Abstract
Proton pumping NADH:ubiquinone oxidoreductase (complex I) is the largest and remains by far the least understood enzyme complex of the respiratory chain. It consists of a peripheral arm harbouring all known redox active prosthetic groups and a membrane arm with a yet unknown number of proton translocation sites. The ubiquinone reduction site close to iron-sulfur cluster N2 at the interface of the 49-kDa and PSST subunits has been mapped by extensive site directed mutagenesis. Independent lines of evidence identified electron transfer events during reduction of ubiquinone to be associated with the potential drop that generates the full driving force for proton translocation with a 4 H+/2e− stoichiometry. Electron microscopic analysis of immuno-labelled native enzyme and of a subcomplex lacking the electron input module indicated a distance of 35–60 Å of cluster N2 to the membrane surface. Resolution of the membrane arm into subcomplexes showed that even the distal part harbours subunits that are prime candidates to participate in proton translocation because they are homologous to sodium/proton antiporters and contain conserved charged residues in predicted transmembrane helices. The mechanism of redox linked proton translocation by complex I is largely unknown but has to include steps where energy is transmitted over extremely long distances. In this review we compile the available structural information on complex I and discuss implications for complex I function.
Keywords: complex I, NADH dehydrogenase, NDH-1, mitochondria, proton pumping, ubiquinone, iron-sulfur cluster, electron microscopic single particle analysis
Introduction
Proton pumping NADH:ubiquinone oxidoreductase (complex I) is a very large and complicated integral membrane protein complex [1]. Coupling the transfer of two electrons from NADH to ubiquinone to the vectorial translocation of four protons across the inner mitochondrial membrane or the plasma membrane of many bacteria, it generates a major portion of the proton motive force that drives ATP synthesis. Mitochondrial complex I from a number of species can undergo a so called active/deactive (A/D) transition [2]. Exposure of complex I to elevated temperature (30–37°C) induces the deactive (D) form that exhibits a characteristic lag phase in the onset of catalytic activity. Catalytic turnover converts complex I back to the metastable active form.
Genetic defects in human complex I manifest as severe degenerative encephalomyopathies, typically diagnosed as Leigh or Leigh-like syndrome [3]. Malfunction of complex I seems to be involved in the pathogenesis of many neurodegenerative diseases [4] and the biology of aging in general [5].
The eucaryotic enzyme has a total mass in the range of 1 MDa. Complex I from bovine comprises 45 [6] and complex I from the aerobic yeast Yarrowia lipolytica 40 different proteins [7] which can be categorised as “central” or “accessory” subunits. The 14 central subunits are conserved in prokaryotes and eukaryotes and harbour all bioenergetic core functions, while the accessory subunits present in eukaryotes only are not associated directly with energy conservation. The central subunits can be divided into seven hydrophilic and seven hydrophobic polypeptides. In most eukaryotes the central hydrophobic subunits (ND1 to ND6 and ND4L) are encoded by mitochondrial DNA [1]. At least a subset of these subunits must be involved in proton pumping but even the number of proton translocation sites is not known. On the other hand the hydrophilic central subunits contain one FMN and eight canonical iron-sulfur clusters as redox active cofactors. These subunits are structurally well described by X-ray crystallography of a subcomplex of a bacterial enzyme [8]. Of the accessory subunits only the structure of the heterologously expressed human accessory subunit B8 has been determined by NMR spectroscopy [9]. To date, no high resolution structure of the holo-enzyme or any of the hydrophobic subunits is available.
The prokaryotic minimal form of complex I has been characterized from the species Escherichia coli [10], Paracoccus denitrificans [11], Thermus thermophilus [12] and Aquifex aeolicus [13]. To establish a yeast genetic approach to complex I, our group has used the obligate aerobic yeast Yarrowia lipolytica [14], since this enzyme is absent in fermentative yeasts like Saccharomyces cerevisiae [15]. Further eucaryotic model organisms in complex I research include Bos taurus [16;17], fungi like Neurospora crassa [18], Podospora anserina [19] and Aspergillus niger [20], plants like Arabidopsis thaliana [21;22], Solanum tuberosum [23] and Oryza sativa [24], and many more. Chloroplasts and cyanobacteria possess a complex I like enzyme, which however has a different electron input module [25–27]. Here we compile the available information on complex I structure, the localization of functional domains and discuss the implications of their spatial arrangement for complex I function.
Overall structure
An L-shaped overall architecture with two arms of approximately equal length was typically found by electron microscopy of complex I from eukaryotes and prokaryotes [13;28–30], despite the large number of accessory subunits present only in the mitochondrial enzyme. A consistent structure between N. crassa and E. coli complex I was reported and the two arms of the bacterial enzyme were found to be only slightly shorter [28].
The identity of the two arms could be established by analysis of subcomplexes from N. crassa complex I [31]: A hydrophilic so-called small form of the enzyme was assembled under conditions where mitochochondrial protein biosynthesis was blocked. A 3D reconstruction of this fragment was generated by electron microscopy of 2D crystals and showed features that allowed an assignment to the “vertical” arm of complex I. Treatment of the holo-enzyme with NaBr specifically removed the part of the enzyme protruding from the membrane phase. The structure of the hydrophobic segment of complex I determined by analysis of single particles and membrane crystals was found to correspond to the “horizontal” arm of complex I. 3D reconstructions of the hydrophilic and the hydrophobic fragments were assembled to match the L-shaped structure evident from 2D average images of single particles. Docking of the hydrophilic part to the membrane intrinsic part was, however, arbitrary.
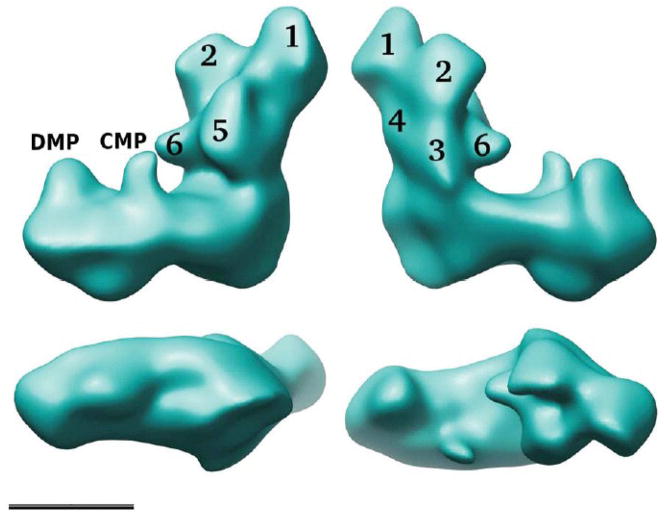
3D reconstructions of complex I are challenging because positional and conformational variability have to be taken into account. For complex I from e.g. E. coli [28], N. crassa [32] and Y. lipolytica [29] the experimental approach to deal with this problem was collection of tilt images and application of the random conical tilt reconstruction method [33]. Extensive classification and 3D reconstruction allowed monitoring detailed substructures within the two arms of complex I from Y. lipolytica (Fig. 1; [29]). In the peripheral arm that extends about 145 Å above the membrane, six distinct domains could be discerned. The membrane arm that has a total length of about 220 Å shows two major protrusions (distal membrane arm protrusion, DMP, and central membrane arm protrusion, CMP) on the matrix side. Hot spots of structural variability could be identified by comparing reconstructions of a number of different classes. CMP appeared in a large number of different sizes and shapes and in some reconstructions indications became visible for possible connections to either DMP or domain 5 of the peripheral arm.
Figure 1. Reconstruction of complex I from Y. lipolytica.
The overall structure combining the five major classes at 24 Å resolution is shown [29]; top, two side views; bottom left, a view of the intermembrane space side of the membrane arm; bottom right, view from the matrix side; scale bar 10 nm. In the peripheral arm six domains labelled 1–6 can be discerned; DMP, distal membrane arm protrusion, CMP, central membrane arm protrusion.
Although determined at different resolution, the 3D reconstructions of complex I from N.crassa and Y. lipolytica are in good agreement. A 3D reconstruction of bovine complex I particles exhibiting rather different structural features was reported [30], but more recently 2D averages of the bovine enzyme were published that look strikingly similar to those of the two fungal enzymes [29;34]. This suggests that the 3D model reported for Y. lipolytica complex I is representative for all mitochondrial enzymes (Fig. 1).
