Abstract
Protein NMR assignments of large proteins using traditional triple resonance techniques depends on double or triple labeling of samples with 15N, 13C, and 2H. This is not always practical with proteins that require expression in non-bacterial hosts. Labeling with isotopically labeled versions of single amino acids (sparse labeling) often is possible, however resonance assignment then requires a new strategy. Here a procedure for the assignment of cross peaks in 15N-1H correlation spectra of sparsely labeled proteins is presented. It relies on the correlation of proton-deuterium amide exchange rates in native and denatured spectra of the intact protein, followed by correlation of chemical shifts in the spectra of the denatured protein with chemical shifts of sequenced peptides derived from the protein. The procedure is successfully demonstrated on a sample of a protein, Galectin-3, selectively labeled with 15N at all alanine residues.
Keywords: sparse labeling, amide exchange, denatured proteins, chemical shifts
Introduction
Structure determination of moderate sized proteins using NMR methods has traditionally relied on uniform isotopic labeling with 15N and 13C1. As molecular size increases various modifications to this labeling strategy have been adopted, including uniform labeling with 2H to reduce spin relaxation rates and improve resolution2. Assignment of the myriad of cross-peaks that appear in multi-dimensional spectra is typically accomplished using triple resonance experiments that pass spin coherence from one isotopically labeled site to another, and assignment is followed by the detection and interpretation of distance dependent Nuclear Overhauser Effects (NOEs). This procedure has proven very powerful leading to thousands of structural depositions in the Protein Data Bank 3, but it is also limited in several respects. As proteins become large (>40 kDa) the number of cross-peaks, and increased widths of those cross-peaks, make assignments based on triple resonance experiments challenging. Also, certain types of proteins (glycosylated proteins, for example) are typically expressed in host cells where uniform isotopic enrichment can be very expensive and deuteration can be very detrimental to cell growth. In these cases sparse labeling with one or two selected amino acids provides a means of avoiding these problems, and at the same time, reducing the number of cross-peaks and limiting assignment options to the selected amino acid types.4–7
Single amino acid labeling can be done easily in E. coli host cells, however consideration must be given to control of metabolic pathways that can cause isotope scrambling.5 While protein expression itself in eukaryotic hosts requires more effort, additional complications due to isotope scrambling appear to be less severe.8–11 However with successful single amino acid labeling, a new problem arises in that sequential assignment of cross-peaks by triple resonance strategies is no longer possible.
In what follows a new approach for the sequential assignment of cross-peaks in 15N-1H single quantum coherence (HSQC) spectra will be demonstrated. It is based on correlation of amide hydrogen exchange rates measured from spectra on the folded protein with amide exchange rates measured from spectra of denatured aliquots sampled at various time points during exchange. Cross-peaks in the latter spectra are found to superimpose with the single cross-peaks in spectra of isolated and sequenced peptides producing a sequence specific assignment for the native protein HSQC spectrum.
While sparse labeling may prohibit the use of NOEs as a primary source of structural data, it is important to realize that there are other types of data that can lead to useful structural models for proteins. Conventional NOE derived distance constraints can be replaced or supplemented with longer range distance constraints from paramagnetic perturbations,11–14 and orientational constraints from residual dipolar couplings (RDCs) can provide additional useful geometric information.15–17 In most cases the derived constraints apply only to backbone positions and orientations. This is not an issue in cases such as structure determination of multi-subunit or multi-domain proteins where independently determined high resolution structures for single subunits or domains might exist. However, for proteins with previously undetermined structures, producing high resolution structures will become increasingly dependent on computational methods for the proper placement of sidechains. Fortunately, great strides in the computational placement of sidechains have been made over the past few years.18,19 Therefore, an increased use of sparse labeling and sparse constraints for structure determination can be envisioned, particularly if sequence specific assignments of sparsely labeled cross-peaks can be provided. A move in this direction can have a great impact on work with large protein assemblies and with certain classes of proteins such as glycosylated proteins. This latter class is not insignificant, as more than 50% of human proteins are, in fact, glycoproteins.20
There have been attempts at sequential assignments of cross-peaks from sparsely labeled proteins, but primarily in cases where the structure of the labeled protein is known. Given a structure and a sufficiently large set of measured RDCs, possible assignments can be permuted, RDCs back-calculated for each assignment set, and the set with the best agreement with experiment identified.21–23 For the case where a structure is not known ahead of time, there is the possibility of pairwise labeling with one 13C carbonyl labeled amino acid and one 15N labeled amino acid.24 This introduces a 15N-13C coupling to cross-peaks where the pair appears sequentially. However, a new expression much be done for each assignment and the procedure is, thus, quite laborious.
