (Invitrogen), which introduces an 11-amino-acid sequence (MGHHHHHHGSR) at the N-terminus. Mutation of His462 to Gln was performed using a QuikChange II site-directed mutagenesis kit (Stratagene). The DNA sequence of the expression constructs was verified by automated DNA sequencing.
E. coli Top10 cells (Invitrogen) containing the expression construct were grown in Luria–Bertani medium at 37°C until the optical density at 600 nm reached 0.3, and then the expression was induced with 0.006% (w/v) L-arabinose. After 15 h at 25°C, the cells were harvested by centrifugation and suspended in buffer A containing 500 mM NaCl, 50 mM imidazole and 50 mM MOPS, pH 7.0, with ‘complete' protease inhibitor cocktail (Roche, Penzberg, Germany) and lysed by sonication for 2 min. The lysate was incubated at 75°C for 45 min for heat denaturation of intrinsic proteins, and centrifuged at 10 000 × g for 30 min. The supernatant was applied to a Ni Sepharose 6 Fast Flow (GE Healthcare), washed with buffer A and the bound protein was eluted with 200 mM imidazole. The eluted protein was subjected to gel filtration on a Superdex75 column (GE Healthcare). The purified CopA-PN was dialysed against 150 mM NaCl, 1 mM DTT and 10 mM MOPS, pH 7.0, and concentrated using Centriprep-30 (Amicon) to a final concentration of approximately 20 mg/ml. For preparation of the SeMet-labelled protein, E. coli cells were grown in LeMaster medium containing 100 mg/ml L-SeMet and the protein was purified by the same procedure using buffers including 1 mM DTT.
Crystallisation and data collection
The protein solution used for crystallisation contains 150 mM NaCl, 1 mM DTT, 10 mM MOPS, pH 7.0, 4 mM MgCl2 (or MnCl2), 2 mM AMPPCP and 12 mg/ml protein. Crystals were grown by vapour diffusion, with a reservoir solution containing 5% (w/v) PEG 6000, 1.25–1.5 M NaCl and 100 mM MES, pH 5.5–5.75, in a drop made by mixing 2 μl of protein solution and 2 μl of reservoir solution at 10°C for up to 3 days. Mutant crystals were generated similarly, but with 14 mM AMPPCP and 20 mM MgCl2 at 20°C. Before flash freezing in cold nitrogen gas, crystals were soaked in the reservoir solution supplemented with AMPPCP, MgCl2 (or MnCl2) and 30% (v/v) ethyleneglycol. ADP-Mg2+-bound crystals were prepared by soaking the AMPPCP-Mg2+-bound crystals in a solution containing 4 mM MgCl2 and 2 mM ADP for 1 day at 4°C.
All the diffraction data were collected from crystals cooled to 100 K at BL41XU of SPring-8 with an ADSC Quantum 315 CCD detector, and processed with Denzo and Scalepack (Otwinowski and Minor, 1997). The crystals belonged to the space group P4322 (a=b=90.8 Å, c=191.8 Å), with two protomers in the asymmetric unit.
Modelling and refinement
The crystal structure was determined by molecular replacement using the program MOLREP in CCP4 (Collaborative Computational project, 1994). The atomic model for CopA-PN without bound nucleotide (Sazinsky et al, 2006b) (PDB entry 2B8E) was used as the search model. The angle between the P- and N-domains was adjusted manually. The atomic models were refined with CNS (Brünger et al, 1998) and REFMAC including TLS refinement (Winn et al, 2001). Statistics of the diffraction data and refinement are listed in Table I.
Isothermal titration calorimetry
Protein samples were dialysed extensively at 4°C against 50 mM imidazole–HCl, pH 7.0. Protein and AMPPCP solutions were degassed and filtered before each titration experiment. All measurements were performed at 25°C using an iTC200 microcalorimeter (MicroCal Inc). Nineteen injections (at 180 s intervals) of 2 μl of AMPPCP solution were performed into CopA-PN protein in the ITC cell. The syringe stirring speed was set to 1000 r.p.m, and the reference power at 5 μcal/s. The thermal power was monitored every 5 s. Baseline data were measured by titration of the AMPPCP solution into the buffer without protein, and these were subtracted from the experimental data. Thermogram analysis was performed with the Origin 7.0 data analysis software provided by MicroCal.