The peripheral arm
Solving the X-ray structure of the peripheral arm fragment of complex I from Thermus thermophilus at 3.3 Å resolution [8] has been a major step forward in the structural characterization of complex I. The overall structure of the Y-shaped peripheral arm fragment has a height of approximately 140 Å. This structure contains all seven hydrophilic central subunits. In addition, there is one extra subunit not commonly found in complex I that contains a putative Fe binding site and is structurally related to frataxin [35]. With FMN and eight iron-sulfur clusters, all redox active prosthetic groups are resolved in this structure. One additional iron-sulfur cluster (N7) present in the structure is only found in some bacteria.
There is an ongoing debate about the assignment of EPR signatures N4 and N5 to clusters seen in the structure [36;37]. Discussing this controversy would go beyond the scope of this review and therefore no definite assignment is given here for four of the tetranuclear clusters. FMN and presumably also the substrate NADH are bound by an atypical Rossman fold in the 51-kDa subunit (bovine subunit nomenclature will be used throughout if not indicated otherwise). Two iron-sulfur clusters are located within electron transfer distance to the flavin. The tetranuclear cluster N3 resides in the same subunit and is the starting point of a 90 Å long linear electron transfer chain that leads from the primary electron acceptor FMN to iron-sulfur cluster N2 which is the immediate electron donor to quinone. While electron transfer chains of T. thermophilus [38] and some other bacterial species function with menaquinone the more common electron acceptor for complex I is ubiquinone. The binuclear cluster N1a in the 24-kDa subunit is located on the opposite side of the flavin forming a dead end for electron transfer of unclear function (see below).
The electron transfer chain between FMN and ubiquinone is formed by seven iron-sulfur clusters. Following N3 there are binuclear cluster N1b and two tetranuclear clusters in the chain that reside in the 75-kDa subunit. The N-terminal third of the subunit is structurally related to [FeFe] hydrogenases, while the rest of the subunit is similar to molybdopterin guanine dinucleotide (MGD) cofactor containing proteins like nitrogenase. In some bacteria the latter domain harbours cluster N7 that probably has a structural but no electron transfer function [39]. The two tetranuclear clusters of the TYKY subunit are bound in a ferredoxin like arrangement and the electron transfer chain terminates with cluster N2 in the PSST subunit. Two of the cysteine ligands of cluster N2 are adjacent but the functional consequences of this unusual arrangement are unclear [40]. The PSST subunit is very closely associated with the 49-kDa subunit and the two polypeptides are related to the small and large subunit of soluble [NiFe] hydrogenases, respectively [41].
On the basis of X-ray structures of [NiFe] hydrogenases that were solved long before the structure of the peripheral arm of complex I, we had proposed evolutionary conservation of the structural fold around the active sites for these distantly related enzymes: the hydrogenase [NiFe] site is structurally conserved in the 49-kDa subunit of complex I to form a significant part of the ubiquinone binding pocket [42]. This concept was also prompted by the then surprising finding that a screen for mutants resistant to the classical complex I inhibitor piericidin A had identified the hydrophilic 49-kDa subunit to be involved in inhibitor binding [43]. A set of mutations resulting in loss of activity or pronounced inhibitor resistance were found to cluster in this domain. This confirmed our hydrogenase based structural model [42]. The X-ray structure of the peripheral arm [8] revealed a pocket formed by the PSST- and the 49-kDa subunit and a striking degree of conservation of the structural fold. Systematic mutagenesis of the residues lining this pocket demonstrated that it comprises indeed the ubiquinone binding domain (Fig. 2; [44]). Mutations resulting in severely reduced ubiquinone reductase activity identified an extended entry path leading from the N-terminal antiparallel β-sheet of the 49-kDa subunit to cluster N2 in the PSST subunit. Tyrosine 144 (Y. lipolytica numbering) in the immediate vicinity of the cluster seems essential for catalytic function as several substitutions even as subtle as the change to phenylalanine (M. A. Tocilescu, unpublished observation) abolished activity completely [44] without severely affecting the EPR spectrum of cluster N2 [42].
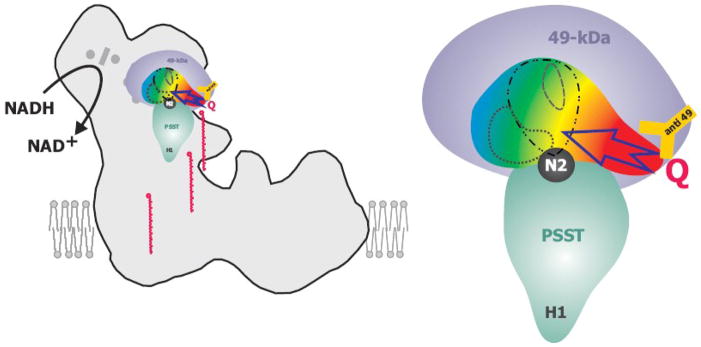
Figure 2. Schematic drawing of the ubiquinone binding pocket.
The 49-kDa subunit (in violet) and the PSST subunit (in cyan) assemble the ubiquinone binding pocket located next to iron-sulfur cluster N2 (in black). The position of the subunits in the peripheral arm has been modelled according to fit 1 [51] shown in figure 3. The pocket is coloured with a color gradient indicating the severity of point-mutations on complex I activity compared to the parental strain. Red: Almost complete loss of complex I activity for all mutations in this region. Yellow: Strong decrease of complex I activity. Green: Some decrease of complex I activity. Blue: essentially no effects on activity for any mutation in this region. A possible ubiquinone access path in the red region is indicated by an arrow. Within the ubiquinone binding cavity, binding sites of different classes of complex I inhibitors are indicated: ··· C12E8; -·· - rotenone and DQA; --- DQA only. The orange Y represents the anti 49-kDa antibody and marks the position of the corresponding epitope. Ubiquinone-9 molecules are depicted in red at various intermediate positions along a hypothetical “access ramp”, illustrating that the hydrophobic substrate has to leave the membrane bilayer to gain access to its reduction site.
Complex I is inhibited by a plethora of hydrophobic compounds which can be grouped into three classes based on their behaviour in enzyme kinetics [45]. Inhibitor binding studies had revealed that inhibitors from these three classes bind to distinct but overlapping binding sites [46], a view that we confirmed recently by an extensive structure guided mutagenesis of the pocket around the ubiquinone binding domain [47]: while the binding sites for DQA (class I/type A) and rotenone (class II/type B) were largely overlapping, the inhibitory detergent C12E8 (type C) penetrated into a subdomain where mutations did not affect binding of the two other inhibitors (Fig. 2).
Knowing that the catalytic centres for the redox reaction reside in the peripheral arm fragment, the functionally much more critical question arises where they are located within the structure of the holo-complex. When the X-ray structure of the peripheral arm was solved, it appeared obvious to assume an orientation that would bring the ubiquinone and inhibitor binding pocket around cluster N2 as close as possible to the hydrophobic membrane phase [8]. A corresponding manual fit of the T. thermophilus fragment into a low resolution 3D model of E. coli complex I was proposed [34]. However, the results of electron microscopic single particle analysis led us to suggest that the spatial arrangement is in fact quite different. Because of the enormous size of complex I, subcomplexes have been especially useful for investigating the subunit architecture of the enzyme [48;49]. We biochemically generated a subcomplex lacking specifically the 51-kDa and 24-kDa subunits harbouring the electron input module of the enzyme [50]. 3D reconstructions of single particles unequivocally identified domain 1 at the very distal end of the peripheral arm (Fig. 1) as the mass which was lost concomitantly with the two subunits [51].
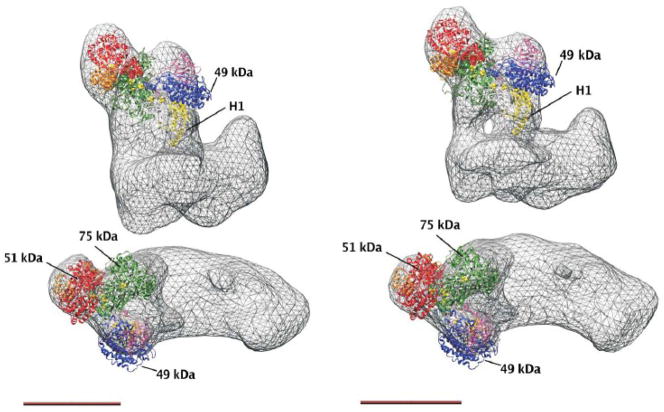
Using domain 1 in the Y. lipolytica complex I as an anchor point allowed five different fits of the peripheral arm X-ray structure into the electron microscopic structure of the complete enzyme [51]. None of these fits placed cluster N2 less than 35 Å away from the membrane. Moreover, 2D reconstructions of antibody labelled complex I showed a distal position of the 49-kDa and the tightly associated 30-kDa subunit [52]. This finding clearly favoured fits with an even longer distance between cluster N2 and the membrane surface. Fit 1 with a position of N2 60 Å outside of the predicted plane of the membrane is in good agreement with the results from immuno-electron microscopy and is shown in Fig. 3 for two different 3D reconstructions of complex I from Y. lipolytica [51].