In an effort to devise a less laborious procedure our laboratory has previously explored ways of correlating mass spectrometry derived sequence information with NMR cross-peaks positions using amide hydrogen-deuterium exchange (HDX).25 In these initial studies, the intensity loss in HSQC cross-peaks from the native protein (as protons are replaced with deuteriums from solvent) was correlated with the loss of peaks in spectra of isolated peptides that could be sequenced by mass spectrometry. Assignment for a single phenylalanine peak in the small protein, Galectin-3 was used to illustrate the procedure. However, there are extreme demands placed on the procedure because the protein must be digested, peptides separated, and NMR spectra collected, all before incorporated deuteriums have a chance to back-exchange. In the procedure to be illustrated here the protein digestion and peptide separation steps are moved to a point where back-exchange is inconsequential. The new approach uses deuterium exchange only to correlate cross-peaks in native and denatured spectra; the cross-peaks in the denatured spectra are subsequently assigned by chemical shift correlation between cross-peaks from the denatured intact protein and cross-peaks from the denatured separated peptides. Denaturation can be very fast, as can collection of spectra on the denatured aliquots. The subsequent correlation between chemical shifts in denatured aliquots and derived peptides is not under any severe time constraints.
As in previous work, the target chosen to illustrate the procedure is Galectin-3. This is a 15 kDa carbohydrate-binding protein; its resonances have been previously assigned using conventional triple resonance methods. (BioMagResBank accession # 4909 and PDB # 1A3K).26 For the present study the protein has been labeled with 15N alanine producing a set of six HSQC cross-peaks for assignment. Assignments using the current method are shown to be consistent with previous assignments, documenting the future applicability to proteins whose structure cannot be approached with traditional methods.
Experimental Section
Protein Expression and purification
The plasmid for the C-terminal carbohydrate binding domain of Galectin-3 was obtained from Dr. Hakon Leffler at Lund University, Sweden, and inserted into a pET-9-a vector (Novagen) by heat shock transformation. Over-expression of the protein was done using E. coli BL21-Gold (DE3) competent cells (Stratagene) in 1 L M9 minimal medium containing NH4CI and glucose at natural abundance. All 19 amino acids except for alanine were added as natural abundance materials at 0.1 g per liter of culture and growth was done in a 250 rpm shaker at 37°C. At an optical cell density (OD600) ~ 0.4, 0.3 g of 15N Ala (Cambridge Isotopes Laboratories) was added to the medium. At OD600 ~ 1.1, 1 ml of 1 mM IPTG was added to induce protein expression. The culture was then grown overnight to OD600 λ ~1.8 at 18° C. The cells were harvested by centrifugation and lysed in 30 ml buffer containing 75 mM KH2PO4, 75 mM NaCl, 5 mM EDTA, 5 mM DTT, and 5 mM NaN3. 1 M NaOH was used to adjust pH to 7.1. After centrifugation the supernatant containing Galectin-3 was loaded onto a lactosyl agarose 1.8 cm × 11 cm column (Sigma) equilibrated with the phosphate buffer. The protein was eluted using the phosphate buffer above, but also containing 300 mM lactose at pH 7.1. The protein yield was ~ 50 mg per L. Protein was concentrated using 10 K centricons (Millipore) and stored in the lactose containing phosphate buffer above. Mass spectrometric analysis showed that Ala is > 90%27 15N labeled in the protein.
Amide proton exchange measurements on the native protein
A portion of the stored solution containing 13.5 mg of protein was lyophilized. 477 µL of 99.9% D2O (Cambridge Isotope Laboratories) was added to the fully protonated dry protein. ID 15N filtered HSQC spectra (Varian biopack sequence gNhsqc with the d1 delay set to 1 sec) were acquired in order to map the protein exchange. Sequential acquisition was done every 4 min 56 sec (nt=256 ss=4) beginning at an initial time point 3 min 30 sec after mixing. In later time points, acquisition spectra where obtained in a time series with geometrically increasing time delays. NMR data were acquired using a Varian 900 MHz spectrometer system equipped with a triple resonance cold probe and a pulsed field gradient unit. Data were processed using VnmrJ version 6.1 and integrated to provide a time-course for loss of intensity in each cross-peak. The time resolved intensities for each peak were fit to the equation, I = I0*exp (−kext), using an in-house curve fitting script (Dr. Yizhou Liu). Errors reported are from a least squares fit to this equation. They are similar to deviation of duplicate runs made on selected samples.
