Abstract
Streptomyces sp. US 24 and Streptomyces sp. TN 58, two strains producing interesting bioactive molecules, were successfully transformed using E. coli ET12567 (pUZ8002), as a conjugal donor, carrying the integrative plasmid pSET152. For the Streptomyces sp. US 24 strain, two copies of this plasmid were tandemly integrated in the chromosome, whereas for Streptomyces sp. TN 58, the integration was in single copy at the attB site. Plasmid pSET152 was inherited every time for all analysed Streptomyces sp. US 24 and Streptomyces sp. TN 58 exconjugants under nonselective conditions. The growth, morphological differentiation, and active molecules production of all studied pSET152 integrated exconjugants were identical to those of wild type strains. Consequently, conjugal transfer using pSET152 integration system is a suitable means of genes transfer and expression for both studied strains. To validate the above gene transfer system, the glucose isomerase gene (xylA) from Streptomyces sp. SK was expressed in strain Streptomyces sp. TN 58. Obtained results indicated that heterologous glucose isomerase could be expressed and folded effectively. Glucose isomerase activity of the constructed TN 58 recombinant strain is of about eighteenfold higher than that of the Streptomyces sp. SK strain. Such results are certainly of importance due to the potential use of improved strains in biotechnological process for the production of high-fructose syrup from starch.
1. Introduction
At the end of the last century, in addition to the conventional screening of naturally occurring bioactive compounds, a strategy was developed to construct desired novel hybrid antibiotics by combinatorial biosynthesis, by specifically combining genes from various biosynthetic pathways originating from different antibiotics producing organisms. In Streptomyces species, the most important producers of bioactive molecules, genetic approaches to improve secondary metabolite production are generally hampered by various phenomena such as restriction barriers, absence of an efficient gene transfer systems, and lack of suitable cloning vectors. Recently, in order to circumvent these problems, there has been considerable interest in the use of intergeneric conjugation as an efficient means of gene transfer and expression in Streptomyces species. This technique allows the construction and the manipulation of recombinant plasmids in E. coli and their subsequent transfer to a recipient of interest. The initial protocol for intergeneric transfer of plasmids from E. coli to Streptomyces species was developed by Mazodier et al. [1]. Since then, this method has been successfully applied to several Streptomyces species [2–4] via methylation-deficient E. coli strains, such as strain ET12567 [5], as a DNA donor to avoid methylated DNA restriction systems of actinomycetes [6]. Several cloning vectors that could be transferred from E. coli to Streptomyces species by conjugation method have been constructed [7, 8]. Among them, pSET152 is a nonreplicative plasmid in Streptomyces that carries the attP site and integrase gene of φC31 phage and consequently can integrate into the chromosomal attB site of the bacteriophage φC31. Like λ integrases, φC31 integrase catalyses the site-specific recombination between the chromosomal attB site and an attP site to form attL and attR. However, unlike those of λ integrases, the sites are both very small, with attP and attB being just 39 and 34 pb in size, respectively [9]. The DNA sequences of attB were characterized from various Streptomyces species, and the analysis of these sequences revealed the presence of high conserved positions. In this genus of bacteria, the presence of pseudo-attB sites (50%–70.3% nt identity to attB sites) has also been reported, but with an integration frequency 300 times lower than that with the corresponding attB site [10]. However, the absence of this attB site from some actinomycete strains, such as Saccharopolyspora erythraea, has also been reported [11].
We have previously reported the isolation from Tunisian soil of two new actinomycete strains called Streptomyces sp. US 24 [12] and Streptomyces sp. TN 58 [13], producing diverse interesting biological activities, and described the purification and structure elucidation of two and five active molecules from the Streptomyces sp. US 24 [14] and the Streptomyces sp. TN 58 [15], respectively. The active molecules produced by Streptomyces sp. US 24 are the 3-indolethanol and a diketopiperazine (DKP) derivative, the Cyclo (L-Phe, L-Pro) diketopiperazine. DKP active molecules form a very important family because of their many potential uses as antibacterial, fungicidal, herbicidal, antiviral, immune-suppressor, and antitumor agents, and therefore they are very attractive for the production of novel hybrid active compounds by combinatorial biosynthesis.
Among the five active molecules characterised from the Streptomyces sp. TN 58, two belong to the rhamnopyranoside family: the 1-O-(2-Aminobenzoyl)-α-L-rhamnoside and the 4-Hydroxybenzoyl α-L-rhamnopyranoside. These two active compounds possess inhibitory activity towards 3α-hydroxysteroid dehydrogenase (3α-HSD), a useful target for anti-inflammatory and antiphlogistic drugs. It should be noted that until now, the Streptomyces sp. TN 58 is the only described strain which produces these two molecules directly without any supplement addition into the culture media [15]. Identification and expression of genes involved in biosynthetic pathways of these molecules constitute a real opportunity for the comprehension of their natural biosynthesis and subsequently the production of hybrid molecules. The aim of this work was thus to succeed in carrying out intergeneric conjugation between E. coli ET12567 and Streptomyces sp. US 24 and TN 58 strains, to analyse the integration sites of the integrative vector pSET152 into the chromosome of these two Streptomyces strains, to study whether the site-specific integration of pSET152 affects essential functions such as the production of active molecules in these two studied strains, and to verify the expression of a heterologous gene in Streptomyces sp. TN 58 strain using this system.