Figure 3. X-ray structure of the peripheral arm fragment from T. thermophilus fitted to 3D reconstructions of complex I from Y. lipolytica, fit 1 from [51].
Left panel, Y. lipolytica complex I at 24 Å resolution as shown in figure 1; right panel, Y. lipolytica complex I class 8_2 with 16.5 Å resolution. At higher resolution the connection of the peripheral arm to the membrane arm via two separate stalks becomes visible. The positions of the largest central hydrophilic subunits and of helix H1 of the PSST subunit are indicated (see text), scale bar 10 nm.
The membrane arm
As there is no high resolution structure available yet and no spectroscopically detectable redox groups are present, much less is known about the membrane arm. Recently, a projection map of the membrane arm from E.coli at 8 Å resolution allowed an initial structural characterization [53] but we still rely mainly on information based on the analysis of subcomplexes, structure prediction, and topology modelling (Fig. 3, Table 1).
Table 1.
| Bos taurus | Prediction TMHa | Y. lipolytica | Prediction TMHa | TMH by experimental evidence/comments |
|---|---|---|---|---|
| subset of subunits of bovine subcomplex Iαj not presentin subcomplex I λj | ||||
| ND6 | 5 | NU6M | 5 | 5 [59] |
| 42 kDab,c | 0 | - | - | |
| 39 kDab | 3g | NUEM | 0 | in hydrophilic subcomplex in Y. lipolytica [67] |
| MWFE | 1 | NIMM | 1 | |
| PGIV | 0 | NUPM | 0 | |
| SDAPd | 0 | ACPM 1or 2 | 0 | acyl carrier protein |
| (B15)d,e | 1 | NB5M | 1 | |
| B14 | 0 | NB4M | 0 | |
| B9 | 1 | NI9M | 1 | |
| 15 kDa | 1 | - | - | |
|
| ||||
| subunits present in bovine subcomplexes Iα and Iλ but with predicted TMH | ||||
| B14.7 | 3 | NUJM | 3 | |
| B16.6 | 1 | NB6M | 1 | |
|
| ||||
| bovine subcomplex Iβ | ||||
| ND4 | 13 | NU4M | 13 | 12 + large inside domain [54] |
| ND5 | 18 | NU5M | 18 | 14 + large inside domains [54] |
| AGGG | 1 | - | - | |
| ASHI | 1 | NIAM | 1 | |
| ESSS | 1 | NESM | 1 | position determined by monoclonal antibody [67] |
| MNLL | 0f | - | - | |
| PDSW | 0 | NIDM | 0 | |
| SDAPd | 0 | ACPM 1or 2 | 0 | acyl carrier protein |
| SGDH | 1 | - | - | |
| B22 | 0 | NI2M | 0 | |
| B18 | 0 | NB8M | 0 | |
| B17 | 1 | - | - | |
| (B15)d,e | 1 | NB5M | 1 | |
| (B14.5b)e | 1 | - | - | |
| B12 | 1 | NB2M | 1 | |
|
| ||||
| bovine subcomplex Iγh | ||||
| ND1 | 8 | NU1M | 10 | 8 + large inside domains [56] |
| ND2i | 10 | NU2M | 14 | 12 + large inside domain [54] |
| ND3 | 3 | NU3M | 3 | 3 + large inside domain [61] |
| ND4L | 2 | NULM | 2 | 3 [58] |
| 42 kDab,c | 0 | - | - | - |
| 39 kDab | 3g | NUEM | 0 | in hydrophilic subcomplex in Y. lipolytica [67] |
| PFFD | 0 | NIPM | 0 | |
| KFYI | 1 | - | - | |
|
| ||||
| accessory Y. lipolytica subunits with predicted TMH not present in bovine subcomplexes | ||||
| - | - | NUNM | 1 | |
| - | - | NUXM | 3 | |
prediction of transmembrane helices (TMH) using algorithm hmmtop [112]
present in subcomplexes Iα and Iγ (but see h)
the 42 kDa subunit is only loosely associated with bovine complex I
present in subcomplexes Iα and Iβ
Parentheses indicate that the subunit is present in minor amounts
one transmembrane helix was predicted by algorithm TMHMM [113]
the prediction of transmembrane helices is ambiguous because it is not confirmed by algorithm TMHMM [17]
the composition and structural integrity of this subcomplex is questionable
ND2 is N-terminally truncated in bovine complex I
for complete composition of subcomplexes Iα and Iλ see e.g. [17]
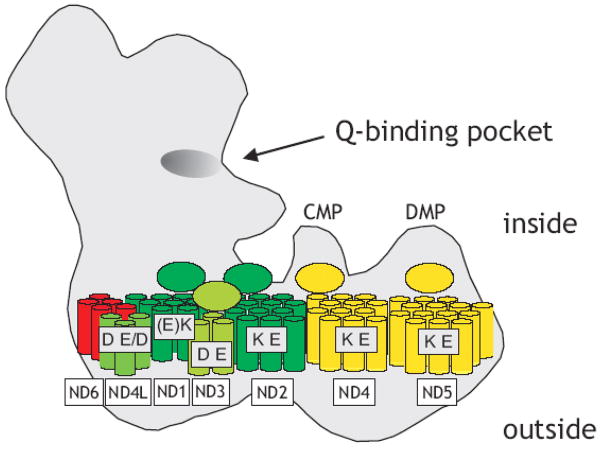
The hydrophobic central subunits ND1 to ND6 and ND4L together have a mass of about 260 kDa. Some 60 transmembrane helices have been predicted, but the exact number depends on the algorithm employed. Moreover it has been shown for example by alkaline phosphatase fusions that some of the predicted transmembrane helices actually seem to form extramembraneous domains [54].
ND5 is the largest hydrophobic subunit of complex I and was predicted to contain up to 18 transmembrane helices [7;54]. However, by analysis of alkaline phosphatase fusion proteins of the bacterial ND5 homologue NUOL [54] it was found that the two putative membrane intrinsic helices X and XI instead form a large domain facing the bacterial cytoplasm that corresponds to the mitochondrial matrix side. In the C-terminal part of the subunit the topology appears to be less clear because two sequence stretches are not unequivocally recognized as transmembrane helices by different prediction algorithms and can therefore also be regarded as extramembraneous [54]. Taken together the topology model for NUOL [54] suggests 14 transmembrane helices and two extramembraneous domains of approximately 8 kDa each facing the cytoplasmic side of the membrane (Fig, 4). ND2 and ND4 are homologous to ND5 but are C-terminally truncated. It seems reasonable to assume that the topology is conserved which would result in 12 transmembrane helices and one large inside domain each for ND2 and ND4 (Fig, 4). It is interesting to note that a mutation responsible for the majority of cases of Leber’s hereditary optic neuropathy (LHON) is located in the putative inside domain of the human ND4 subunit (ND4-11778) [54;55].
Figure 4. Subcomplexes and central hydrophobic subunits of the membrane arm.
Subunits assigned to subcomplex Iα, red; Iγ green; Iβ, yellow. Note that the structural integrity of subcomplex Iγ is a matter of debate (indicated by different shades of green). Ellipses denote putative extramembraneous domains on the inside (note that the topology for ND3 is a matter of debate); highly conserved charged residues in transmembrane helices are assigned to individual subunits (see text), Brackets indicate that residues are close to the ends of putative membrane spanning segments.
Again using alkaline phosphatase fusions subunit ND1 was determined to have 8 transmembrane helices with extended extramembraneous domains facing the inside [56]. As for subunit ND4 one of the putative inside domains in human ND1 is the site of a frequent LHON mutation (ND1-3460; [57]). In topology studies using maltose binding protein fusions subunits ND4L [58] and ND6 [59] were shown to have 3 and 5 transmembrane helices respectively (Fig, 4).