Deuterium content measurements on aliquots of denatured protein
A parallel sample under the same conditions as above was prepared and incubated in 99.9% D2O. Eleven 53 µL aliquots of partially deuterated protein were sampled following an approximately geometrically increasing set of time delays as follows: 1, 16, 32 mins, 1, 2, 4, 8, 16, 32, 64, and 128 hrs. Each aliquot was placed in an Eppendorf tube and rapidly quenched using 100 µL of a cold acidic deuterated urea solution to produce samples 8.3 M in urea and at pD 3.1 (pD = pH(meter) + 0.4). These samples were quickly frozen using liquid nitrogen and stored at −80°C.
Each quenched aliquot was later thawed and quickly transferred to a 3mm NMR tube for analysis using a 600 MHz Varian spectrometer system equipped with a 3 mm cold probe. 2D pulsed field gradient 15N filtered HSQC spectra (Varian biopack sequence gNhsqc) were acquired over 3 min 39 sec (nt=2 ss=2 ni=32). The data were processed with NMRPipe and cross peak volumes integrated in NMRDraw to quantitate the deuterium content of each 15N alanine residue in each aliquot sampling point. The time resolved intensities for each peak were fit to the same exchange equation as used for the native protein.
Spectral analysis of digested protein
A 1.5 mg sample of Galectin-3 was lyophilized, then dissolved in 900 µl of 50 mM ammonium bicarbonate at pH 8. 20 µg of sequencing grade modified trypsin (Promega), was also dissolved in 300 µl of 50 mM ammonium bicarbonate. The trypsin solution was added to the protein sample such that the final volume was 1.5 ml, making a protease to protein ratio of 1:25. The protein enzyme solution was incubated overnight at 37°C. The peptides in the digested protein sample were separated by HPLC on a C18 column (Agilent Technologies, 4.6mm × 150mm 5µm, 300 Å) using a gradient running from 95% buffer A (0.1% TFA in H2O) and 5% buffer B (0.1% TFA in ACN) to 60% buffer B, at a flow rate of 1 ml/min over 25 min.
The fractions collected were analyzed by MALDI/TOF mass spectrometry, α-cyano-4-hydroxycinnamic acid (Sigma Aldrich) was used as the MALDI matrix on a 4700 Proteomics Analyzer (Applied Biosystems) using a mass range of 560 to 4000 Da, and a laser intensity of 5000. Analysis of 15N content was done using the 4700 Data Explorer program and ISOTOPICA27 by comparing isotopomer profiles of labeled peptides to those for previously analyzed unlabeled peptides. Previous identification of unlabeled tryptic fragments was done using the PROWL28 Proteinlnfo search engine.
The HPLC fractions containing 15N labeled peptides were lyophilized to remove the HPLC solvents and each fraction was dissolved back into solution using 140 µl of 8 M protonated urea at pH 2.4. 2D pulsed field gradient 15N-1H HSQC spectra were acquired as in the denatured protein to map the chemical shifts of each identified peptide to cross-peaks of the denatured protein.
Results
Amide proton exchange measurements in the native protein
