Abstract
A method of identifying the best structural model for a protein of unknown structure from a list of structural candidates using unassigned 15N-1H residual dipolar coupling (RDC) data and probability density profile analysis (PDPA) is described. Ten candidate structures have been obtained for the structural genomics target protein PF2048.1 using ROBETTA. 15N-1H residual dipolar couplings have been measured from NMR spectra of the protein in two alignment media and these data have been analyzed using PDPA to rank the models in terms of their ability to represent the actual structure.
A number of advantages in using this method to characterize a protein structure become apparent. RDCs can easily and rapidly be acquired, and without the need for assignment, the cost and duration of data acquisition is greatly reduced. The approach is quite robust with respect to imprecise and missing data. In the case of PF2048.1, a 79 residue protein, only 58 and 55 of the total RDC data were observed. The method can accelerate structure determination at higher resolution using traditional NMR spectroscopy by providing a starting point for the addition of NOEs and other NMR structural data.
Keywords: unassigned NMR data, residual dipolar coupling, PDPA, protein structure modeling, powder pattern analysis, structural genomics
1. Introduction
One of the objectives of the protein structure initiative has been the production of a sufficient number of experimental structures to allow computational modeling of the proteins coded by the thousands of new gene sequences deposited in sequence data bases each month. While there have been tremendous advances in computational modeling tools in terms of reliability and ease of use [1–3], confidence in modeled structures still lies well short of confidence in experimental structures. In fact, during computational protein folding, it has become the practice to present a number of ranked models for a new protein to assure that a model matching experimental data will fall within the top 5 to 10 models [4]. Methods that rely on a minimum set of experimental data to confirm or reject computationally hypothesized structures, could boost confidence and potentially reduce the cost (time and money) of protein structure determination. Recent studies have, in fact, shown significant improvements in the quality of computationally modeled protein structures when a small amount of experimental data is incorporated [5,6]. Among the more useful sources of data has been NMR data, such as residual dipolar couplings (RDCs) and long range paramagnetic constraints [7]. However, use of these data usually requires assignment of resonances, one of the most time consuming steps in the study of macromolecules by NMR spectroscopy. A method for using NMR data (RDCs in particular) for the selection among computational models without the necessity of assigning resonances is presented here. The method employs a statistical evaluation of distributions of RDCs (powder patterns) referred to as probability density profile analysis (PDPA).
Previously, PDPA was introduced as a method for the rapid classification of an unknown protein to a fold family [8] using unassigned RDC data. The approach used just a single set of 1H-15N RDC data and was evaluated only by simulation, assuming all RDCs would be observed and measured with high precision. The present work puts PDPA to an experimental test in which data are subject to experimental uncertainties and subsets of data are missing due to peak overlap and dynamic broadening of certain crosspeaks. Analysis has been extended to multiple sets of 1H-15N RDC data (acquired on the same protein in different media) and data sets have been combined to partially take advantage of correlation among data sets. Rather than attempt to classify folds in this more difficult situation, we have chosen to use the analysis to select the best model from among a set of models posed by the program ROBETTA[9].
A target protein of unknown function, PF2048.1, selected initially for structure determination by Southeast Structural Genomics Collaboratory (SECSG) and subsequently adopted by the Northeast Structural Genomics Consortium (NESG – target ID PfG2) has been subjected to RDC data collection and analysis by PDPA. PF2048.1 is found in the genome of the hyperthermophilic archaeon, Pyrococcus furiosus. It encodes a 8.2 kDa acidic protein (pI = 5.0) rich in glutamate (12 of 71 residues). In the P. furiosus genome PF2048.1 is one of four closely linked genes (≤15 bp apart). Three of the genes encode proteins (PF2050, PF2049, and PF2048.1), all of which are annotated as conserved hypothetical [10]. The fourth gene is small (55 nts) and lies between PF2049 and PF2050 and encodes an RNA (snoRNA-45) [11]. This 4-gene arrangement is also found in the genomes of the closely related species, P. horikoshii, P. abyssi, and Thermococcus kodakaraensis, all of which have at least one similar snoRNA sequence overlapping with the ORF homologous to PF2048.1. As of yet, there is no indication of the function of these three putative proteins. They may be involved in processing the snoRNAs, although their role is not fully understood [11,12].