The topology of the ND3 subunit has been discussed controversially. Initially, an orientation was described where the extended loop connecting the first two out of three transmembrane helices faced the periplasmic/intermembrane space side [60]. This report was based on the reactivity of antibodies against the N- and C-terminus of the protein and on the accessibility of Cys47 (P. denitrificans numbering) in the large hydrophilic loop towards alkylating agents. However, this cysteine was recently identified in subunit ND3 from B. taurus [61] as the residue specifically exposed only in the active form of complex I and shown to reside on the matrix side of the membrane [62]. This seems plausible, because the conformational change associated with the A/D transition that was monitored by the exposure of the cysteine residue is much more likely to take place at the functionally critical interface between the two arms than on the outside [61]. In addition, alkaline phosphatase fusion experiments place the C-terminal end of subunit ND3 on the cytoplasmic/matrix side of the membrane (C. Hägerhall, personal communication) also contradicting the original findings [60]. Because of this conflicting evidence the topology of ND3 remains unclear at this stage. However, there is rather strong indirect evidence that seems to favour the topology placing the large extramembrane loop on the inside: pathogenic mutations have been identified in three positions in this loop near the unique cysteine [63–65]. Moreover, subunit ND3 could be cross-linked to the PSST subunit [66] of the peripheral arm which is much better to reconcile with an orientation of the large loop facing the inside.
The membrane arm of eukaryotic complex I contains a number of accessory subunits with predicted transmembrane helices. None of them is larger than 25 kDa and the additional mass appears to be evenly distributed around a core formed by the central subunits [28]. The NESM subunit of complex I from Y. lipolytica that could be localized in the membrane arm by a monoclonal antibody [67] is the prototype of a single transmembrane domain (STMD) subunit. STMD subunits are small proteins that have one transmembrane helix and highly hydrophilic domains on either side with a high proportion of charged residues [67]. Eight predicted STMD subunits were found in Y. lipolytica complex I [7] and as much as 14 in the bovine enzyme [17]. The function of STMD subunits is not clear but it has been proposed that they may assist assembly and are important for maintaining structural integrity [68]. In addition, accessory subunit B14.7 (from bovine heart, corresponding to NUJM of Y. lipolytica) and the fungi-specific NUXM protein have three and two predicted transmembrane helices, respectively.
Overall a total number of 57 transmembrane helices is most likely for bacterial complex I to which 12–16 transmembrane segments are added from accessory subunits in the mitochondrial enzymes (Fig, 4).
First clues on how the membrane integral subunits are arranged in the membrane arm came from the biochemical characterization of subcomplexes (Table 1). Bovine complex I was split into subcomplexes Iα and Iβ by exposure to the harsh detergent LDAO [16;17;48;49]. The hydrophobic subcomplex Iβ contained the central subunits ND4 and ND5 (Fig, 4) and a number of accessory subunits. In contrast, subcomplex Iα consisted predominantly of hydrophilic subunits but also harboured a section of the membrane arm as indicated by the presence of the hydrophobic central subunit ND6. Subcomplex Iα releases the even more hydrophilic subcomplex Iλ that contains almost exclusively hydrophilic subunits [69]. In Table 1 we have used the presence of a subunit in Iα but not in Iλ as an admittedly not stringent criterion to assume an association of the specific subunit with the membrane arm. The hydrophobic subunits ND1, ND2, ND3 and ND4L have been assigned to a third type of subcomplex named Iγ [49]. However, it has been questioned whether Iγ is in fact a stable structural unit. Therefore conclusions from the association of subunits with Iγ have to be viewed with caution [17;49]. The authors of [49] concluded that at least ND1 and ND2 are tightly linked and must be situated in a different domain of the membrane arm than subunits ND4 and ND5 (Fig, 4). Resolution of complex I from E.coli into subcomplexes [70] and analysis by electron microscopy showed that subunits ND4 and ND5 are located in the distal part of the membrane arm [71]. It follows that subunits ND1 and ND2 should be located in the proximity of the peripheral arm, which can be assumed also for subunit ND3 from crosslinking studies (Fig, 4; [66;72]).
Very little is known about the question which and how many subunits and transmembrane segments form the proton pumping elements of complex I. It can be assumed that proton translocation requires protonable residues in transmembrane segments of the hydrophobic central subunits. Subunits ND2, ND4 and ND5 contain several acidic and basic residues in predicted transmembrane helices but only one glutamate and one lysine are highly conserved (Fig, 4; [73;74]). These subunits may be indeed involved in proton pumping, because they are homologous to a specific type of Na+/H+ antiporters found in many bacterial species. These antiporters are involved in pH homeostasis and Na+-resistance under alkaline growth conditions and their genes are typically organized in an operon together with other transport proteins [54;75]. The hetero-oligomeric assembly in the membrane and tight functional interaction of the different polypeptides coded by the mrpA-G (multiple resistance and pH) operon has been demonstrated [76]. Three central hydrophobic complex I subunits are homologous to antiporters of the mrp type. ND5, the largest hydrophobic central subunit of complex I corresponds to MrpA while ND2 and ND4 correspond to MrpD. Interestingly, also the smallest central subunit ND4L is related to the MrpC protein of this operon [77] and also contains two conserved membrane intrinsic acidic residues suggesting that also ND4L might be involved in proton translocation [78]. However, this does not exclude the other subunits from being part of the proton pumping machinery, because also subunit ND3 features two acidic residues [79] and ND1 one lysine in putative transmembrane helices. Three glutamates in the ND1 subunit are located in hydrophobic regions but their position at the ends of predicted membrane spanning helices renders their contribution to proton translocation questionable [80].
Connections between the peripheral and the membrane arm
Considering that all redox chemistry seems to be confined to the peripheral arm while the proton pumping machinery must operate in the membrane arm, the connections between these two parts of complex I are of particular interest for understanding its mechanism. It has been known for a long time that bacterial complex I is much less stable than the eukaryotic enzyme and easily falls apart at this junction [10;12]. This local instability eventually allowed to crystallize the peripheral arm fragment of complex I from T. thermophilus and to determine its structure [81]. The comparison of electron microscopic structures of complex I from N. crassa and E. coli showed the largest mass differences at the interface region of the two arms [28] which in the prokaryotic enzyme seemed to be connected only via a thin stalk. However, other 3D reconstructions of E. coli complex I did not reproduce this structural feature [34;82]. Depending on the resolution and surface threshold applied, the peripheral arm of complex I from Y. lipolytica appears to be attached to the membrane arm via two separate stalk-like connections (Fig. 3; [29]). In line with previous suggestions [28] we therefore hypothesize that one of the two connections in the mitochondrial enzyme is formed by accessory subunits, possibly the 39 kDa subunit of the peripheral arm [67]. This would explain a single thin connection in bacterial enzymes that is common to all versions of complex I and would then have to be formed by domains from central subunits. In our model for the orientation of the peripheral arm that places cluster N2 35–60 Å above the membrane arm, the question arises which domains are available to form this common stalk. Based on the published structure of the peripheral arm and its most likely orientation (Fig. 3) only helix H1 of the PSST subunit extends towards the membrane arm and seems to just reach it. However, there are disordered parts of the peripheral arm not resolved in the X-ray structure that could contribute to the connection: 34 N-terminal amino acids of the 49-kDa subunit and 32 N-terminal amino acids of the PSST subunit are likely to be located below the ubiquinone binding pocket. Furthermore, as discussed above the ND1 subunit has a significant portion of extramembraneous domains with more than 100 amino acids on the matrix side. Following the topology model proposed in [61] the ND3 subunit exposes one large loop with approximately 25 amino acids to the inside. Moreover, assuming that the subunits related to the sodium/proton antiporters have similar topologies some 70 additional amino acids of subunit ND2 could potentially contribute to the interface region of the two arms (Fig. 4). In summary, there is clearly enough protein mass available from central subunits of both the peripheral and the membrane arm to form the common stalk, but its composition and structure remains obscure.
Implications of the structure for complex I function
Although a high resolution structure of the holo-complex is still missing the structural information outlined above has provided a framework to get insight into the redox linked proton translocation reaction by mitochondrial complex I:
The reaction is reversible and in the presence of a proton motive force backward electron transfer from succinate to NAD+ is observed (reviewed in [2]).
A striking difference between complex I and the other complexes of the respiratory electron transfer chain is that all known redox groups are located in the membrane extrinsic part of the enzyme and, although it has been suggested that there may be redox active cofactors in the membrane arm [83], a thorough analysis of the complex I proteome did not provide any supporting evidence for this [84].