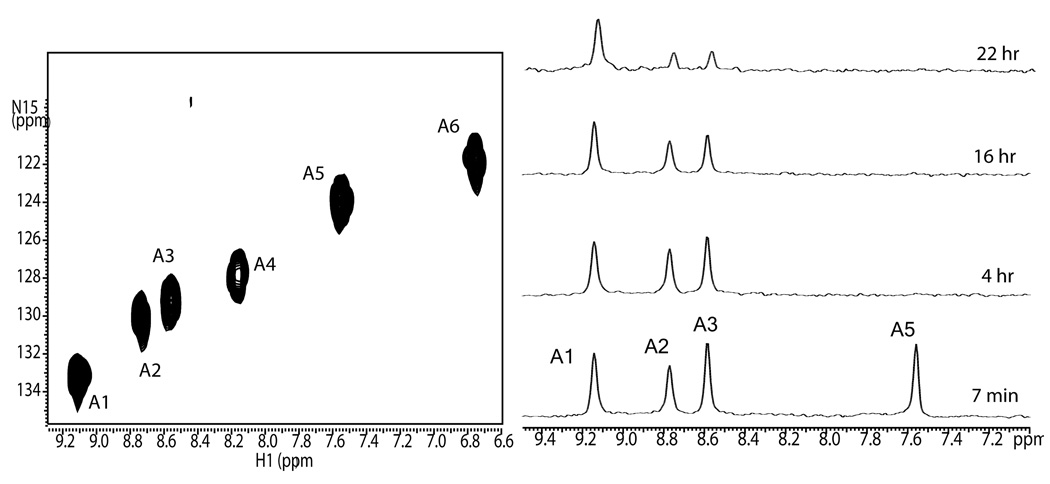
The initial step in the assignment procedure involves monitoring deuterium incorporation as a function of time into a natively folded, fully protonated, 15N sparsely labeled protein. This is normally accomplished using a time series of 2D HSQC spectra, but can also be done using 1D 15N-filtered spectra if resolution is adequate. Figure 1a shows an HSQC spectrum of Galectin-3 labeled with 15N at all alanine sites. The peaks are arbitrarily labeled A1–A6. The sample was lyophilized and at zero time re-dissolved in an equivalent amount of 2H2O. Figure 1b shows a time course of ID 15N filtered spectra undergoing exchange at pH 7.1 and 25°C, running from 7 min to 22 hrs. At the earliest point two sites are already fully exchanged and peaks A4 and A6 are not observed. The remaining peaks disappear with vastly different exchange rates. Peak A1 retains more than half its intensity even at 22 hrs. The exchange rates of each of the residues are obtained by fitting the equation, I = I0*exp (−kext), to peak areas as a function of time.
Figure 1.
900 MHz spectra of 15N Ala Galectin-3; ~0.8 mM, pH 7.1, 25°C. (a) 2D HSQC spectrum of fully protonated protein with all six Ala residues labeled, (b) 1D HSQC exchange spectra of folded 15N Ala Galectin-3, showing deuterium incorporation as a function of time.
The resulting rate constants are reported in column 2 of Table 1, along with constants derived from data collected on samples at pH 5.8 and 15°C to allow measurement of rates for peaks A4 and A6. Additional spectra were also collected at pH 8.2 and 25°C to distinguish exchange rates for A2 and A3 that exchange at a fairly similar rate at pH 7.1.
Table 1.
Exchange rates obtained from plotting the HSQC intensities versus time*.
| Cross-peak number | kex native samples (s−1) | kex denatured samples (s−1) | MS-based peptide identity | Triple resonance assignment |
|---|---|---|---|---|
| A1 | < 3.8 × 10−7 | < 3.8 × 10−7 | VA212VNDA216HLLQYNHR | A212 |
| A2 | 6.5 × l0−6 ± 3.0 × 10−7 | 7.0 × 10−6 ± 4.9 × 10−6 | GNDVA156FHFNPR | A156 |
| A3 | 8.4 × l0−6 ± 1.6 × l0−6 | 8.7 × l0−6 ± 4.7 × 10−7 | IA146LDFQR***** | Al46 |
| A4 | 3.1 × l0−2 ± 3.0 × l0−3** | > 1.0 × l0−2 | LGISGDIDLTSA245SYTMIC*** | A245**** |
| A5 | 1.0 × l0−3 ± 1.4 × l0−4 | 1.3 × l0–3 ± 3.4 × l0−5 | VA212VNDA216HLLQYNHR***** | A216 |
| A6 | 3.0 × l0−2 ± 4.0 × l0−3** | > 1.0 × 10−2 | MLITILGTVKPNA142NR | A142**** |
Exchange data were collected at pH 7.1, 25° C except as noted. Errors are standard deviations taken from best fits to time course data.
Exchange data were collected for these residues at pH 5.8 at 15° C, but scaled by a factor of 13 to correct for the ten-fold change in the exchange rates with each pH unit.
Peptide not found by mass spectrometry after HPLC analysis but predicted for cleavage by trypsin.
Ambiguous assignments because of similar rates.
Ambiguous assignments because of end-group effects.