2. Theory
2.1 Orientation Dependence of RDC
Residual dipolar Couplings (RDCs) originate from a through-space dipolar interaction, which is dependent on the angle between an internuclear vector and the magnetic field. These normally average to zero in solution NMR samples, but if a molecule is dissolved in a dilute liquid crystalline medium it becomes partially aligned. As a result, the dipolar couplings are not completely averaged to zero and lead to a small contribution to splittings of NMR signals. The angular dependence of these couplings can provide valuable structural information. In partially ordered systems, residual dipolar couplings are given by Eq. [1] where Skl contains the orientation information and directional cosines relate various vectors to an arbitrarily chosen molecular frame. Dmax is defined in Eq. [2] where γij are the gyro magnetic ratios of nuclei i and j and rij is the internuclear distance between the two nuclei.
| (1) |
| (2) |
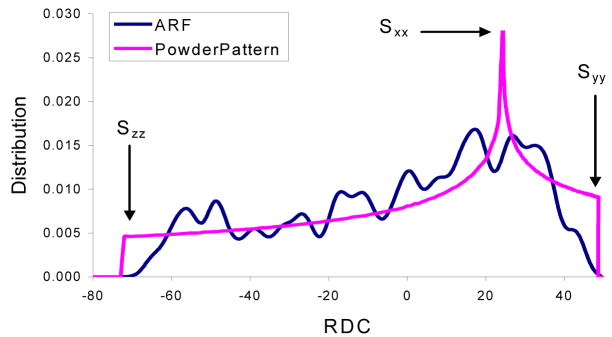
It can be shown that the distribution of dipolar couplings for a large number of uniformly distributed vectors within a sphere will converge to the relatively featureless powder pattern shown in (Figure 1). The theoretical basis of this behavior is well documented and an analytical form for this phenomenon can be derived [13–18]. While no particularly useful structural information can be obtained from this powder pattern, the three principal order parameters can be obtained by examining the extreme points of this distribution. Within the context of our work these three parameters in the principal alignment frame are designated as S′zz, S′yy, and S′xx based on the following relationship: |S′zz| ≥ |S′yy| ≥ |S′xx|.
Figure 1.
A calculated powder pattern in pink is for a large number of uniformly distributed NH and the PDP for ARF (PDB code 1HUR) using principal order parameters of −71.1, 47.4 and 23.7 in units of Hz, while the pattern in blue is the powder pattern based on the calculated order parameters using the same RDC value.
2.2 PDP Analysis
Probability Density Profile Analysis (PDPA) is founded on the simple observation that proteins appropriate in size for NMR spectroscopy neither contain a large number of vectors (of a specific type such as backbone Cα-Hα or N-H) nor sample the entire space uniformly. Figure 1 illustrates a powder pattern of theoretically generated RDC data for a large number of uniformly distributed N-H vectors with an arbitrarily selected principal order parameters of 0.001, 0.002 and −0.003 (−71.1, 47.4 and 23.7 respectively in units of Hz for backbone N-H vectors). The blue line in this figure represents the distribution of the backbone N-H RDC data of a 20 kDa protein (the ADP ribosylating factor, PDB code 1HUR) using the same principal order parameters and an assumed orientation of the principal order frame. This line deviates significantly from the ideal powder pattern. We define probability-density-profile (PDP) as the distribution of an observed set of RDC data which can also be viewed as a structural finger print. PDPs are sensitive to structural variation and can possibly reflect the number and type of secondary structures given in a protein.