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
E.coli strain ET12567 (pUZ8002) [2, 5], used as the donor in intergeneric conjugation, is a methylation-defective strain (dam-13:: Tn9 dcm-6 hsdM Cmr). E. coli strain DH5α [16] (F− ϕ80 dlacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1hsdR17 (r−k, m+k) deoR thi-1 susE44 λ− gyrA96 relA1) and E. coli HB101 (F− hsdS2 recA13 ara−14 pro A2 lacY1 galk2 rps L20 xyl− 5 mtl sup E44) were used as host strain, and E. coli ATCC 8739 and Micrococcus luteus LB 14110 were used as indicator micro-organisms for the antibacterial activity assays.
The two Streptomyces strains US 24 [12] and TN 58 [13] were used as recipient for intergeneric conjugation. Streptomyces SK strain is a thermostable glucose isomerase producer [17]. CBS4 strain was the glucose isomerase deficient Streptomyces violaceoniger in which the xylA gene of the Streptomyces SK strain, under the control of the ermE-up constitutive promoter, was integrated in its chromosome via the integrative vector pTS55 [18].
Plasmid pUZ8002 is a derivative of RK2 with a mutation in oriT [19]. pSET152 is an integrative plasmid carrying φC31 int and att functions and an apramycin resistance gene for selection in Streptomyces and E. coli [2]. pGEM-T Easy vector is a TA cloning vector AmpR (Promega) used for cloning of the PCR products.
2.2. Media and Culture Conditions
E. coli strains were grown in Luria broth (LB) medium. Ampicillin (50 μg mL−1), chloramphenicol (25 μg mL−1), nalidixic acid, and kanamycin (50 μg mL−1) were added to growth media when required and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (40 μg mL−1) when appropriate [20]. Transformation of E. coli DH5α or HB101 with pGEM-T pSET152 derivatives was carried out according to the manufacturer's instructions (Promega).
Streptomyces sp. US 24 and TN 58 strains were grown in Tryptone soya broth (TSB: 30 g tryptic soy broth plus 5 g yeast extract per 1000 mL distilled water) medium for the preparation of genomic DNA [21] and in R2YE [22] plates for the preparation of spore stocks. TSB supplemented with 30 μg of apramycin mL−1 was used to grow exconjugants for the preparation of genomic DNA and to maintain them.
AS1 medium [23] (1 g Yeast extract, 5 g Soluble starch, 0.2 g L-alanine, 0.2 g L-arginine, 0.5 g L-asparagine, 2.5 g NaCl, 10 g Na2SO4 and 20 g agar, pH adjusted at 7.5 with KOH) was used for conjugation experiments.
For bioactive molecules production, Streptomyces sp. US 24 strain and the corresponding exconjugants were grown in TSB medium, supplemented at 1% (w/v) with starch and with a trace element solution: (7.5 mL L−1 final; 0.4 g L−1 ZnCl2; 2 g L−1 FeSO4·7H2O; 0.065 g L−1 H3BO3 and 0.135 g L−1 MoNa2O4·2H2O). Streptomyces TN 58 strain and corresponding exconjugants were grown in TSB medium, supplemented with glycerol at 1% (w/v) and with potassium at 1 mmol L−1. For antibacterial activity determination, indicator microorganisms were grown overnight in LB medium at 30°C for M. luteus LB14110 and at 37°C for E. coli ATCC 8739, then diluted 1 : 100 in LB medium and incubated for 5 hours under constant agitation at 200 rpm at the appropriate temperature.
For growth study of the two bacteria: Streptomyces TN 58/xylA and CBS4, spores at 107 mL−1 of each strain were used to inoculate 1000 mL Erlenmeyer flasks with four indents, containing 200 mL of minimum liquid medium ([NH4]2SO4 2.5 g L−1 , MgSO4 0.3 g L−1, CaCl2 0.2 g L−1, KH2PO4 1 g L−1, yeast extract 1 g L−1 and 1 mL of trace element solution) in presence of xylose as solely carbon source at 10 g L−1. Biomass of the two studied Streptomyces strains was determined at different growth times by measurement by weighing to constant dry weight after drying at 105°C.
2.3. Intergeneric Conjugation
Intergeneric conjugation between E. coli and Streptomyces US 24 and TN 58 strains was performed as described previously by Flett et al., [24] with minor modifications. E. coli ET12567 (pUZ8002/pSET152) was grown to an absorbance of 0.4–0.6 at 600 nm. The cells were pelleted by centrifugation, washed twice in an equal volume of LB, pelleted again, and finally resuspended in 1/10 volume of LB. Aliquots of the two studied Streptomyces strain spores suspension stored at −20°C were used as recipients. Spores (eq108) were washed in 2 × yeast extract tryptone medium [25], resuspended in 500 μL of 2 × yeast extract tryptone medium, and incubated at 50°C for 10 minutes to induce germination. Donor cells (500 μL approximately 108 cells) were added to the treated spores, the mixture was pelleted by centrifugation, and finally the pellets were resuspended in the residual liquid, and the undiluted mixtures were plated. The mating mixtures were spread on AS1 plates containing 10 mM MgCl2 and incubated for 18 hours at 37°C for the Streptomyces US 24 and at 30°C for the Streptomyces TN 58 strain. The plates were overlaid each with 1 mL of water containing 500 μg of nalidixic acid and 1 mg of apramycin, incubated further for 5 days at the appropriate temperature, and the exconjugants were counted. Cultures prepared from some clones were used for further studies. Control experiments were performed as described above but without the addition of E. coli donor cells. The viable count of the donor culture was determined by spreading the cells on LB agar plates supplemented with appropriate antibiotics.