Amide proton exchange in the denatured protein
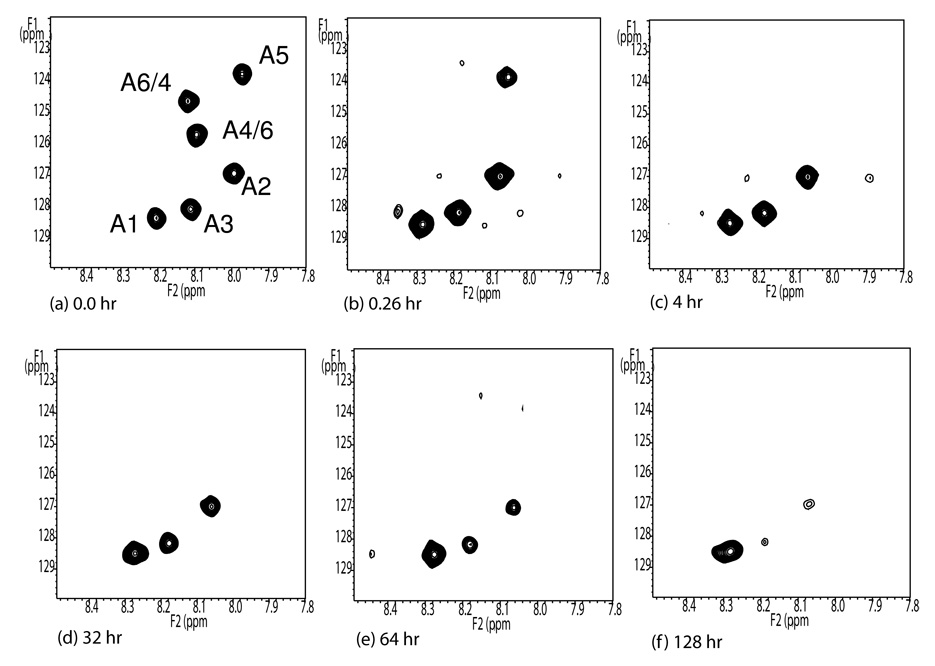
The second step in the procedure involves collection of a similar time course from HSQC spectra of aliquots pulled from an exchanging sample and quickly denatured before observation. The conditions of the exchanging sample approximate those described above (0.6 mM protein in 75 mM phosphate buffer, pH 7.1, 25°C). 53 µL aliquots were transferred to Eppendorf tubes, diluted with 100 µL of cold 12.8 M urea in 2H2O at pD 3.1 and quickly frozen for later analysis. After thawing aliquots were transferred to 3mm NMR tubes and fast HSQC spectra were collected (approximately 2 min preparation time and 3 min acquisition time). Examples of the resulting HSQC spectra are shown in Figure 2. The dispersion of the cross-peaks is much less than in the native protein, but the resonances are much sharper. All resonances are resolved and of full intensity at zero time (an aliquot made from a fully protonated sample). Aliquots pulled at later times show a progressive loss of peak intensities. Having these intensities accurately reflect deuterium content at the point of aliquot extraction required that denaturing be fast and that exchange under denatured conditions occur in a time long compared to spectral acquisition times. The former is supported by observation of a fully denatured spectrum at the first time point taken. The latter is supported by collecting a series of HSQCs on a given sample once it is thawed and placed in the magnet. These show continued exchange to occur with half times greater than 30 min for all observed peaks, a time much greater than that for acquisition of a single spectrum. Exchange rates were therefore calculated from the first time point for each of the six Ala residues using the formula given above. These are reported in column 3 of Table 1. With the exception of one pair where correlation is ambiguous, it is clear that one can pair cross-peaks in the denatured spectra with cross-peaks in the native spectrum based on matching of exchange rates. Peaks in the zero time denatured spectrum (Fig. 2a) have been labeled based on this correlation.
Figure 2.
Fully protonated denatured 15N Ala Galectin-3 (a) and partially deuterated 15N Ala Galectin-3 (b–f) The HSQC spectra are acquired in a 600 MHz spectrometer at 5°C in ~ 8 M protonated urea pH 2.4 (a) and pD 3.1 (b–f) respectively. The small offsets apparent in the proton dimension between the fully protonated sample (a) and the exchanging spectra (b-f) may arise differences in lock frequencies for HDO (10% D2O) vs D2O (~100% D2O).