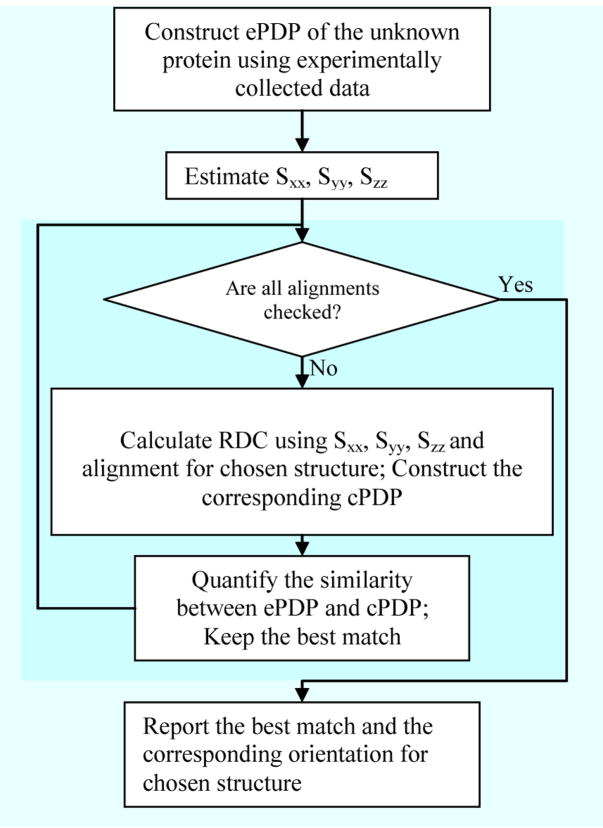
Here we first introduce the concepts of “query” and “subject” proteins in order to facilitate further discussions. A query protein is the protein for which experimental data have been acquired and structural information is sought. A subject protein is the protein for which a detailed atomistic description of structure already exists, as a candidate structure from modeling or as a representative of a fold family. The PDP of a query protein can be obtained using experimental data (denoted as ePDP). The PDP of a subject protein can be obtained using RDCs computed from the structure of the protein and a given order matrix (denoted as cPDP). A comparison of ePDP and cPDP can provide a measure of structural similarity between the query and subject proteins. The process of utilizing PDPs to obtain structural similarity between two proteins is referred to as Probability Density Profile Analysis (PDPA). The flowchart in (Figure 2) illustrates the proposed process of choosing a structure based on the similarity between the experimental and calculated PDPs. The program can be downloaded from the following website, http://ifestos.cse.edu.
Figure 2.
General flowchart operation of PDPA
A number of impediments rooted in innate properties of RDC data stand in the way of simply comparing two PDPs in order to ascertain structural homology. First, PDPs depend on preferred orientation of protein structures, that is, a given structure can produce completely different PDPs when aligned differently with respect to the external magnetic field B0. Second, it is possible that two completely different structures produce identical PDPs if elements in the two structures are related by certain symmetry operations (such as 180° rotations). The first impediment can be resolved by an exhaustive exploration of all possible orientations of the subject protein. Therefore, any structure similar to the true structure should produce at least one instance of a PDP similar to the experimental one at some orientation of the subject protein. The second impediment is simply rooted in symmetric properties of RDC data and has been previously addressed [19]. Collection of RDC data from a second independent alignment medium, which is simple to obtain, should discriminate between two structures that may appear similar from the perspective of the first alignment medium. While it is possible that a structure in a second alignment medium could share the structural degeneracies of the first alignment medium, occurrence of this phenomenon in two alignment media should be unlikely if the RDCs in the two media differ by more than a simple scaling factor.
In general, a PDP of any given structure depends on three components: its tertiary structure, its principal order parameters and the orientational alignment of the protein. Therefore, a thorough approach to ascertaining structural homology is the construction of an algorithm that conducts a search over all structures, order parameters, and possible orientations of each structure. However, the search over the entire space of principal order parameters can be confined by estimation of order parameters from the experimentally observed PDP (or the ePDP). The attainment of the principal order parameters from an unassigned list of RDC data has been previously demonstrated [13,14,20]. In this report, the minimum and maximum values of the observed RDC data have been used to estimate Sxx, Syy and Szz. The search over all protein structures is limited to a finite list of structures obtained from structure modeling tools within the context of our proposed approach. The current implementation of PDPA utilizes a grid search over all possible alignments parameterized by three Euler rotations. The resolution of the grid search can be selected based on the available computational resources and the exact objective of the search. Under the objective of validating a single structure, a grid search with a resolution of 1° can be implemented.
Selection of an appropriate metric in quantifying the similarity of two PDP maps is very critical. We have considered a large number of different metrics, such as correlation coefficient, root-mean-squared-deviation (rmsd), Manhattan, and Euclidian distance, which have been used successfully in other fields [21,22]. Based on this consideration, we have selected a modified χ2 scoring scheme for our studies. The conventional χ2 score is not appropriate, because it does not produce a symmetric report of the distance between two patterns; that is, for patterns A and B, χ2(A,B) ≠ χ2(B,A). The main goal of our modification is to eliminate this lack of symmetry while reducing the harsh penalty of missing data. Eq. [3] and Eq. [4] define the scoring mechanism used in this research. The term S(cPDP, ePDP) in Eq. [3] denotes the final comparison score between cPDP and ePDP. The summation index M denotes the number of points that are sampled in comparing the two PDPs. Entities ci and ei indicate the values of computed and experimentally determined PDPs at the location i, respectively. The distance at any given position of two PDPs is determined by χ2(c,e) as defined in Eq. [4] where T is a small threshold value.