2.4. DNA Isolation and Manipulation
Total DNA preparation was carried out from Streptomyces strains according to [21]. Small-scale plasmid preparations from E. coli were performed as described by [20].
Digestion with restriction endonucleases, and separation of DNA fragments by agarose gel electrophoresis, alkaline calf intestinal phosphatase, ligation of DNA fragments, and transformation were done according to [20] for E. coli and [21] for Streptomyces.
For blot manipulations [26], DNA was transferred to Hybond N nylon membranes (Amersham). About 1 μg and 50 ng were loaded for the digested genomic DNA and plasmids DNA, respectively. The hybridization conditions and subsequent detection were in accordance with the manufacturer's instructions. 32P-labeled probes were prepared using the Random prime labelling system (Amersham).
PCR amplification of the attB sites of the two studied Streptomyces strains was performed using the two primers ATTB1 and ATTB2 previously described by [10]. Approximately 200 ng genomic template DNA was used with 100 pmol of each primer per 50 μL reaction volume. To improve the denaturation of the DNA, 5% (v/v) DMSO was added to the reaction mixture. Amplifications were performed in a Gene AmpR PCR System 2700 (Applied Biosystems) using 1U Pfu DNA polymerase (Stratagene) and the recommended buffer system according to the following amplification profile: 94°C (5 minutes) followed by 45 cycles of denaturation at 94°C (30 seconds), annealing at 60°C (1 minute) and extension at 72°C (1 minute). The PCR reaction mix was analysed by agarose gel electrophoresis, and the products having the expected sizes were purified then cloned into pGEM-T Easy vector.
Nucleotide sequences were determined on both strands using the dideoxy chain-termination method [27]. Reactions were performed with a thermo sequenase cycle sequencing kit (Amersham) and specific primers. Homology search was performed using Blast Search algorithm [28].
2.5. Extraction of Active Compounds and Biological Assay of Antimicrobial Activities
For the extraction of the active molecules from the Streptomyces sp. US 24 and TN 58 strains and their corresponding exconjugants (four exconjugants randomly chosen for each strain), spores at 107 mL−1 were used to inoculate 500 mL Erlenmeyer flasks with four indents containing 100 mL of culture medium. After incubation for 24 hours in an orbital incubator with shaking at 250 rpm at the appropriate temperature, the preculture was used to inoculate (5% v/v) a total volume of 1000 mL culture medium having the same composition of the preculture. After three days incubation at 37°C or 30°C for Streptomyces US 24 and TN 58 strains, respectively, and their corresponding exconjugants, in an orbital incubator with shaking at 250 rpm, the culture broths were filtered to separate mycelium and supernatant. Each supernatant was extracted twice with an equal volume of ethyl acetate and then evaporated on a Rotavapor (Laborata 4000), and obtained crude extract was dissolved in 1 mL ethyl acetate and used for antibacterial activities as follows: a paper disk was impregnated with 80 μL of the corresponding sample and then laid on the surface of an agar plates containing 3 mL of top agar inseeded by 40 μL of a 5-hour old culture of M. luteus LB 14110 or E. coli ATCC 8739. After 2 hours at 4°C, plates containing M. luteus were incubated at 30°C and those inoculated with E. coli at 37°C, all for overnight. Plates were examined for evidence of antibacterial activities represented by a zone of inhibition of growth of the corresponding indicator micro-organisms around the paper disk. Thin layer chromatography (TLC) was performed on silica gel plates SiO2 (Merck). Visualization of active compounds was realised with anisaldehyde vapour and Ehrlich's reagent.
2.6. Expression of the xylA Gene in Streptomyces sp. TN 58
The BglII fragment carrying the insert xylA gene of Streptomyces sp. SK placed under the control of the ermE-up constitutive promoter [29] from pMM6 [18] was cloned into the Streptomyces integrative vector pSET152 linearised by BamHI, leading to the pSS6 vector construct. This plasmid was firstly obtained within E. coli HB101 strain and then transferred to E. coli ET12567/pUZ8002 for the construction of the Streptomyces TN 58/xylA strains by conjugal transfer. Exconjugants were selected on the basis of apramycin resistance.
Preparation of the cell-free lysate of the Streptomyces SK, TN 58, TN 58/pSET152, TN 58/pSS6, and CBS4 (used as positive control) strains was achieved as follows. The cells were grown in TSB medium for 48 hours and were harvested by centrifugation at 8000 rpm for 10 minutes, and the pellets were suspended in TE-buffer with 10 mM MgCl2 and 1 mM CoCl2. After incubation for an hour on ice in the presence of 5 mgmL−1 lysozyme, 100 μgmL−1 PMSF, and 1 μgmL−1 Pepstatin A, cells were disrupted by sonication at 4°C for 6 minutes (pulsations of 3 seconds, amplify 90) using a vibra cell Sonicator (Fisher-Bioblock Scientific), and debris were removed by centrifugation at 20 000 rpm for 30 minutes.