In the peripheral arm there are one FMN and eight canonical iron-sulfur clusters. The spectroscopic properties of cluster N1a are a matter of discussion [37;85] and a special function associated with protection against ROS formation at the flavine site has been suggested [8;81;86–88]. All other EPR detectable clusters except N2 display a roughly similar midpoint potential of around −250 mV [89] but also broader ranges of midpoint potentials were reported for purified enzyme preparations e.g. [90]. Cluster N2 the direct reductant of ubiquinone exhibits several features that are distinct from the other clusters. With typical values around −150 mV (pH 7) for the mitochondrial enzyme, cluster N2 has the most positive redox midpoint potential of all iron-sulfur clusters of complex I [89]. The finding that the redox midpoint potential of cluster N2 from bovine heart complex I exhibits a strong pH-dependence between pH 6 and 8.5 [91] provoked mechanistic models that assigned cluster N2 and its associated redox-Bohr group a key function in proton translocation (reviewed in [92]). Replacing histidine-226 by methionine in the 49-kDa subunit of Y. lipolytica complex I completely abolished this redox-Bohr effect and shifted the midpoint potential of cluster N2 below −200 mV. However, this did not affect catalytic activity or proton translocation stoichiometry excluding a role of this protonable group and the associated redox-Bohr effect in proton pumping [93]. Measuring real time electron transfer in complex I from E. coli using an ultrafast freeze-quench approach, Verkhovskaya and coworkers found that transfer of the two electrons donated by NADH is bifurcated at the FMN site to reduce cluster N2 and cluster N1a at the submillisecond time scale [94]. Intriguingly, reduction of cluster N2 occurred with a time constant very close to the one predicted from electron tunnelling theory [95] but no electron transfer to ubiquinone was observed. The consistent conclusion of the analysis of mutant H226M and the very fast electron transfer through the iron-sulfur clusters was that only the redox chemistry occurring after reduction of cluster N2 i.e. the two electron reduction of the substrate ubiquinone can be associated with energy conversion [93;94].
The midpoint potential of the Q/Q.− redox couple in the rather non-physiological environment of 80 % ethanol is −240 mV and was estimated to be below −300 mV when bound to complex I under the conditions of the freeze quench experiment [94]. Thus essentially the full potential gap between NADH and ubiquinone of about 400 mV is still available at this point to drive proton pumping with the observed 2 H+/e− stoichiometry.
The interaction of complex I with ubiquinone is a key issue that has been addressed by many different groups. Iron-sulfur cluster N2 displays paramagnetic interactions with SQNf. This fast relaxing semiquinone species formed during catalytic turnover, which is only found in the presence of ΔμH+ and is sensitive to the complex I inhibitors rotenone and piericidin A [96]. From the analysis of dipolar interaction of the two paramagnetic species, a distance between SQNf and cluster N2 of about 12 Å was calculated [96;97]. SQNs, a second semiquinone species characterized by slow spin relaxation, was detected by the same group. SQNs is not sensitive to the membrane potential. Because it was not found to interact paramagnetically with cluster N2, SQNs was proposed to reside at a significantly longer distance to cluster N2 [89]. However, other reasons why this interaction is not detectable cannot be excluded.
The ubiquinone reduction site at the interface between the PSST and 49-kDa subunit has been characterized in detail by site-directed mutagenesis [44] and there is no doubt that SQNf must be found at this site. The fact that the accumulation of this semiquinone species is linked to the presence of a membrane potential is consistent with a central role in proton pumping. It is more difficult to understand the SQNs signal. Based on the paramagnetic interactions with N2 it has been proposed that the two semiquinone species are intermediates of two ubiquinone molecules bound at different sites of complex I [96;97]. Indeed putative ubiquinone binding motifs are found in the antiporter like subunits of the membrane arm [98]. However, site directed mutagenesis of all histidine residues possibly involved in ubiquinone binding in the ND4 subunit did not show any significant effect on kinetic parameters of ubiquinone reduction and on the IC50 for the competitive inhibitor capsaicin [73]. Also the fact that the ND4 subunit of complex I from E. coli was labelled with an azido-ubiquinone derivative [99] does not present strong evidence for a quinone binding site in this subunit, because the identification of specific and functionally relevant ubiquinone binding by affinity labelling is difficult and may be misleading as has been found in the case of the QPC subunit of the bc1 complex [100]. Moreover, the localization of subunit ND4 in the distal part of the membrane arm would place this ubiquinone site at a distance too far away from the ubiquinone binding pocket near cluster N2 to allow for electron transfer. We therefore consider it more likely that the two semiquinone species that can be observed by EPR spectroscopy reflect the same semiquinone intermediate bound to the now well characterized ubiquinone binding pocket but in different conformational states of the complex. The change in spin relaxation properties and paramagnetic interaction could then be explained by a change in the environment of the semiquinone and possibly a significant difference in the distance to cluster N2. The conformational state resulting in the SQNf signature would be favoured by the presence of a membrane potential. It should be noted that the sum of the spin concentrations of the two semiquinone species was never found to exceed one per complex I. Still, the nature of SQNs and its relationship to SQNf remains unclear and it seems obvious that solving this issue will be prerequisite to understand the proton pumping mechanism of complex I.
Inhibitors have been very valuable tools to differentiate between the two ubiquinone binding sites of the cytochrome bc1 complex [101]. In contrast, it has been shown by equilibrium binding studies that there is only one extended binding pocket with partially overlapping binding sites in complex I [46]. Inhibitor resistant mutants for all classes of complex I inhibitors were mapped at the interface between the PSST and 49 kDa subunits [43;44;47], in line with labelling of subunit PSST with a photoactive pyridaben derivative [102]. The same compound was found to bind to subunit ND1 but competition experiments and dose dependent correlation of inhibition and labelling showed the inhibitory action to be confined to subunit PSST [102;103]. In contrast, the ND1 subunit was labelled exclusively by an acetogenin derivative and binding competition with other complex I inhibitors indicated that this subunit contributes to the common inhibitor binding pocket in a functionally relevant way [104]. Based on the evidence summarized here it can not be excluded with certainty that complex I contains multiple functionally relevant ubiquinone interaction sites as postulated e.g. in [105]. However, we think that there is also no compelling evidence against our working hypothesis that the catalytic mechanism of complex I involves only one ubiquinone molecule that is reduced at the interface of the PSST/49-kDa subunits.
Our structural model of the peripheral arm of complex I places the ubiquinone reduction site at a significant distance to the membrane domain. It follows that the hydrophobic ubiquinone and also hydrophobic inhibitors have to leave the lipid bilayer at least partially. We have speculated that this is facilitated via a hydrophobic ramp or channel [52] in the common stalk connecting the two arms of complex I. Changed kinetic parameters of ubiquinone reduction in site directed mutants of residues located in a putative surface helix of ND1 [106] that might be part of the connection between the two arms and inhibitor binding to this subunit [104] is consistent with this view. Even if we assume the shortest possible distance of N2 to the membrane of 35 Å we have to face the fact that the power stroke of proton translocation is initiated in the peripheral arm and that the energy for proton translocation has to be transmitted to the membrane arm.
As discussed above, we still do not know how many and which of the hydrophobic subunits participate in proton translocation. A number of mutants in hydrophobic subunits aimed at identifying proton translocation sites have been generated. It was however found that many of them affected ubiquinone reductase activity, but because of the loss of activity it was not possible to measure proton translocation [73;74;78;79]. Loss of catalytic activity caused by mutations far away from the ubiquinone reduction site may reflect interference with the proton pumping machinery and therefore the tight coupling between electron transfer and proton translocation that is also illustrated by the full reversibility of the complex I reaction. However, also global and therefore unspecific structural changes can explain such effects. As a consequence it seems difficult to provide positive evidence for the contribution of individual subunits to proton translocation by mutagenesis. However, the homology to antiporters and the presence of conserved charged residues in transmembrane segments strongly suggests that even the subunits in the distal part of the membrane arm are involved in proton pumping. This means that energy transmission in complex I has to be exerted over distances as long as up to several hundreds of Ångstroms and at present it can only be speculated how this is achieved. Electron microscopy of complex I from E. coli in the presence of nucleotides has shown only limited structural changes upon addition of NADH at low resolution [34] but there are a number of observations of redox linked differences in cross linking patterns and protease sensitivity [107–109] in line with a conformational coupling mechanism [110]. A cascade of electrostatic interactions between conserved lysine residues in subunits ND1, ND2, ND4 and ND5 triggered by formation of the semiquinone anion was suggested recently as another possible mechanistic principle of coupling [74].
Long range conformational coupling was described in detail for other ion translocating membrane proteins like the Ca2+ pump of the sarcoplasmatic reticulum [111]. However for complex I there is no structural information even at modest resolution that would allow linking the mechanism of the enzyme to distinct conformational states. Our electron microscopic 3D reconstructions of many different classes of complex I particles give some idea about the pronounced structural flexibility of this giant protein complex [29]. Future work will address the question if and how structural changes might be related to complex I function.