| (3) |
| (4) |
2.3 Integration of RDC data from different alignment media
Collection of RDC data from more than one alignment medium is often times recommended [19,23–25]. This practice has been established to address some limitations of RDC data such as inherent insensitivity to 180° rotations and varying sensitivity as a function of position within the principal alignment frame (PAF) [15,26–29]. It is for these reasons that we insist on utilizing RDC data from two alignment media even though data from a single alignment medium may be adequate in some instances. Alteration of alignment can take place by selecting a second medium that aligns based on differing principles such as steric interactions versus electrostatic interactions with a protein, or simply by addition of salts or charged amphiphiles to perturb the electrostatic component of a medium having a mixed origin of interaction [19,30]. Although data collected from different alignment media can be used independently to carry out PDP analysis and classify structures, there is actually value in recognizing that the data are correlated. Positions of the cross-peaks in HSQC spectra, from which RDCs are measured, change very little on alignment in different media. Hence, one can be reasonably certain that RDCs measured from a given cross-peak in two different media pertain to the same H-N vector. The frequencies of observation for any pair of RDC measurements could then be represented on a 2D plot instead of a 1D histogram. The generation of modeled 2D plots for comparison to experiment is, however, computationally demanding since the orientation of a model must be searched independently for the two media (an N cubed problem). This would not be the case if two vectors (H-N and Cα-Hα) in the same medium were measured, but this requires a more complex protein labeling scheme and a more complex data acquisition. Protein sample which is only 15N labeled is more cost effective. What we do here is to partially recognize the correlation by noting that the pair wise sum of RDCs from two media can be used as a third data set and the three sets independently compared to 1D histograms calculated for a model (a 3N problem).
Inclusion of even unpaired data should be useful since it will in principle eliminate any accidental similarity between two structures by 180° rotation about any axes of the principal alignment frame. Moreover, it is likely that vectors that had accidentally oriented in the direction of lower sensitivity in one medium are found to be oriented in a more advantageous orientation in the second alignment medium. The correct or homolog structure should exhibit the same degree of similarity of the PDPs in any frame under any independent alignment condition, as well as the PDP for the paired sum of RDCs. The final score can simply be calculated as the weighed sum of all three PDPA scores where the appropriate weights are determined based on completeness and quality of data. An appropriate scoring mechanism (discussed in the Result section) will take into account all of these factors.
The improvised approach to take advantage of the pairing information with a minimal addition of the computation time is shown in Eq. [5] below. This represents the paired knowledge of RDC data for one vector from 3 alignment media (note that in these equations a constant multiplier is omitted for brevity). Here denotes the RDC value observed for the ith vector (no relation to the location in the sequence) from the mth alignment medium and denotes the ijth element of the order tensor describing the alignment within the mth alignment medium. The entities x, y and z corresponds to the Cartesian coordinates of the normalized interacting vector. Assuming the structure of the unknown protein remains unchanged across different alignment media, Eq [6] can be created by simply averaging equations from Eq [5] (given for a simple pair of media). In this equation, denotes the average value of RDCs observed across three different alignment media and S̄ij denotes the ijth element of the average order tensor describing the average alignment of the unknown protein. Note that the resulting average order tensor will have the necessary traceless and symmetric properties of a valid order tensor. Hence, there will also be a set of unique orientations for a correct model that can reproduce the properly paired averages of RDCs. The PDPA analysis of this approach can proceed by averaging the RDC data. This procedure has been applied to PF2048.1 and the results are shown in the subsequent section.
| (5) |
| (6) |
3. Materials and Methods
3.1 Protein expression
PCR primers were designed based on the Pyrococcus furiosus genome sequence obtained from NCBI GBank. The gene sequence was annotated from PF2048.1 as described by Poole et al [31] and is hence denoted as PF2048.1. The PCR product was cloned using standard techniques into the expression vector pET-14b (with His-tag MAHHHHHHGS- at the N-terminus) and it has been modified to include a Hind III restriction site. The amplified PCR product was cloned into a modified version of the expression vector pET24d (EMD Biosciences, Madison, WI) called pET24dBam as described [32, 33], which creates an amino terminal affinity tag (M)AHHHHHHGS-, where the N-terminal methionine residue is cleaved in the expression strain.