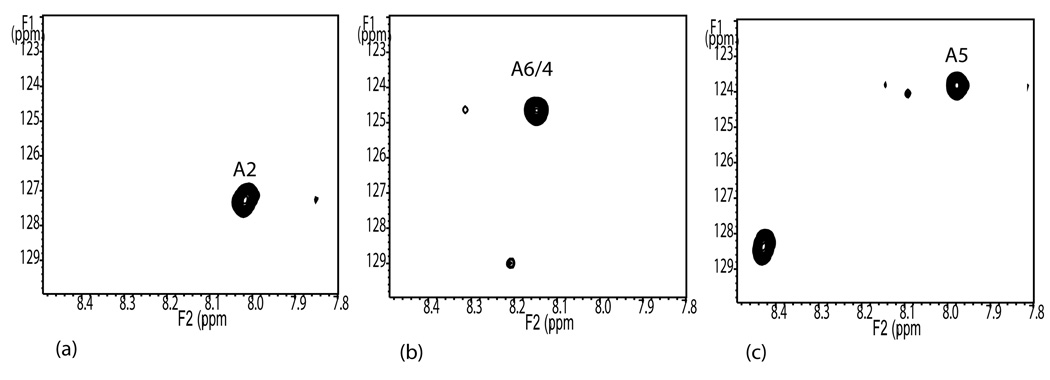
The third step in the procedure requires correlation of cross-peaks in the HSQC spectra of denatured aliquots with cross-peaks in HSQC spectra of isolated and sequenced peptides. Correlation depends on an assumption that chemical shifts of labeled alanines in the denatured proteins will depend on only the local sequence and hence chemical shifts of alanines in isolated peptides of sufficient length will be identical to chemical shifts of those alanines in the denatured protein. A fully protonated aliquot of the protein was treated with trypsin and peptides were isolated by elution on an HPLC column. The peptides in the fractions were identified by mass spectrometry and three fractions containing peptides having alanines more than two residues from the peptide ends were selected for analysis. HSQC spectra of these peptides were run under the same denaturing conditions as the intact protein. Spectra are shown in panels a, b, and c of Figure 3. The peaks closely superimpose on three peaks of the denatured protein as displayed in Figure 2. The labels, (A2, A5 and A6/4) have been transferred on that basis. The spectrum in Figure 3c has two peaks corresponding to the two alanines in its sequence. The second peak (8.45 ppm by 128.4 ppm) does not overlap with any peak in the spectrum of the denatured protein. This clearly belongs to A212 which is just one amino acid removed from the N-terminus of the peptide. Such shifts for residues near the terminus are not unexpected29,30. Since the peptides are sequenced we can assign the three peaks A2, A5, and A6/4 to A156, A216, and A142 respectively. These assignments have been entered into the fourth column of Table 1, and the assignments are compared with the earlier triple resonance assignments as entered in column 5.
Figure 3.
600 MHz HSQC spectra of isolated, mass spectrometry sequenced, and denatured peptides. Spectra obtained at 5°C in 8 M protonated urea pH 2.4 (a) 1274 m/z - GNDVA156FHFNPR, (b)1641 m/z - MLITILGTVKPNA142NR, (c) 1650 m/z -VA212VNDA216HLLQYNHR
Discussion
Table 1 shows that we have been able to uniquely assign three cross-peaks in the spectrum of the denatured protein to the three Ala residues, A156, A216, A142. Differences in amide proton exchange rates have allowed a unique correlation for two of these to cross peaks in the HSQC spectrum of the folded protein (A2 to A156 and A5 to A216). The third cross peak, A142, because of similarities in exchange rates, must be correlated with either A6 or A4. These assignments agree with those previously made using triple resonance experiments (A142 is assigned to A6).26 While there is some ambiguity in assignment, the effort required for assignment is minimal. Monitoring amide proton exchange in folded proteins is a high sensitivity experiment. Analysis of denatured aliquots proceeds with even higher sensitivity because of the narrower lines and lesser dispersion of cross-peaks. The digestion of an aliquot, followed by isolation and analysis of peptides is more time consuming but employs methodology frequently used in the mass spectrometry community. Most importantly, labeling with selected amino acids in a variety of hosts is possible and usually significantly less expensive than uniform labeling with 15N and 13C.
The actual amount of spectrometer time used in the assignment procedure can be very small. For monitoring exchange in the native protein, 5 min acquisitions over 12 time points would require just 1 hr of spectrometer time, assuming the sample could be removed from the spectrometer between acquisitions. Realistically the initial 6, more closely spaced, time points would be taken without removing the sample from the spectrometer (3 hrs) and the remaining 6 time points would require some spectrometer set-up (30 min × 6 = 3hrs). For analysis of the denatured aliquots, if we were to use just a single initial spectrum on each denatured aliquot and conduct exchange at just one pH, 12 samples, using 3 ½ min per acquisition, amount to just 42 min of collection time. The process of melting samples, getting them into the spectrometer, and optimizing field homogeneity, of course takes additional time, but much of this can be automated (perhaps 2 hrs total).