Acknowledgments
The excellent technical assistance of Gudrun Beyer and Karin Siegmund to the work of our group presented in this article is gratefully acknowledged. This work has been supported by grants of the Deutsche Forschungsgemeinschaft (SFB 472 Projekt P2) to V.Z. and U.B. and of the National Institute of Health (NIH RO1 GM068650) to M.R..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brandt U. Energy converting NADH:quinone oxidoreductase (Complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 2.Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (Complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta. 1998;1364:169–185. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 3.Smeitink J, Van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 4.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 5.Lenaz G, Bovina C, Castelluccio C, Fato R, Formiggini G, Genova ML, Marchetti M, Merlo Pich M, Pallotti F, Parenti Castelli G, Biagini G. Mitochondrial Complex I defects in aging. Mol Cell Biochem. 1997;174:329–333. [PubMed] [Google Scholar]
- 6.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 7.Morgner N, Zickermann V, Kerscher S, Wittig I, Abdrakhmanova A, Barth HD, Brutschy B, Brandt U. Subunit mass fingerprinting of mitochondrial complex I. Biochim Biophys Acta. 2008;1777:1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 9.Brockmann C, Diehl A, Rehbein K, Strauss H, Korn B, Kuhne R, Oschkinat H. The oxidized subunit B8 from human complex I adopts a thioredoxin fold. Structure. 2004;12:1645–1654. doi: 10.1016/j.str.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 10.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 11.Yano T, Yagi T. H+-translocating NADH-Quinone Oxidoreductase (NDH-1) of Paracoccus denitrificans. J Biol Chem. 1999;274:28606–28611. doi: 10.1074/jbc.274.40.28606. [DOI] [PubMed] [Google Scholar]
- 12.Yagi T, Honnami K, Ohnishi T. Purification and Characterization of Two Types of NADH-Quinone Reductase from Thermus thermophilus HB-8. Biochem. 1988;27:2008–2013. doi: 10.1021/bi00406a030. [DOI] [PubMed] [Google Scholar]
- 13.Peng G, Fritzsch G, Zickermann V, Schägger H, Mentele R, Lottspeich F, Bostina M, Radermacher M, Huber R, Stetter KO, Michel H. Isolation, characterization and electron microscopic single particle analysis of the NADH:ubiquinone oxidoreductase (complex I) from the hyperthermophilic eubacterium Aquifex aeolicus. Biochem. 2003;42:3032–3039. doi: 10.1021/bi026876v. [DOI] [PubMed] [Google Scholar]
- 14.Kerscher S, Dröse S, Zwicker K, Zickermann V, Brandt U. Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochim Biophys Acta. 2002;1555:83–91. doi: 10.1016/s0005-2728(02)00259-1. [DOI] [PubMed] [Google Scholar]
- 15.Büschges R, Bahrenberg G, Zimmermann M, Wolf K. NADH: ubiquinone oxidoreductase in obligate aerobic yeasts. Yeast. 1994;10:475–479. doi: 10.1002/yea.320100406. [DOI] [PubMed] [Google Scholar]
- 16.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 18.Marques I, Duarte M, Assuncao J, Ushakova AV, Videira A. Composition of complex I from Neurospora crassa and disruption of two “accessory” subunits. Biochim Biophys Acta. 2005;1707:211–220. doi: 10.1016/j.bbabio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Gredilla R, Grief J, Osiewacz HD. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp Gerontol. 2006;41:439–447. doi: 10.1016/j.exger.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Weidner U, Nehls U, Schneider R, Fecke W, Leif H, Schmiede A, Friedrich T, Zensen R, Schulte U, Ohnishi T, Weiss H. Molecular genetic studies of complex I in Neurospora crassa, Aspergillus niger and Escherichia coli. Biochim Biophys Acta. 1992;1101:177–180. [PubMed] [Google Scholar]
- 21.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol. 2005;350:263–277. doi: 10.1016/j.jmb.2005.04.062. [DOI] [PubMed] [Google Scholar]
- 23.Eubel H, Heinemeyer J, Braun HP. Identification and characterization of respirasomes in potato mitochondria. Plant Physiol. 2004;134:1450–1459. doi: 10.1104/pp.103.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heazlewood JL, Howell KA, Millar AH. Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim Biophys Acta. 2003;1604:159–169. doi: 10.1016/s0005-2728(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 25.Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prommeenate P, Lennon AM, Markert C, Hippler M, Nixon PJ. Subunit composition of NDH-1 complexes of Synechocystis sp PCC 6803 - Identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J Biol Chem. 2004;279:28165–28173. doi: 10.1074/jbc.M401107200. [DOI] [PubMed] [Google Scholar]
- 27.Arteni AA, Zhang P, Battchikova N, Ogawa T, Aro EM, Boekema EJ. Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim Biophys Acta. 2006;1757:1469–1475. doi: 10.1016/j.bbabio.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 28.Guenebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Mol Biol. 1998;276:105–112. doi: 10.1006/jmbi.1997.1518. [DOI] [PubMed] [Google Scholar]
- 29.Radermacher M, Ruiz T, Clason T, Benjamin S, Brandt U, Zickermann V. The three-dimensional structure of complex I from Yarrowia lipolytica: A highly dynamic enzyme. J Struct Biol. 2006;154:269–279. doi: 10.1016/j.jsb.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J Mol Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- 31.Hofhaus G, Weiss H, Leonard K. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (complex I) J Mol Biol. 1991;221:1027–1043. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 32.Guenebaut V, Vincentelli R, Mills D, Weiss H, Leonard K. Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction. J Mol Biol. 1997;265:409–418. doi: 10.1006/jmbi.1996.0753. [DOI] [PubMed] [Google Scholar]
- 33.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. Journal of Electron Microscopy Technique. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 34.Morgan DJ, Sazanov LA. Three-dimensional structure of respiratory complex I from Escherichia coli in ice in the presence of nucleotides. Biochim Biophys Acta. 2008;1777:711–718. doi: 10.1016/j.bbabio.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 35.Hinchliffe P, Carroll J, Sazanov LA. Identification of a novel subunit of respiratory complex I from Thermus thermophilus. Biochem. 2006;45:4413–4420. doi: 10.1021/bi0600998. [DOI] [PubMed] [Google Scholar]
- 36.Yakovlev G, Reda T, Hirst J. Reevaluating the relationship between EPR spectra and enzyme structure for the iron-sulfur clusters in NADH: quinone oxidoreductase. Proc Natl Acad Sci USA. 2007;104:12720–12725. doi: 10.1073/pnas.0705593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohnishi T, Nakamaru-Ogiso E. Were there any “misassignments” among iron-sulfur clusters N4, N5 and N6b in NADH-quinone oxidoreductase (complex I)? Biochim Biophys Acta. 2008;1777:703–710. doi: 10.1016/j.bbabio.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sazanov LA. Respiratory complex I: Mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochem. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 39.Pohl T, Bauer T, Dorner K, Stolpe S, Sell P, Zocher G, Friedrich T. Iron-sulfur cluster N7 of the NADH:ubiquinone oxidoreductase (Complex I) is essential for stability but not involved in electron transfer. Biochem. 2007;46:6588–6596. doi: 10.1021/bi700371c. [DOI] [PubMed] [Google Scholar]
- 40.Gurrath M, Friedrich T. Adjacent cysteines are capable of ligating the same tetranuclear iron-sulfur cluster. Proteins. 2004;56:556–563. doi: 10.1002/prot.20155. [DOI] [PubMed] [Google Scholar]
- 41.Böhm R, Sauter M, Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 42.Kashani-Poor N, Zwicker K, Kerscher S, Brandt U. A central functional role for the 49-kDa subunit within the catalytic core of mitochondrial complex I. J Biol Chem. 2001;276:24082–24087. doi: 10.1074/jbc.M102296200. [DOI] [PubMed] [Google Scholar]
- 43.Darrouzet E, Issartel JP, Lunardi J, Dupuis A. The 49-kDa subunit of NADH-ubiquinone oxidoreductase (complex I) is involved in the binding of piericidin and rotenone, two quinone-related inhibitors. FEBS Lett. 1998;431:34–38. doi: 10.1016/s0014-5793(98)00719-4. [DOI] [PubMed] [Google Scholar]