The vector carrying PF2048.1 was transformed into E. coli BL21 (DE3) cells and the cells were grown using M9 minimal media [34]. The media used 0.3 % w/v glucose as the carbon source and 0.1 % (w/v) ammonium-15N chloride (Isotec, Miamisberg, OH) as the nitrogen source. The sample for the present study was 13C labeled as well as 15N labeled for other reasons. However, a C1/C2-13C glucose strategy was used that resulted in just 16% 13C labeling. This allowed spectroscopic acquisitions similar to a sample labeled only with 15N. Kanamycin and chloramphenicol were added to final concentrations of 100 μg/mL and 25 μg/mL, respectively. A 100mL flask was grown overnight while shaking at 37° C. The following day 25mL of the 100mL culture was used to inoculate 1L of M9 media, which was further grown at 37° C while shaking for about 5 hours. The culture was then monitored for OD600 until the OD600 = ~0.7; it was then induced with IPTG (0.5 – 1.0 mM). The 1L flask was moved to a 22° C incubator/shaker, where it was allowed to grow overnight. The cells were harvested on the following day and ready for protein preparation or storage at −80° C.
3.2 Purification of recombinant protein
After harvesting the cells, the cells were re-suspended in 50 mL of 50mM Tris-MOPs, 500mM KCl, 0.2% Sodium Cholate pH 8.0, and then 0.1mM PMSF (protease inhibitor) was added. The re-suspended cells were then lysed by sonication. This was then centrifuged at 44,000 rpm for 30 minutes at 4° C. The supernatant was added to a Ni2+ affinity column. The column was first washed with 25mL of the lysis buffer, and then the protein was eluted with 5 mL of 50mM Tris-MOPs, 500mM KCl, 0.2% Sodium Cholate, 300mM imidazole at pH 8.0. This protein was further dialyzed overnight at 4°C into 20mM Tris, 100mM KCl, pH 8.0, and after dialysis it was concentrated down to 1mL (~2mM). Concentration of the protein sample was determined by UV spectroscopy.
3.3 NMR sample preparation, including alignment
For measurements under isotropic conditions a sample of PF2048.1 was prepared at a concentration of 1.6 mM in 20 mM Tris and 70 mM NaCl at pH 7. All samples also contained 2 mM DTT, 0.02% azide, 1 mM DSS and 10% D2O. An anisotropic sample is required for the measurement of RDCs. After isotropic data collection, the PF2048.1 sample was used to prepare two partially aligned samples to satisfy this requirement. A sample with pf1 phage as the alignment medium [35] was prepared which contained 0.88 mM PF2048.1 and 48 mg/mL phage in Tris buffer. After equilibration at room temperature for 10mins at 25 °C the sample showed a deuterium splitting of 8.8 Hz when placed in the magnet. A second aligned sample was prepared in a 5mm Shigemi tube using positively charged poly-acrlylamide compressed gels [36]. This sample contained approximately 0.77 mM PF2048.1. After equilibration at 4°C for 7–8 hrs the sample showed uniform swelling of the gel which is compressed vertically.
3.4 NMR data collection
NMR data were collected on a Varian Unity Inova 600 MHz spectrometer at 298K using a conventional z-gradient triple resonance probe or a z-gradient triple resonance cryogenic probe (Varian Inc., Palo Alto, CA). The experiments were run using the conventional probe for measurement of residual dipolar couplings: 15N IPAP-HSQC [37]. Data were acquired for the isotropic and the two aligned samples to provide a complete set of 15N-1HN, residual dipolar couplings. Data collection for the 15N IPAP-HSQC included 256 t1 points, and 2048 t2 points collected over 12 h. Residual dipolar couplings were calculated as the difference of the coupling measured in the aligned and isotropic conditions.
3.5 NMR data processing and analysis
All data were processed using NMRPipe and visualized using NMRDraw [38]. Peaks were picked using the automatic picking procedure in NMRDraw. Arbitrary assignments were automatically transferred in from the HSQC and the splittings (J or J+D) calculated using a series of Tcl scripts modified from NMRDraw. A table of RDCs was generated from the difference between splittings in aligned and isotropic datasets.