Glucose isomerase activity was determined using fructose as substrate by quantitatively measuring the glucose production. The activity was assayed in a reaction mixture containing the enzyme (100 μL of an appropriate diluted purified or crude extract) with 10 mM MgCl2, 1 mM CoCl2 and 15% fructose, in a volume of 400 μL. In standard condition, assays were incubated for 30 minutes at 80°C, and the reaction was stopped by cooling the tubes on ice. The amount of glucose generated was determined by glucose-oxidase (GOD-PAP) enzyme system, and A650 was measured after 40 minutes at room temperature. One unit of glucose isomerase activity is defined as the amount of enzyme needed to produce 1 μmol of product per minute under the assay conditions. Protein concentration was determined by Bradford's method [30] using the Bovine Serum Albumin as standard.
3. Results and Discussion
3.1. Conjugal Transfer of pSET152 and φC31-Directed, Site-Specific Recombination in Streptomyces sp. US 24 and Streptomyces sp. TN 58 Strains
The plasmid pSET152 was mobilized from E. coli ET12567 (pUZ8002) into Streptomyces sp. US 24 and TN 58 strains. Exconjugants were obtained at a frequency of approximately 5 × 10−5 and 3 × 10−4 per recipient spore of Streptomyces sp. US 24 and TN 58 strains, respectively, which showed a high frequency of exconjugants. Indeed, Nikodinovic et al., [31] consider that the transformation efficiency (5 × 10−5 exconjugants recipient−1) of the Streptomyces nodosus by conjugal transfer of DNA from E. coli was a high transformation frequency.
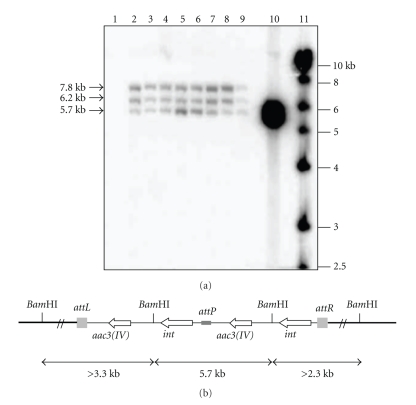
The integrative plasmid pSET152 cannot replicate in Streptomyces; so stable exconjugants should be obtained only if pSET152 has been integrated into the chromosome of studied strain. In order to identify the copy number of pSET152 in the recipient Streptomyces sp. US24 and Streptomyces sp. TN 58, eight arbitrary chosen exconjugants were studied for each strain. Chromosomal DNAs were extracted, digested by BamHI and probed with the whole linearised vector pSET152 in a blot analysis. Since pSET152 had a single BamHI site, DNA from exconjugants was expected to show hybridisation of two fragments to probe in the case of single copy integration.
For Streptomyces sp. US24 exconjugants, obtained hybridisation patterns were identical and showed the presence of three bands of about 5.7, 6.2, and 7.8 Kb (Figure 1(a)). The same probe did not hybridise to any DNA fragment of the wild type genomic DNA. This result implied that the plasmid was presumably integrated at the same locus in all corresponding exconjugants. However, among the three obtained bands, the one of 5.7 kb corresponds exactly to the size of the linearized vector pSET152. This fact can be explained via the tandem integration of two copies of the pSET152 vector in the chromosome of Streptomyces sp. US24 strain (Figure 1(b)). The presence of tandemly repeated copies of pSET152 vector in the attB site can be explained by the property of pSET152 to integrate at attR or attL sites via attP/attR or attP/attL recombination. Normally, the integration of pSET152 vector will be via attB/attP recombination. So, in certain cases, we can assist to the multiple independent recombination events, and pSET152 can integrate at attL or attR sites via attP/attL or attP/attR. As was hypothesised by Combes et al. [10], if this event occurs, this reaction must be specific to the attL and attR sites as tandem repeats of pSET152 in the pseudo-attB sites were not generally observed. Alternatively, the generation of tandem repeats of pSET152 in the attB may be a consequence of the rate of integration. If the rate of recombination into the attB site is rapid, integration occurs early during the mating period, and when the hyphae containing integrated pSET152 receives a further copy of pSET152, this last one may, generally, integrate by homologous recombination to generate a tandem repeat.
Figure 1.
(a) Southern blot of Streptomyces sp. US24 BamHI-digested total DNA hybridised with 32P-labeled pSET152 plasmid. Lane 1, untransformed wild type Streptomyces sp. US24; lanes 2–9, the eight studied exconjugants; lane 10, BamHI-digested pSET152 plasmid DNA (5.7 Kb); lane 11, the 1 kb ladder used as DNA marker. (b) Schematic representation of tandemly insertion of the pSET152 into the Streptomyces sp. US24 attB site. Chromosomal DNA hybridising is represented by thin lines and pSET152 by thick line. The integrase gene (int), and the apramycin resistance gene [aac3(IV)] from pSET152 attP, attL, and attR sites are also shown.