- 44.Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U. Exploring the ubiquinone binding cavity of respiratory complex I. J Biol Chem. 2007;282:29514–29520. doi: 10.1074/jbc.M704519200. [DOI] [PubMed] [Google Scholar]
- 45.Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochim Biophys Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 46.Okun JG, Lümmen P, Brandt U. Three classes of inhibitors share a common binding domain in mitochondrial complex I (NADH:ubiquinone oxidoreductase) J Biol Chem. 1999;274:2625–2630. doi: 10.1074/jbc.274.5.2625. [DOI] [PubMed] [Google Scholar]
- 47.Fendel U, Tocilescu MA, Kerscher S, Brandt U. Exploring the inhibitor binding pocket of respiratory complex I. Biochim Biophys Acta. 2008;1777:660–665. doi: 10.1016/j.bbabio.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 48.Finel M, Skehel JM, Albracht SPJ, Fearnley IM, Walker JE. Resolution of NADH:ubiquinone oxidoreductase from bovine heart mitochondria into two subcomplexes, one of which contains the redox centers of the enzyme. Biochem. 1992;31:11425–11434. doi: 10.1021/bi00161a022. [DOI] [PubMed] [Google Scholar]
- 49.Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochem. 2000;39:7229–7235. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- 50.Zickermann V, Zwicker K, Tocilescu MA, Kerscher S, Brandt U. Characterization of a subcomplex of mitochondrial NADH:ubiquinone oxidoreductase (complex I) lacking the flavoprotein part of the N-module. Biochim Biophys Acta. 2007;1767:393–400. doi: 10.1016/j.bbabio.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Clason T, Zickermann V, Ruiz T, Brandt U, Radermacher M. Direct localization of the 51 and 24 kDa subunits of mitochondrial complex I by three-dimensional difference imaging. J Struct Biol. 2007;159:433–442. doi: 10.1016/j.jsb.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zickermann V, Bostina M, Hunte C, Ruiz T, Radermacher M, Brandt U. Functional implications from an unexpected position of the 49 kDa subunit of complex I. J Biol Chem. 2003;278:29072–29078. doi: 10.1074/jbc.M302713200. [DOI] [PubMed] [Google Scholar]
- 53.Baranova EA, Holt PJ, Sazanov LA. Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 angstrom resolution. J Mol Biol. 2007;366:140–154. doi: 10.1016/j.jmb.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 54.Mathiesen C, Hagerhall C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane- bound hydrogenases and bona fide antiporters. Biochim Biophys Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 55.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 56.Roth R, Hägerhäll C. Transmembrane orientation and topology of the NADH:quinone oxidoreductase putative quinone binding subunit NuoH. Biochim Biophys Acta. 2001;1504:352–362. doi: 10.1016/s0005-2728(00)00265-6. [DOI] [PubMed] [Google Scholar]
- 57.Huoponen K, Vilkki J, Aula P, Nikoskelainen EK, Savontaus ML. A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am J Hum Genet. 1991;49:939–950. [PMC free article] [PubMed] [Google Scholar]
- 58.Kao MC, Di Bernardo S, Matsuno-Yagi A, Yagi T. Characterization of the membrane domain Nqo11 subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochem. 2002;41:4377–4384. doi: 10.1021/bi025525d. [DOI] [PubMed] [Google Scholar]
- 59.Kao MC, Di Bernardo S, Matsuno-Yagi A, Yagi T. Characterization and topology of the membrane domain Nqo10 subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochem. 2003;42:4534–4543. doi: 10.1021/bi034166z. [DOI] [PubMed] [Google Scholar]
- 60.Di Bernardo S, Yano T, Yagi T. Exploring the membrane domain of the reduced nicotinamide adenine dinucleotide-quinone oxidoreductase of Paracoccus denitrificans: Characterization of the NQO7 subunit. Biochem. 2000;39:9411–9418. doi: 10.1021/bi0006619. [DOI] [PubMed] [Google Scholar]
- 61.Galkin A, Meyer B, Wittig I, Karas M, Schagger H, Vinogradov A, Brandt U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem. 2008;283:20907–20913. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gostimskaya IS, Cecchini G, Vinogradov AD. Topography and chemical reactivity of the active-inactive transition-sensitive SH-group in the mitochondrial NADH: ubiquinone oxidoreductase (Complex I) Biochim Biophys Acta. 2006;1757:1155–1161. doi: 10.1016/j.bbabio.2006.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarzi E, Brown MD, Lebon S, Chretien D, Munnich A, Rotig A, Procaccio V. A novel recurrent mitochondrial DNA mutation in ND3 gene is associated with isolated complex I deficiency causing Leigh syndrome and dystonia. Am J Med Genet A. 2007;143:33–41. doi: 10.1002/ajmg.a.31565. [DOI] [PubMed] [Google Scholar]
- 64.Taylor RW, Singh-Kler R, Hayes CM, Smith PE, Turnbull DM. Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann Neurol. 2001;50:104–107. doi: 10.1002/ana.1084. [DOI] [PubMed] [Google Scholar]
- 65.McFarland R, Kirby DM, Fowler KJ, Ohtake A, Ryan MT, Amor DJ, Fletcher JM, Dixon JW, Collins FA, Turnbull DM, Taylor RW, Thorburn DR. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- 66.Di Bernardo S, Yagi T. Direct interaction between a membrane domain subunit and a connector subunit in the H+-translocating NADH-quinone oxidoreductase. FEBS Lett. 2001;508:385–388. doi: 10.1016/s0014-5793(01)03111-8. [DOI] [PubMed] [Google Scholar]
- 67.Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schägger H, Kerscher S, Brandt U. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Biophys Acta. 2004;1658:148–156. doi: 10.1016/j.bbabio.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 68.Brandt U, Abdrakhmanova A, Zickermann V, Galkin A, Dröse S, Zwicker K, Kerscher S. Structure–function relationships in mitochondrial complex I of the strictly aerobic yeast Yarrowia lipolytica. Biochem Soc Trans. 2005;33:840–844. doi: 10.1042/BST0330840. [DOI] [PubMed] [Google Scholar]
- 69.Finel M, Majander AS, Tyynelä J, de Jong AMP, Albracht SPJ, Wikström MKF. Isolation and characterisation of subcomplexes of the mitochondrial NADH:ubiquinone oxidoreductase (complex I) Eur J Biochem. 1994;226:237–242. doi: 10.1111/j.1432-1033.1994.tb20046.x. [DOI] [PubMed] [Google Scholar]
- 70.Holt PJ, Morgan DJ, Sazanov LA. The location of NuoL and NuoM subunits in the membrane domain of the Escherichia coli complex I - Implications for the mechanism of proton pumping. J Biol Chem. 2003;278:43114–43120. doi: 10.1074/jbc.M308247200. [DOI] [PubMed] [Google Scholar]
- 71.Baranova EA, Morgan DJ, Sazanov LA. Single particle analysis confirms distal location of subunits NuoL and NuoM in Escherichia coli complex I. J Struct Biol. 2007;159:238–242. doi: 10.1016/j.jsb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 72.Cardol P, Boutaffala L, Memmi S, Devreese B, Matagne RF, Remacle C. In Chlamydomonas, the loss of ND5 subunit prevents the assembly of whole mitochondrial complex I and leads to the formation of a low abundant 700 kDa subcomplex. Biochim Biophys Acta. 2008;1777:388–396. doi: 10.1016/j.bbabio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 73.Torres-Bacete J, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1 - Conserved charged residues essential for energy-coupled activities. J Biol Chem. 2007;282:36914–36922. doi: 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- 74.Euro L, Belevich G, Verkhovsky MI, Wikström M, Verkhovskaya M. Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I) Biochim Biophys Acta. 2008;1777:1166–1172. doi: 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 75.Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005;9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- 76.Morino M, Natsui S, Swartz TH, Krulwich TA, Ito M. Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J Bacteriol. 2008;190:4162–4172. doi: 10.1128/JB.00294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathiesen C, Hagerhall C. The ‘antiporter module’ of respiratory chain Complex I includes the MrpC/NuoK subunit - a revision of the modular evolution scheme. FEBS Lett. 2003;549:7–13. doi: 10.1016/s0014-5793(03)00767-1. [DOI] [PubMed] [Google Scholar]