3.6 Modeling of the structure of PF2048.1
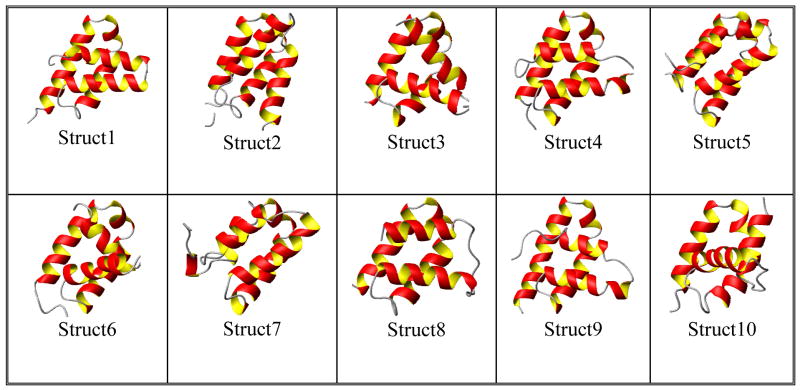
PF2048.1 is a 9.16 kDa, 79 residue, (including His-tag) monomeric protein with less than 20% sequence identity to any structurally characterized protein. To obtain starting structural models of PF2048.1, the protein threading program ROBETTA [9] was used to find structural homologs. The input to ROBETTA is just the amino acid sequence of PF2048.1. The program was run on a server available through the web (http://robetta.bakerlab.org). An ensemble of ten structures has been obtained and is shown in (Figure 3). In ROBETTA, structural models are generated by either comparative modeling or de novo structure prediction methods. In the presence of a decent match (using BLAST, PSI-BLAST etc) to a protein of known structure, the matching structure is used as a template for comparative modeling. In the absence of any match, structures are predicted using the de novo Rosetta fragment insertion method.
Figure 3.
The top 10 structures reported by ROBETTA for PF2048.1. It is seen that structure 5 has lowest PDPA scores.
3.7 PDPA of PfG2 (PF2048.1)
Three principal order parameters Sxx, Syy and Szz are estimated based on the extrema of the distribution of experimental data. The conversion from units of Hz to unitless values of the order parameters was performed based on the following equation:
| (7) |
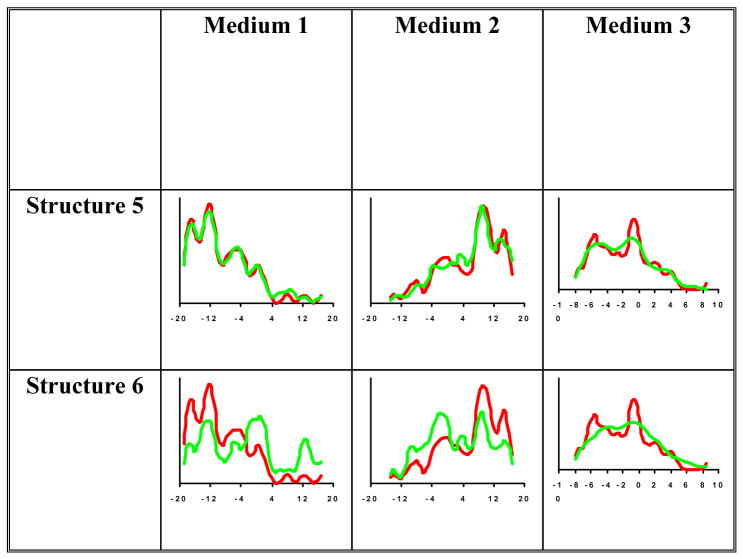
In this equation 24350 corresponds to the maximum observable value possible for the N-H interaction and 1.01 Å corresponds to a typical N-H bond length reported by the Amber 97 force-field. Backbone N-H RDC data have been acquired from two separate alignment media (phage and compressed gel). In total 58 and 55 individual RDCs were observed from the two alignment media respectively. Note that these quantities of data correspond to 73% and 69% of the complete set of data and should serve as a demonstration of the tolerance of PDPA to missing data. In general the collected RDCs spanned an approximate range of −20 to 20 Hz. During the PDPA analysis an experimental error near 5% of the range of RDC data has been assumed (±2 Hz) even though the true experimental error might have been much smaller. This expansion of the experimental error is necessary in order to accommodate structural noise such as an imperfect N-H bond length. PDPA was applied to the set of 10 structures and the corresponding best match PDPs are shown in (Figure 4).
Figure 4.