Protein data bank accession codes
Coordinates and structural factors have been deposited in the Protein Data Bank under accession codes 3A1C (wild-type AMPPCP-Mg2+ form), 3A1D (wild-type ADP-Mg2+ form) and 3A1E (His462Gln mutant AMPPCP-Mg2+ form).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 1 Legend
Supplementary Figure 2
Supplementary Figure 2 Legend
Supplementary Figure 3
Supplementary Figure 3 Legend
Supplementary Figure 4
Supplementary Figure 4 Legend
Supplementary Figure 5
Supplementary Figure 5 Legend
Supplementary Tables I
Supplementary Tables II
Acknowledgments
We thank Drs N Shimizu and M Kawamoto for data collection at BL41XU of SPring-8, and Y Kondou for structure analysis. We are indebted to Dr DB McIntosh for his help in improving the paper. This work is a part of an ongoing long-term project (2006B0013) at SPring-8 and was supported in part by a Grant-in-Aid for Young Scientists (B) (to TT) and a Creative Science Project Grant and Special Promoted Project Grant (to CT) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Argüello JM (2003) Identification of ion-selectivity determinants in heavy-metal transport P-1B-type ATPases. J Membr Biol 195: 93–108 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101 [DOI] [PubMed] [Google Scholar]
- Bèrneche S, Roux B (2000) Molecular dynamics of the KcsA K+ channel in a bilayer membrane. Biophys J 78: 2900–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig KD, Wunderli-Ye H, Duda PW, Solioz M (2001) Structure-function analysis of purified Enterococcus hirae CopB copper ATPase: effect of Menkes/Wilson disease mutation homologues. Biochem J 357: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L (2006) Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 361: 1003–1034 [DOI] [PubMed] [Google Scholar]
- Cappello V, Tramontano A, Koch U (2002) Classification of proteins based on the properties of the ligand-binding site: the case of adenine-binding proteins. Proteins 47: 106–115 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cox DW, Moore SD (2002) Copper transporting P-type ATPases and human disease. J Bioenerg Biomembr 34: 333–338 [DOI] [PubMed] [Google Scholar]
- Das SK, Ray K (2006) Wilson's disease: an update. Nat Clin Pract Neurol 2: 482–493 [DOI] [PubMed] [Google Scholar]
- Dmitriev O, Tsivkovskii R, Abildgaard F, Morgan CT, Markley JL, Lutsenko S (2006) Solution structure of the N-domain of Wilson disease protein: distinct nucleotide-binding environment and effects of disease mutations. Proc Natl Acad Sci USA 103: 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guerrero M, Argüello JM (2008) Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci USA 105: 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y, Hirata A, Toyoshima C, Lewis D, Pilankatta R, Inesi G (2008) Intermediate phosphorylation reactions in the mechanism of ATP utilization by the copper ATPase (CopA) of Thermotoga maritima. J Biol Chem 283: 22541–22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y, Majima E, Tsuda T, Toyoshima C (2007) Domain organization and movements in heavy metal ion pumps: papain digestion of CopA, a Cu+-transporting ATPase. J Biol Chem 282: 25213–25221 [DOI] [PubMed] [Google Scholar]
- Haupt M, Bramkamp M, Heller M, Coles M, Deckers-Hebestreit G, Herkenhoff-Hesselmann B, Altendorf K, Kessler H (2006) The holo-form of the nucleotide binding domain of the KdpFABC complex from Escherichia coli reveals a new binding mode. J Biol Chem 281: 9641–9649 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Kühlbrandt W (2004) Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol 5: 282–295 [DOI] [PubMed] [Google Scholar]
- Lübben M, Guldenhaupt J, Zoltner M, Deigwelher K, Haebel P, Urbanke C, Scheidig AJ (2007) Sulfate acts as phosphate analog on the monomeric catalytic fragment of the CPx-ATPase CopB from Suffolobus solfataricus. J Mol Biol 369: 368–385 [DOI] [PubMed] [Google Scholar]
- Lübben M, Portmann R, Kock G, Stoll R, Young MM, Solioz M (2009) Structural model of the CopA copper ATPase of Enterococcus hirae based on chemical cross-linking. Biometals 22: 363–375 [DOI] [PubMed] [Google Scholar]
- Mandal AK, Argüello JM (2003) Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochemistry 42: 11040–11047 [DOI] [PubMed] [Google Scholar]
- Marchand A, Winther AM, Holm PJ, Olesen C, Montigny C, Arnou B, Champeil P, Clausen JD, Vilsen B, Andersen JP, Nissen P, Jaxel C, Møller JV, le Maire M (2008) Crystal structure of D351A and P312A mutant forms of the mammalian sarcoplasmic reticulum Ca2+-ATPase reveals key events in phosphorylation and Ca2+ release. J Biol Chem 283: 14867–14882 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Clarke DM, Fujii J, Inesi G, Loo TW, MacLennan DH (1989) Functional consequences of alterations to amino acids located in the catalytic center (Isoleucine 348 to Threonine 357) and nucleotide-binding domain of the Ca2+ -ATPase of sarcoplasmic reticulum. J Biol Chem 264: 13038–13042 [PubMed] [Google Scholar]