- 78.Kervinen M, Patsi J, Finel M, Hassinen IE. A pair of membrane-embedded acidic residues in the NuoK subunit of Escherichia coli NDH-1, a counterpart of the ND4L subunit of the mitochondrial complex I, are required for high ubiquinone reductase activity. Biochem. 2004;43:773–781. doi: 10.1021/bi0355903. [DOI] [PubMed] [Google Scholar]
- 79.Kao MC, Di Bernardo S, Perego M, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T. Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J Biol Chem. 2004;279:32360–32366. doi: 10.1074/jbc.M403885200. [DOI] [PubMed] [Google Scholar]
- 80.Kurki S, Zickermann V, Kervinen M, Hassinen IE, Finel M. Mutagenesis of Three Conserved Glu Residues in a Bacterial Homologue of the ND1 Subunit of Complex I Affects Ubiquinone Reduction Kinetics but Not Inhibition by Dicyclohexylcarbodiimide. Biochem. 2000;39:13496–13502. doi: 10.1021/bi001134s. [DOI] [PubMed] [Google Scholar]
- 81.Hinchliffe P, Sazanov LA. Organization of iron-sulfur clusters in respiratory complex I. Science. 2005;309:771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- 82.Böttcher B, Scheide D, Hesterberg M, Nagel-Steger L, Friedrich T. A novel, enzymatically active conformation of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2002;277:17970–17977. doi: 10.1074/jbc.M112357200. [DOI] [PubMed] [Google Scholar]
- 83.Friedrich T, Brors B, Hellwig P, Kintscher L, Rasmussen T, Scheide D, Schulte U, Mäntele W, Weiss H. Characterization of two novel redox groups in the respiratory NADH:ubiquinone oxidoreductase (complex I) Biochim Biophys Acta. 2000;1459:305–309. doi: 10.1016/s0005-2728(00)00165-1. [DOI] [PubMed] [Google Scholar]
- 84.Carroll J, Fearnley IM, Walker JE. Definition of the mitochondrial proteome by measurement of molecular masses of membrane proteins. Proc Natl Acad Sci USA. 2006;103:16170–16175. doi: 10.1073/pnas.0607719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reda T, Barker CD, Hirst J. Reduction of the iron-sulfur clusters in mitochondrial NADH:Ubiquinone oxidoreductase (Complex I) by Eu-II-DTPA, a very low potential reductant. Biochem. 2008;47:8885–8893. doi: 10.1021/bi800437g. [DOI] [PubMed] [Google Scholar]
- 86.Galkin A, Brandt U. Superoxide radical formation by pure complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica. J Biol Chem. 2005;280:30129–30135. doi: 10.1074/jbc.M504709200. [DOI] [PubMed] [Google Scholar]
- 87.Kussmaul L, Hirst J. The mechanism of superoxide production by NADH: ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci USA. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Esterhazy D, King MS, Yakovlev G, Hirst J. Production of reactive oxygen species by complex I (NADH:ubiquinone oxidoreductase) from Escherichia coli and comparison to the enzyme from mitochondria. Biochem. 2008;47:3964–3971. doi: 10.1021/bi702243b. [DOI] [PubMed] [Google Scholar]
- 89.Ohnishi T. Iron-sulfur clusters semiquinones in complex I. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 90.Euro L, Bloch DA, Wikstrom M, Verkhovsky MI, Verkbovskaya M. Electrostatic interactions between FeS clusters in NADH:Ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochem. 2008;47:3185–3193. doi: 10.1021/bi702063t. [DOI] [PubMed] [Google Scholar]
- 91.Ingledew WJ, Ohnishi T. An analysis of some thermodynamic properties of iron-sulfur centres in site I of mitochondria. Biochem J. 1980;186:111–117. doi: 10.1042/bj1860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brandt U. Proton-translocation by membrane-bound NADH:ubiquinone-oxidoreductase (complex I) through redox-gated ligand conduction. Biochim Biophys Acta. 1997;1318:79–91. doi: 10.1016/s0005-2728(96)00141-7. [DOI] [PubMed] [Google Scholar]
- 93.Zwicker K, Galkin A, Dröse S, Grgic L, Kerscher S, Brandt U. The redox-Bohr group associated with iron-sulfur cluster N2 of complex I. J Biol Chem. 2006;281:23013–23017. doi: 10.1074/jbc.M603442200. [DOI] [PubMed] [Google Scholar]
- 94.Verkhovskaya ML, Belevich N, Euro L, Wikström M, Verkhovsky MI. Real-time electron transfer in respiratory complex I. Proc Natl Acad Sci USA. 2008;105:3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 96.Magnitsky S, Toulokhonova L, Yano T, Sled VD, Hagerhall C, Grivennikova VG, Burbaev DS, Vinogradov AD, Ohnishi T. EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH- ubiquinone oxidoreductase (complex I) in situ. J Bioenerg Biomembr. 2002;34:193–208. doi: 10.1023/a:1016083419979. [DOI] [PubMed] [Google Scholar]
- 97.Yano T, Dunham WR, Ohnishi T. Characterization of the ΔH+-sensitive ubisemiquinone species (SQNf) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochem. 2005;44:1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- 98.Fisher N, Rich PR. A motif for quinone binding sites in respiratory and photosynthetic systems. J Mol Biol. 2000;296:1153–1162. doi: 10.1006/jmbi.2000.3509. [DOI] [PubMed] [Google Scholar]
- 99.Gong X, Xie T, Yu L, Hesterberg M, Scheide D, Friedrich T, Yu CA. The ubiquinone-binding site in NADH:ubiquinone oxidoreductase from Escherichia coli. J Biol Chem. 2003;278:25731–25737. doi: 10.1074/jbc.M302361200. [DOI] [PubMed] [Google Scholar]
- 100.Usui S, Yu L, Yu CA. The small molecular mass ubiquinone-binding protein (QPc-9,5 kDa) in mitochondrial ubiquinol-cytochrome c reductase: Isolation, ubiquinone-binding domain, and immunoinhibition. Biochem. 1990;29:4618–4626. doi: 10.1021/bi00471a017. [DOI] [PubMed] [Google Scholar]
- 101.Link TA, Haase U, Brandt U, von Jagow G. What information do inhibitors provide about the structure of the hydroquinone oxidation site of ubihydroquinone:cytochrome c oxidoreductase? J Bioenerg Biomembr. 1993;25:221–232. doi: 10.1007/BF00762584. [DOI] [PubMed] [Google Scholar]
- 102.Schuler F, Yano T, Di Bernardo S, Yagi T, Yankovskaya V, Singer TP, Casida JE. NADH-quinone oxidoreductase: PSST subunit couples electron transfer from iron-sulfur cluster N2 to quinone. Proc Natl Acad Sci USA. 1999;96:4149–4153. doi: 10.1073/pnas.96.7.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schuler F, Casida JE. Functional coupling of PSST and ND1 subunits in NADH: ubiquinone oxidoreductase established by photoaffinity labeling. Biochim Biophys Acta. 2001;1506:79–87. doi: 10.1016/s0005-2728(01)00183-9. [DOI] [PubMed] [Google Scholar]
- 104.Murai M, Ishihara A, Nishioka T, Yagi T, Miyoshi H. The ND1 subunit constructs the inhibitor binding domain in bovine heart mitochondrial complex I. Biochem. 2007;46:6409–6416. doi: 10.1021/bi7003697. [DOI] [PubMed] [Google Scholar]
- 105.Ohnishi T, Salerno JC. Conformation-driven and semiquinone-gated proton-pump mechanism in the NADH-ubiquinone oxidoreductase (complex I) FEBS Lett. 2005;579:4555–4561. doi: 10.1016/j.febslet.2005.06.086. [DOI] [PubMed] [Google Scholar]
- 106.Zickermann V, Barquera B, Wikström MKF, Finel M. Analysis of the pathogenic human mitochondrial mutation ND1/3460, and mutations of strictly conserved residues in its vicinity, using the bacterium Paracoccus denitrificans. Biochem. 1998;37:11792–11796. doi: 10.1021/bi9810555. [DOI] [PubMed] [Google Scholar]
- 107.Yamaguchi M, Belogrudov G, Hatefi Y. Mitochondrial NADH-ubiquinone oxidoreductase (complex I). Effect of substrates on the fragmentation of subunits by trypsin. J Biol Chem. 1998;273:8094–8098. doi: 10.1074/jbc.273.14.8094. [DOI] [PubMed] [Google Scholar]
- 108.Belogrudov G, Hatefi Y. Catalytic sector of complex I (NADH:Ubiquinone oxidoreductase): subunit stoichiometry and substrate-induced conformation changes. Biochem. 1994;33:4571–4576. doi: 10.1021/bi00181a018. [DOI] [PubMed] [Google Scholar]
- 109.Mamedova AA, Holt PJ, Carroll J, Sazanov LA. Substrate-induced conformational change in bacterial complex I. J Biol Chem. 2004;279:23830–23836. doi: 10.1074/jbc.M401539200. [DOI] [PubMed] [Google Scholar]
- 110.Brandt U, Kerscher S, Dröse S, Zwicker K, Zickermann V. Proton pumping by NADH:ubiquinone oxidoreductase. A redox driven conformational change mechanism? FEBS Lett. 2003;545:9–17. doi: 10.1016/s0014-5793(03)00387-9. [DOI] [PubMed] [Google Scholar]
- 111.Olesen C, Picard M, Winther AML, Gyrup C, Morth JP, Oxvig C, Moller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–10U5. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 112.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 113.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]