PDPA results of the best structure (Structure5), worst structure (Structure 6) using data from all three alignment media. Red pattern corresponds to the ePDP and green patterns correspond to the best cPDPs
4. Results and discussion
An ensemble of ten structures For PF2048.1 has been obtained using the modeling program ROBETTA [9]. The resulting models are shown in (Figure 3). These structures exhibit pair-wise backbone rmsds ranging from 3.3 to 9.39 Å over the entire length of the protein and 1.77 to 5.67 Å over residues 10–60. It is clear that there is higher consistency among the models for the central core of the protein. The models are ranked according to the probability of their correctly representing an experimental structure. However, examination of modeling competitions such as CASP would suggest that the best model may be anyone of the top 5.
4.1 Probability Density Profile Analysis of PF2048.1 Structures
PDPA as described before [8], was applied to the ten modeled structures of PF2048.1 using the order parameters obtained as described in the previous section. The search for the orientation component of the alignment tensor was conducted in a grid fashion between 0°–80° in steps of 3°. cPDPs of the ten modeled structures were constructed and a comparison was made with that of experimental PDP of PF2048.1. The best scores for each alignment medium corresponding to each structure are shown in Table I. Results of PDPA from the first alignment medium clearly suggest that Structures 5 and 8 are the closest modeled structures to that of the real structure. Note that the ePDP (red pattern) and cPDPs (green pattern) in (Figure 4) obtained from these structures exhibit an obvious similarity. The results from medium 2 are also listed in table I where the top two structures are Structure 1, and 5. Table I also shows the results from the third virtual medium which is obtained by averaging the individual pairs of RDC observables from two media as discussed in section 3.3. The information content of this third medium is relatively low as the number of data points is less i.e. 49 data points. This is attributed to the fact that only those “pairs” that include RDC data from both media 1 and 2 can be utilized for this approach. Despite this reduction in the total number of data points, there is still useful information in the ePDP for the third virtual medium. Although at first glance, analysis of the this medium may appear to be redundant, it does provide independent information that is not available through independent analysis of data from each medium. The RDC data from different alignment media can be assigned to the same interacting pair of nuclei (based on chemical shifts) without any knowledge of the location of the interacting vector within the sequence. This correlation of data can therefore be utilized as additional restraints in order to improve the results of our proposed analysis. Currently, our proposed method of analyzing the sum of RDC data is the most computationally efficient way of incorporating the correlation information between two (or many) sets of data.
Table 1.
Results of PDPA applied to RDCs for media 1, 2 and the addition of media 1 and 2 for all 10 model structures
| Medium1 | Medium 2 | Media 1+ Media 2 | Weighted average | ||||
|---|---|---|---|---|---|---|---|
| Structure No. | Score | Structure No. | Score | Structure No. | Score | Structure No. | Score |
| 5 | 0.04269 | 1 | 0.03943 | 9 | 0.0128806 | 5 | 0.0273455 |
| 8 | 0.08519 | 5 | 0.04658 | 8 | 0.0186911 | 8 | 0.038599 |
| 7 | 0.09235 | 2 | 0.05501 | 4 | 0.0200126 | 4 | 0.0438387 |
| 4 | 0.10133 | 7 | 0.05602 | 1 | 0.0251499 | 7 | 0.044505 |
| 2 | 0.10790 | 8 | 0.06090 | 10 | 0.025688 | 1 | 0.047496 |
| 1 | 0.13656 | 4 | 0.06541 | 5 | 0.0302152 | 2 | 0.0500132 |
| 9 | 0.15343 | 9 | 0.08371 | 7 | 0.044275 | 9 | 0.059249 |
| 10 | 0.20898 | 10 | 0.08603 | 2 | 0.0537394 | 10 | 0.0759467 |
| 3 | 0.48253 | 3 | 0.17055 | 3 | 0.0971687 | 3 | 0.176722 |
| 6 | 0.72813 | 6 | 0.23317 | 6 | 0.125283 | 6 | 0.2566985 |
The top two structures resulted from this virtual data are 8 and 9. To account for the different information content of the various media a final score has been calculated using a weighted average in which the weights are given by the relative number of data points. Based on the average scores shown in table I, the top two structures are structure 5 and 8. This result coincides with the PDPA scores from independent media as well where structure 5 is the top structure from Medium 1 and has the second best scores in Medium 2. Also Structure 8 is one of the top two structures from Medium 3. Considering the difference in the average PDPA scores between structures 5 and 8 from all three media, structure 5 is identified as the model best representing the true structure of PF2048.1 Validation of this prediction awaits deposition of further experimental data on PF2048.1.