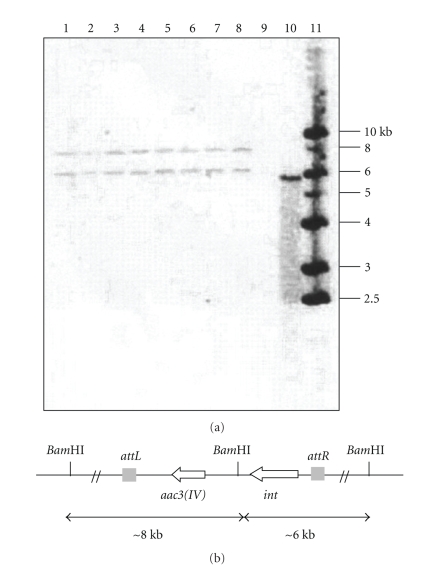
Concerning Streptomyces TN 58, two identical hybridisation signals were obtained for all eight analysed exconjugants corresponding to two DNA fragments of about 6.8 and 8 Kb (Figure 2(a)). The same probe did not hybridise to any DNA fragment of untransformed Streptomyces. sp TN 58 genomic DNA. This result implied that a single copy of the pSET152 plasmid was integrated at the same locus in all the exconjugants in strain Streptomyces sp. TN 58. We can suggest that the chromosome of this strain contains a single functional attB site for vectors that integrate site specifically using the bacteriophage φC31 att/int system (Figure 2(b)). All together, our data strongly suggest the presence of an efficiently recognised attB site in the chromosome of the two studied Streptomyces strains.
Figure 2.
(a) Southern blot of Streptomyces sp. TN 58 BamHI-digested total DNA hybridised with 32P-labeled pSET152 plasmid. Lanes 1–8, the eight studied exconjugants; lane 9, untransformed wild type Streptomyces sp. TN 58; lane 10, BamHI-digested pSET152 plasmid DNA (5.7 Kb); lane 11, the 1 kb ladder used as DNA marker. (b) Schematic representation of the integration of plasmid pSET152 in the chromosome of Streptomyces sp. TN 58 by site-specific recombination.
3.2. Identification of the ΦC31 attB Sites
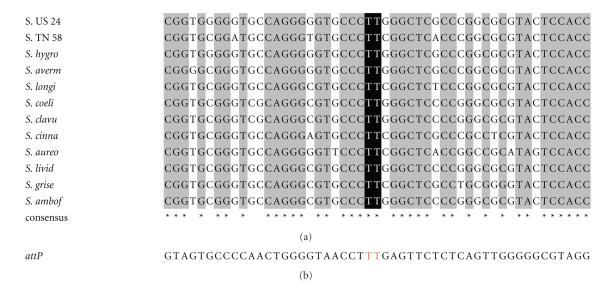
Integration of pSET152 into the Streptomyces sp. US 24 and TN 58 strains chromosomes implies the existence of one attB site recognised by the ΦC31 integrase. Taking advantage of the two PCR primers (ATTB1 and ATTB2) described by Combes et al., [10], we recovered a unique PCR product at the expected size of approximately 0.3 Kb from the chromosomal DNAs of the two studied strains. These fragments were cloned into the pGEM-T Easy vector yielding pSS4 and pSS5 plasmids for US 24 and TN 58 strains, respectively. The insert of three randomly chosen clones for pSS4 and pSS5 plasmids was sequenced. All three nucleotide sequences were identical for each plasmid insert. The two inserts possess a size of 292 bp and 293 bp for US 24 and TN 58 strains, respectively, and a high nucleotide identity of 86% (Figure 3). These two amplified DNA fragments encode a homolog of the Streptomyces coelicolor A3(2) Sco3798 ORF with a nucleotide identity of 92% and 86% for Streptomyces sp. US 24 and Streptomyces TN 58 strains, respectively. This ORF is a putative chromosome condensation protein, with sequence similarity to a mammalian-encoded protein, pirin (an identified nuclear protein that interacts with Bcl-3 and nuclear factor I), in which the attB site of ΦC31 lies in various Streptomyces species [32].
Figure 3.
Alignment of the attB region of Streptomyces sp. US 24 and TN 58 strains. Solid boxes, identical sequences, underlined sequence, attB site.
For Streptomyces species, all characterised ΦC31 attB sites showed high nucleotide sequence similarity varying from 82.4% to 100%. However, the presence of pseudo-attB sites (50%–70.3% nt identity to attB sites) into which integration occurred at a frequency 300 times lower than into the corresponding attB site [10] has also been reported in this genus of bacteria. The multiple-sequence alignment of attB sites of Streptomyces sp. US 24 and TN 58 with several other Streptomyces species is shown in Figure 4. This alignment indicates that these sequences are conserved, and some positions in the minimal site can tolerate nucleotide changes. Kuhstoss and Rao [33] reported, by comparing the S. ambofaciens attB site with attP, that the core sequence (i.e., the region at which the crossover occurs) is 5′TTG. Combes et al., [10] had shortened this sequence to two nucleotides 5′TT which was present in the attB site of both studied strains Streptomyces sp. US 24 and TN 58.
Figure 4.
Natural variation of ΦC31 attB sites in various species of Streptomyces. (a) Alignment of attB nucleotide sequence of Streptomyces sp. US 24 strain (S. US 24) and Streptomyces sp. TN 58 (S. TN 58) with attB sequences of S. hygroscopicus NRRL5491 (S. hygro); S. avermitilis MA-4680 (S. aver); S. longisporoflavus 83E6 (S. longi); S. coelicolor (S. coel); S. clavugerus (S. clav); S. cinnamonensis (S. cinna); S. aureofaciens (S. aureo); S. lividans 66 TK64 (S. livid); S. griseus ATCC 12475 (S. grise); S. ambofaciens (S. ambof). Dark boxes, identical sequences; black shading sequence at which crossover occurs (TT) for the natural attB and attP sites. (b); Sequence of the attP site in φC31.