- McDonald IK, Thornton JM (1994) Satisfying hydrogen bonding potential in proteins. J Mol Biol 238: 777–793 [DOI] [PubMed] [Google Scholar]
- McIntosh DB, Clausen JD, Woolley DG, MacLennan DH, Vilsen B, Andersen JP (2004) Roles of conserved P-domain residues and Mg2+ in ATP binding in the ground and Ca2+-activated states of sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem 279: 32515–32523 [DOI] [PubMed] [Google Scholar]
- Morgan CT, Tsivkovskii R, Kosinsky YA, Efremov RG, Lutsenko S (2004) The distinct functional properties of the nucleotide-binding domain of ATP7B, the human copper-transporting ATPase: analysis of the Wilson disease mutations E1064A, H1069Q, R1151H, and C1104F. J Biol Chem 279: 36363–36371 [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sørensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P (2007) Crystal structure of the sodium-potassium pump. Nature 450: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Obara K, Miyashita N, Xu C, Toyoshima I, Sugita Y, Inesi G, Toyoshima C (2005) Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc Natl Acad Sci USA 102: 14489–14496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkeri J, Laakkonen L, Haltia T (2004) The nucleotide-binding domain of the Zn2+-transporting P-type ATPase from Escherichia coli carries a glycine motif that may be involved in binding of ATP. Biochem J 377: 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–325 [DOI] [PubMed] [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P (2007) Crystal structure of the plasma membrane proton pump. Nature 450: 1111–1114 [DOI] [PubMed] [Google Scholar]
- Picard M, Jensen AM, Sørensen TL, Champeil P, Møller JV, Nissen P (2007) Ca2+ versus Mg2+ coordination at the nucleotide-binding site of the sarcoplasmic reticulum Ca2+-ATPase. J Mol Biol 368: 1–7 [DOI] [PubMed] [Google Scholar]
- Rice WJ, Kovalishin A, Stokes DL (2006) Role of metal-binding domains of the copper pump from Archaeoglobus fulgidus. Biochem Biophys Res Commun 348: 124–131 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Granillo A, Sedlak E, Wittung-Stafshede P (2008) Stability and ATP binding of the nucleotide-binding domain of the Wilson disease protein: effect of the common H1069Q mutation. J Mol Biol 383: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Sazinsky MH, Agarwal S, Argüello JM, Rosenzweig AC (2006a) Structure of the actuator domain from the Archaeoglobus fulgidus Cu+-ATPase. Biochemistry 45: 9949–9955 [DOI] [PubMed] [Google Scholar]
- Sazinsky MH, Mandal AK, Argüello JM, Rosenzweig AC (2006b) Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J Biol Chem 281: 11161–11166 [DOI] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C (2009) Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature (doi:10.1038/nature07939) [DOI] [PubMed] [Google Scholar]
- Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW (1995) The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet 9: 210–217 [DOI] [PubMed] [Google Scholar]
- Toyoshima C (2008) Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys 476: 3–11 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Mizutani T (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430: 529–535 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nakasako M, Nomura H, Ogawa H (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405: 647–655 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nomura H, Tsuda T (2004) Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432: 361–368 [DOI] [PubMed] [Google Scholar]
- Tsivkovskii R, Efremov RG, Lutsenko S (2003) The role of the invariant His-1069 in folding and function of the Wilson's disease protein, the human copper-transporting ATPase ATP7B. J Biol Chem 278: 13302–13308 [DOI] [PubMed] [Google Scholar]
- Tsivkovskii R, MacArthur BC, Lutsenko S (2001) The Lys1010-Lys1325 fragment of the Wilson's disease protein binds nucleotides and interacts with the N-terminal domain of this protein in a copper-dependent manner. J Biol Chem 276: 2234–2242 [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Wu CC, Rice WJ, Stokes DL (2008) Structure of a copper pump suggests a regulatory role for its metal-binding domain. Structure 16: 976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414: 43–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 1 Legend
Supplementary Figure 2
Supplementary Figure 2 Legend
Supplementary Figure 3
Supplementary Figure 3 Legend
Supplementary Figure 4
Supplementary Figure 4 Legend
Supplementary Figure 5
Supplementary Figure 5 Legend
Supplementary Tables I
Supplementary Tables II