In terms of materials, a 0.8 mM (6.3mg in 500 µL) sample of Galectin-3 was used for monitoring exchange of the folded native protein, 1.5 mg was used for the tryptic digest sample, and 16.5 mg was aliquoted into eleven 1.5 mg fractions for the denatured samples. The total amount of sample used in the experiments presented was just under 25 mg. However, the material required can clearly be reduced by a factor of 2. For the spectra of denatured aliquots one could do with a factor of 2 lower signal to noise ratio, and with a 30 min back-exchange rate one could have signal averaged for 12 min. This would have reduced total sample requirements to about 12 mg. More efficient spectral acquisition procedures, for example, Hadamard HSQCs31 or new fast recovery HSQC sequences applicable to the folded protein32 might reduce requirements further. The total sample requirements could therefore be similar to that used in typical NMR experiments on proteins.
The application illustrated is to a protein only 15 kDa in molecular weight. However, based on resolution in the current spectra, one expects to be able to resolve and assign cross-peaks from denatured proteins of at least two times the size. One also expects to be able to differentiate most peaks based on amide proton exchange rates for similarly sized proteins. In a folded protein, the distribution of exchange rates is not only governed by secondary structure effects but also by the local environment with a reasonably high dependence on amino acid sequence.33 This results in a considerable variation of the exchange rates for different sequential sites. Assuming a protein has a geometrically distributed set of amide proton exchange half times, each differing by a factor of two and the whole set ranging from 1 min to 2 weeks, one could differentiate at least 14 sites. If the 20 amino acids are equally represented in the sequence, 14 sites correspond to a 280 residue or 31 kDa protein.
One of the keys to the success of the procedure described for Galectin-3 is the similarity of chemical shifts for residues in the denatured protein and in the derived peptides. The reason for the lack of similarity in shifts for some of the alanines is well understood. The labeled sites are too close to the ends of the peptide in some of the fragments. There are some options that can remove this limitation in the future. One is simply to use alternate digestion protocols to obtain fragments where more sites are in the interior of the peptides. Digestions with endoproteinases like Glu-C and pepsin are possibilities. Another would be to accurately predict end-group effects on chemical shifts.
Prediction of chemical shifts for random coil proteins and peptides has been undertaken by a number of authors,29,30 These were not undertaken for the purpose of predicting end effects, but instead undertaken to provide a reference set for extracting secondary structure effects from the spectra of folded proteins. The fact that one might be able to predict not just end effects, but the actual chemical shifts of all residues in a denatured protein is intriguing. Using the full protein sequence and a good prediction tool one might be able to predict the spectra shown in Figure 2a and assign cross-peaks without the need to generate and examine spectra of digested peptides. The accuracy of the prediction methods are not currently adequate to do this for all residues in our spectra, but once the 3 definitive assignments were made, ambiguities were, in fact, reduced enough to complete most assignments using predictions described by Wang and Jardetzky.29 Thus, with some improvements in shift prediction algorithms, an extremely simple approach to assignment of HSQC cross-peaks in spectra of sparsely labeled proteins may soon be available.
Conclusion
At this point we have been able to demonstrate a viable assignment strategy for HSQC cross-peaks from a 15 kDa protein labeled using a single amino acid (alanine) enriched in 15N. The method does not depend on having a previously determined structure for the protein and should be applicable to a variety of proteins targeted for structural characterization. Application will be particularly important for proteins that need to be expressed in non-bacterial hosts, proteins that require native glycosylation for example, where use of amino acids as metabolic precursors is common. It is likely that the methods can lead to nearly complete assignments of these proteins up to a size of 30 kDa and to partial assignments of much larger proteins. Effort is currently comparable to that required for traditional assignment methods, but it is easy to envision improvement for the future.
Acknowledgements
This work was supported by grant 5P41RR005351 from the National Institutes of Health. We acknowledge Dr. Fang Tian for suggesting the observation of denatured protein as a step in the assignment process, Laura Morris for computational programming to assist in the chemical shifts referencing work, and Dr. John Glushka, for his productive discussions and NMR assistance.