3.3. Site-Specific Integration Does Not Affect the Essential Functions and Active Molecule Production of Streptomyces sp. US 24 and Streptomyces sp. TN 58 Strains
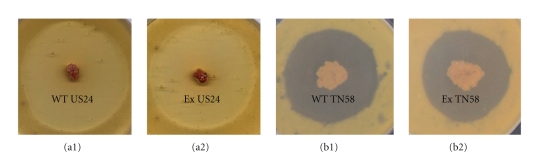
Despite their wide use and clear advantages, it has been reported that integration of the integrative vectors into the φC31 attB site can cause detrimental effects on antibiotic production in some strains [34]. In fact, it should be considered that the decrease in antibiotics production for many Streptomyces strains carrying integrated site specific plasmids such as S. fradiae producer of tylosin, S. kanamyceticus producer of kanamycin and others has been reported [35]. Therefore, while integrative vectors could be used for the development of stable recombinant producers of antibiotics, their possible negative effect on the level of antibiotic production should be considered. Thus, in order to determine whether the site-specific integration of the plasmid pSET152 affected relevant functions of Streptomyces sp. US 24 and Streptomyces sp. TN 58 strains, we have followed the growth of both strain and their corresponding exconjugants carrying plasmid pSET152. All exconjugants grew well in the minimal medium, and there was no difference compared to the wild type strains. The presence of pSET152 had no effects on growth and sporulation of the two strains. We have studied the stability of the pSET152 inheritance in exconjugants. This stability was determined as the proportion of colonies that retained resistance to apramycin (at 50 μg mL−1) after several passages of the exconjugants under nonselective conditions. Plasmid pSET152 was inherited every time for the all studied exconjugants under nonselective conditions for both strains. Meanwhile, antimicrobial activity was determined by observing bacterial growth inhibition of the wild type strains (US 24 and TN 58) and corresponding exconjugants. There was no obvious difference between wild type Streptomyces sp. US 24 and Streptomyces sp. TN 58 and their corresponding exconjugants (Figure 5 and Table 1). In addition, the analysis on TLC plates of the ten studied active crude extracts revealed the presence of the expected active molecules in each strain and its corresponding exconjugants. For the Streptomyces sp. US 24 strain and its four studied exconjugants, two bands were easily detected on TLC plates under UV light at 254 nm after spraying with anisaldehyde/sulphuric acid. The first band gives an orange colouration with a retention factor, Rf = 0.46 (CHCl3/MeOH 10%) and the second one a violet colouration, Rf = 0.37 (CH2Cl2/9% MeOH). According to our previously work [14], these two bands correspond to the 3-indolethanol and the Cyclo (L-Phe, L-Pro) diketopiperazine active molecules, respectively. Concerning the Streptomyces sp. TN 58 strain and its studied exconjugants, the five active molecules (M1 to M5) were present in all five crude extracts. M1 belongs to the acetyltryptamine family and gives a colouration orange and violet after anisaldehyde/sulphuric acid and Erhlich's reagent visualisation. The Rf of this molecule is 0.5 (CHCl3/5% MeOH). M2, Rf = 0.57 (CH2Cl2/1% MeOH), and M3, Rf = 0.47, (CHCl3/5% MeOH) belong to the thiazole and brevinamide families, respectively. M2 gives a chestnut colouration on spraying with anisaldehyde/sulphuric acid and M3 gives a pinkish colouration after Erhlich's reagent visualisation. The two other active molecules (M4 and M5) characterised from the Streptomyces sp. TN 58 strain, and belonging to the rhamnopyranosides family, do not have antimicrobial activities, but they possess inhibitory activity towards 3α-hydroxysteroid dehydrogenase (3α-HSD). Indeed, the rhamnosylated aromatic compounds were easily detected on TLC plates due to their striking yellow/greenish colouration after visualisation with anisaldehyde vapour. For all five analysed crude extracts, the wild type one (from the Streptomyces sp. TN 58 strain) and those from the four corresponding exconjugants, we obtained similar TLC migration profiles and two yellow/greenish bands having a retention factor (CHCl3/15% MeOH) of 0.22 and 0.33 corresponding to the two molecules belonging to the rhamnopyranoside family. For the five active molecules of the Streptomyces sp. TN 58 and its four studied exconjugants, TLC profiles, bands colouration, and retention factors of the different active compounds were in perfect concordance with the results of previously works [15]. These data clearly demonstrated that the morphological differentiation and active molecule production of all studied pSET152 integrated exconjugants were similar to those of wild type Streptomyces sp. US 24 and Streptomyces sp. TN 58 strains. Consequently, conjugal transfer using attP/B site integration can be concluded to be a suitable means of gene transfer and expression for both studied strains.
Figure 5.
Antibacterial activities in solid media against M. luteus of the Streptomyces sp. US 24 strain wild type “WT US24” (a1) and one corresponding exconjugant US24/pSET152 “Ex US24” (a2) and Streptomyces TN 58 strain wild type “WT TN 58” (b1) and one corresponding exconjugant TN 58/pSET152 “Ex TN 58” (b2).
Table 1.
Antibacterial activities in liquid media against M. luteus and E. coli of the two studied Streptomyces strains and their corresponding exconjugants. WT (wild type). Ex (Exconjugant).
| Diameter (mm) of inhibition zones | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Streptomyces sp. US24 | Streptomyces TN 58 | |||||||||
| Indicator microorganisms | WT | Ex.241 | Ex.242 | Ex.243 | Ex.244 | WT | Ex.581 | Ex.582 | Ex.583 | Ex.584 |
| M. luteus | 20 | 19 | 20 | 21 | 20 | 17 | 17 | 16 | 17 | 17 |
| E. coli | 21 | 21 | 20 | 21 | 21 | 15 | 15 | 15 | 16 | 16 |
3.4. Heterologous Expression of xylA Gene in Streptomyces sp. TN 58 Strain
The establishment of a gene transfer system and the stable integration of cloned genes at the specific loci in the genome of Streptomyces sp. TN 58 prompted us to express some heterologous gene in this strain. Xylose isomerase (D-xylose ketol isomerase EC 5.3.1.5) catalyses the reversible isomerisation of D-xylose into D-xylulose. It is also referred to as glucose isomerase (GI) because of its ability to convert D-glucose to d-fructose. This property is widely exploited industrially for the production of high-fructose syrup from starch [36]. Thus, the enhancement of strain productivity is certainly of importance. So we expressed the xylA gene cloned from Streptomyces. sp SK [17], and we attempted to examine whether this gene could express and fold effectively in strain TN 58. It should be noted that the wild type Streptomyces TN 58 strain cannot assimilate the xylose as solely carbon source. Heterologous xylA gene was inserted into pSET152 to yield pSS6 plasmid. Based on intergeneric conjugal transfer, transformation efficiency of strain TN 58 by plasmid pSS6 was achieved at high frequency (10−4 exconjugants recipient−1). The chromosomal structure of three TN 58/xylA strains, arbitrary chosen, was checked by Southern Blot hybridisation using the 800 bp PstI DNA fragment internal to pSET152 as probe, which contain the oriT region. As expected, a unique and identical large BamHI band was obtained for the three TN 58/xylA strains chromosomal DNA, whereas wild type strain TN 58 chromosomal DNA does not hybridise to the probe (data not shown). One of the three recombinant strains studied was subcloned five times on a solid media in the absence of apramycin followed by cultivation for several generations in liquid media in the absence of selection pressure. The plating of this culture on solid media, on both with and without selective pressures, showed that 100% of the colonies were apramycin resistant and able to grow in the presence of xylose as solely carbon source.
Determination of the mycelial intracellular glucose isomerase activity of Streptomyces TN 58, Streptomyces TN 58/pSET152, Streptomyces sp. SK, CBS4, and TN 58/xylA strains shows that glucose isomerase activity of the TN 58/xylA recombinant strain is of about eighteenfold higher than that of the SK strain and comparable to that of CBS4 strain. No activity was detected for the wild type TN 58 strain and the recombinant strain TN 58/pSET152 (Table 2). Study of the growth of the two strains, Streptomyces TN 58/xylA and CBS4, in minimum liquid media containing xylose as solely carbon source showed that the two strains grew well, and for each time, the resulting biomass was quite similar. It should be noted that glucose isomerase activity is correlated with biomass production, and optimum activity has been obtained after 48 hours of incubation.
Table 2.
Glucose isomerase activity of Streptomyces TN 58 wild type (S. TN 58), Streptomyces TN 58/pSET152 (S. TN 58/pSET152), Streptomyces sp. SK (S. SK), CBS4, and Streptomyces TN 58/xylA (S. TN 58/xylA) strains from 48-hour old cultures.
| S. TN 58 | S. TN 58/pSET152 | S. SK | CBS4 | S. TN 58/xylA | |
|---|---|---|---|---|---|
| Specific activity U mg−1 | 0 | 0 | 0.1 | 1.68 | 1.81 |
According to these results, we can deduce that site-specific recombination directed by pSET152 makes it possible to stably insert heterologous DNA in strain TN 58. This should be very important for a number of cases such as genes expression, genetic complementation, and biosynthetic pathway manipulations of the active molecules of this strain especially those belonging to the rhamnopyranoside family.
Acknowledgments
This work was supported by the Tunisian government (Contract Program CBS-LEMP) and the CMCU project (2006–2008) no. 06/S 0901 “MELLOULI/AIGLE”.