References
- 1.Yee A, Gutmanas A, Arrowsmith CH. Current Opinion in Structural Biology. 2006;16:611–617. doi: 10.1016/j.sbi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Kay LE. Journal of Magnetic Resonance. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Billeter M, Wagner G, Wuthrich K. J Biomol NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. J Biomol NMR. 2004;29:289–297. doi: 10.1023/B:JNMR.0000032523.00554.38. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi K, Ng E, Malia TJ, Wagner G. J Biomol NMR. 2007;38:89–98. doi: 10.1007/s10858-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 6.Tong KI, Yamamoto M, Tanaka T. J Biomol NMR. 2008;42:59–67. doi: 10.1007/s10858-008-9264-0. [DOI] [PubMed] [Google Scholar]
- 7.Ohki SY, Kainosho M. Progress in Nuclear Magnetic Resonance Spectroscopy. 2008;53:208–226. [Google Scholar]
- 8.Werner K, Richter C, Klein-Seetharaman J, Schwalbe H. J Biomol NMR. 2008;40:49–53. doi: 10.1007/s10858-007-9205-3. [DOI] [PubMed] [Google Scholar]
- 9.Strauss A, Bitsch F, Cutting B, Fendrich G, Graff P, Liebetanz J, Zurini M, Jahnke W. J Biomol NMR. 2003;26:367–372. doi: 10.1023/a:1024013111478. [DOI] [PubMed] [Google Scholar]
- 10.Chen CY, Cheng CH, Chen YC, Lee JC, Chou SH, Huang W, Chuang WJ. Proteins. 2006;62:279–287. doi: 10.1002/prot.20742. [DOI] [PubMed] [Google Scholar]
- 11.Liu S, Venot A, Meng L, Tian F, Moremen KW, Boons GJ, Prestegard JH. Chem Biol. 2007;14:409–418. doi: 10.1016/j.chembiol.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang T, Lee HS, Imperiali B, Prestegard JH. Protein Sci. 2008;17:1220–1231. doi: 10.1110/ps.034561.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertini I, Luchinat C, Parigi G, Pierattelli R. Dalton Transactions. 2008:3782–3790. doi: 10.1039/b719526e. [DOI] [PubMed] [Google Scholar]
- 14.Pintacuda G, John M, Su XC, Otting G. Accounts of Chemical Research. 2007;40:206–212. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- 15.Bouvignies G, Markwick PRL, Blackledge M. Chemphyschem. 2007;8:1901–1909. doi: 10.1002/cphc.200700353. [DOI] [PubMed] [Google Scholar]
- 16.Bax A, Grishaev A. Current Opinion in Structural Biology. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Prestegard JH, Bougault CM, Kishore AI. Chemical Reviews. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 18.Looger LL, Hellinga HW. Journal of Molecular Biology. 2001;307:429–445. doi: 10.1006/jmbi.2000.4424. [DOI] [PubMed] [Google Scholar]
- 19.Lopes A, Alexandrov A, Bathelt C, Archontis G, Simonson T. Proteins. 2007;67:853–867. doi: 10.1002/prot.21379. [DOI] [PubMed] [Google Scholar]
- 20.Apweiler R, Hermjakob H, Sharon N. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 21.Jung YS, Zweckstetter M. J Biomol NMR. 2004;30:25–35. doi: 10.1023/B:JNMR.0000042955.14647.77. [DOI] [PubMed] [Google Scholar]
- 22.Hus JC, Prompers JJ, Bruschweiler R. Journal of Magnetic Resonance. 2002;157:119–123. doi: 10.1006/jmre.2002.2569. [DOI] [PubMed] [Google Scholar]
- 23.Pintacuda G, Keniry MA, Huber T, Park AY, Dixon NE, Otting G. Journal of the American Chemical Society. 2004;126:2963–2970. doi: 10.1021/ja039339m. [DOI] [PubMed] [Google Scholar]
- 24.Kainosho M, Tsuji T. Biochemistry. 1982;21:6273–6279. doi: 10.1021/bi00267a036. [DOI] [PubMed] [Google Scholar]
- 25.Feng L, Lee HS, Prestegard JH. J Biomol NMR. 2007;38:213–219. doi: 10.1007/s10858-007-9159-5. [DOI] [PubMed] [Google Scholar]
- 26.Umemoto K, Leffler H. JBiomol NMR. 2001;20:91–92. doi: 10.1023/a:1011269008175. [DOI] [PubMed] [Google Scholar]
- 27.Femandez-de-Cossio J, Gonzalez LJ, Satomi Y, Betancourt L, Ramos Y, Huerta V, Amaro A, Besada V, Padron G, Minamino N, Takao T. Nucleic Acids Res. 2004;32:W674–W678. doi: 10.1093/nar/gkh423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beavis R, Fenyo D. Chapter 13. Curr Protoc Bioinformatics. 2004 doi: 10.1002/0471250953.bi1302s7. Unit 13 2. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Jardetzky O. J Am Chem Soc. 2002;124:14075–14084. doi: 10.1021/ja026811f. [DOI] [PubMed] [Google Scholar]
- 30.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. J Am Chem Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 31.Kupce E, Freeman R. JMagn Reson. 2003;163:56–63. doi: 10.1016/s1090-7807(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 32.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]