References
- 1.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. Journal of Bacteriology. 1989;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 3.Matsushima P, Baltz RH. A gene cloning system for ‘Streptomyces toyocaensis’. Microbiology. 1996;142(2):261–267. doi: 10.1099/13500872-142-2-261. [DOI] [PubMed] [Google Scholar]
- 4.Fouces R, Rodríguez M, Mellado E, Díez B, Barredo JL. Conjugation and transformation of Streptomyces species by tylosin resistance. FEMS Microbiology Letters. 2000;186(2):319–325. doi: 10.1111/j.1574-6968.2000.tb09124.x. [DOI] [PubMed] [Google Scholar]
- 5.MacNeil DJ, Occi JL, Gewain KM, et al. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115(1-2):119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 6.Stegmann E, Pelzer S, Wilken K, Wohlleben W. Development of three different gene cloning systems for genetic investigation of the new species Amycolatopsis japonicum MG417-CF17, the ethylenediaminedisuccinic acid producer. Journal of Biotechnology. 2001;92(2):195–204. doi: 10.1016/s0168-1656(01)00360-1. [DOI] [PubMed] [Google Scholar]
- 7.Voeykova T, Emelyanova L, Tabakov V, Mkrtumyan N. Transfer of plasmid pTO1 from Escherichia coli to various representatives of the order Actinomycetales by intergeneric conjugation. FEMS Microbiology Letters. 1998;162(1):47–52. doi: 10.1111/j.1574-6968.1998.tb12977.x. [DOI] [PubMed] [Google Scholar]
- 8.Fouces R, Rodríguez M, Mellado E, Díez B, Barredo JL. Conjugation and transformation of Streptomyces species by tylosin resistance. FEMS Microbiology Letters. 2000;186(2):319–325. doi: 10.1111/j.1574-6968.2000.tb09124.x. [DOI] [PubMed] [Google Scholar]
- 9.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Combes P, Till R, Bee S, Smith MCM. The Streptomyces genome contains multiple pseudo-attB sites for the ϕC31-encoded site-specific recombination system. Journal of Bacteriology. 2002;184(20):5746–5752. doi: 10.1128/JB.184.20.5746-5752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory MA, Till R, Smith MCM. Integration site for Streptomyces phage ϕBT1 and development of site-specific integrating vectors. Journal of Bacteriology. 2003;185(17):5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellouli L, Ben Ameur-Mehdi R, Sioud S, Salem M, Bejar S. Isolation, purification and partial characterization of antibacterial activities produced by a newly isolated Streptomyces sp. US24 strain. Research in Microbiology. 2003;154(5):345–352. doi: 10.1016/S0923-2508(03)00077-9. [DOI] [PubMed] [Google Scholar]
- 13.Mellouli L, Karray-Rebai I, Sioud S, Ben Ameur-Mehdi R, Naili B, Bejar S. Efficient transformation procedure of a newly isolated Streptomyces sp. TN58 strain producing antibacterial activities. Current Microbiology. 2004;49(6):400–406. doi: 10.1007/s00284-004-4292-9. [DOI] [PubMed] [Google Scholar]
- 14.Ben Ameur-Mehdi R, Mellouli L, Chabchoub F, Fotso S, Bejar S. Purification and structure elucidation of two biologically active molecules from a new isolated Streptomyces sp. US 24 strain. Chemistry of Natural Compounds. 2004;40(5):510–513. [Google Scholar]
- 15.Ben Ameur-Mehdi R, Shaaban KA, Karray-Rebai I, Smaoui S, Bejar S, Mellouli L. Five naturally bioactive molecules including two rhamnopyranoside derivatives isolated from the Streptomyces sp. strain TN58. doi: 10.1080/14786410802362352. Natural Product Research. In press. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Srih-Belghith K, Bejar S. A thermostable glucose isomerase having a relatively low optimum pH: study of activity and molecular cloning of the corresponding gene. Biotechnology Letters. 1998;20(6):553–556. [Google Scholar]
- 18.Mezghani M, Borgi MA, Kammoun R, Aouissaoui H, Bejar S. Construction of new stable strain over-expressing the glucose isomerase of the Streptomyces sp. SK strain. Enzyme and Microbial Technology. 2005;37(7):735–738. [Google Scholar]
- 19.Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor σEis required for normal cell wall structure in Streptomyces coelicolor A3(2) Journal of Bacteriology. 1999;181(1):204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Hopwood DA, Bibb MJ, Chater KF, et al. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, UK: The John Innes Foundation; 1985. [Google Scholar]
- 22.Thompson CJ, Ward JM, Hopwood DA. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature. 1980;286(5772):525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- 23.Baltz RH. Genetic recombination by protoplast fusion in Streptomyces. Developments in Industrial Microbiology. 1980;21:43–54. [Google Scholar]
- 24.Flett F, Mersinias V, Smith CP. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiology Letters. 1997;155(2):223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Bittner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics: A Laboratory Manual. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 26.Maniatis TE, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibb MJ, Janssen GR, Ward JM. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1–3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 31.Nikodinovic J, Barrow KD, Chuck J-A. High frequency transformation of the Amphotericin-producing bacterium Streptomyces nodosus. Journal of Microbiological Methods. 2003;55(1):273–277. doi: 10.1016/s0167-7012(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 32.Choi S-U, Lee C-K, Hwang Y-I, Kinoshita H, Nihira T. Intergeneric conjugal transfer of plasmid DNA from Escherichia coli to Kitasatospora setae, a bafilomycin B1 producer. Archives of Microbiology. 2004;181(4):294–298. doi: 10.1007/s00203-004-0654-8. [DOI] [PubMed] [Google Scholar]
- 33.Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage φC31. Journal of Molecular Biology. 1991;222(4):897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 34.Baltz RH. Genetic manipulation of antibiotic-producing Streptomyces. Trends in Microbiology. 1998;6(2):76–83. doi: 10.1016/S0966-842X(97)01161-X. [DOI] [PubMed] [Google Scholar]
- 35.Luzhetskii AN, Ostash BE, Fedorenko VA. Intergeneric conjugation Escherichia coli—Streptomyces globisporus 1912 using integrative plasmid pSET152 and its derivatives. Russian Journal of Genetics. 2001;37(10):1123–1129. [PubMed] [Google Scholar]
- 36.Bhosale SH, Rao MB, Deshpande VV. Molecular and industrial aspects of glucose isomerase. Microbiological Reviews. 1996;60(2):280–300. doi: 10.1128/mr.60.2.280-300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]