5. Conclusions
The results reported here have demonstrated the potential of PDPA in identifying the most homologous structure from a set of computational models using a minimum set of unassigned RDC data. PDPA combined with currently existing protein structure modeling tools represents a new hybrid approach to protein structure determination that successfully combines the cost-effective advantage of the computational methods with some of the reliability of experimental methods. H-N RDC data are among the most easily acquired sets of NMR data and can quickly produce validation of a computational model.
The method as described validates only the backbone structure of a protein. However, it can also provide an efficient and faster route to a more complete structure determination by providing a reliable starting point for the interpretation of more conventional NMR data. NMR based structure determination frequently uses a crude initial experimental structure to resolve ambiguities in assignment of NOE peaks before going on to produce high resolution structures. A correct computational model could serve a similar purpose [39–41]. Backbone folds also can be used in combination with paramagnetic perturbations and RDCs to produce assignment of backbone resonances in the absence of a complete set of triple resonance experiments [7]. The application of PDPA can easily be extended to larger proteins (~15–20kD). In fact the larger proteins will increase the likelihood of properly sampling the RDC space and provide better estimates of the critical values of Syy and Szz.
There are obvious extensions of the approach described. Perhaps the most useful would be a full implementation of correlation among data sets. We have introduced the concept of a ‘virtual’ medium to create a third 1D data set that incorporates some correlation information. However, a full comparison of a 2D histogram would be much more powerful. This can be done in a straightforward way if two sets of RDCs can be collected in a single medium, for example, H-N and Cα-Hα couplings, or H-N couplings and C=O chemical shift anisotropy offsets in a protein where HNCA or HNCO experiments correlate the appropriate pairs of cross-peaks. It may also be possible to implement a more powerful search algorithm for multiple sets of H-N RDCs. We continue our exploration of these alternatives.
Acknowledgments
We would like to acknowledge Clay Baucom for expressing the protein PF2048.1. This work has been funded by NSF grant number MCB-0644195 to Dr. Homayoun Valafar by DOE (FG05-95ER20175) to Dr. Michael W. W. Adams, and funds provided to Dr. James H. Prestegard as a part of the Northeast Structural Genomics Consortium, NIH grant U54-GM-074958.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xu Y, Xu D, Crawford OH, Einstein JR, Larimer F, Uberbacher E, Unseren MA, Zhang G. Protein threading by PROSPECT: a prediction experiment in CASP3. Protein Engineering. 1999;12:899–907. doi: 10.1093/protein/12.11.899. [DOI] [PubMed] [Google Scholar]
- 2.Jones DT. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. Journal of Molecular Biology. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 3.Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins-Structure Function and Genetics. 1999:171–176. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- 4.Lattman EE. Sixth Meeting on the Critical Assessment of Techniques for Protein Structure Prediction. Proteins-Structure Function and Bioinformatics. 2005;61:1–2. doi: 10.1002/prot.21849. [DOI] [PubMed] [Google Scholar]
- 5.Rohl CA, Baker D. De novo determination of protein backbone structure from residual dipolar couplings using rosetta. Journal of the American Chemical Society. 2002;124:2723–2729. doi: 10.1021/ja016880e. [DOI] [PubMed] [Google Scholar]
- 6.Mayer KL, Qu Y, Bansal S, LeBlond PD, Jenney FE, Brereton PS, Adams MWW, Xu Y, Prestegard JH. Structure determination of a new protein from backbone-centered NMR data and NMR-assisted structure prediction. Proteins-Structure Function and Bioinformatics. 2006;65:480–489. doi: 10.1002/prot.21119. [DOI] [PubMed] [Google Scholar]
- 7.Pintacuda G, Keniry MA, Huber T, Park AY, Dixon NE, Otting G. Fast structure-based assignment of 15N HSQC spectra of selectively 15N-labeled paramagnetic proteins. J Am Chem Soc. 2004;126:2963–70. doi: 10.1021/ja039339m. [DOI] [PubMed] [Google Scholar]
- 8.Valafar H, Prestegard JH. Rapid classification of a protein fold family using a statistical analysis of dipolar couplings. Bioinformatics. 2003;19:1549–1555. doi: 10.1093/bioinformatics/btg201. [DOI] [PubMed] [Google Scholar]
- 9.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Nikolskaya AN, Orchard S, Orengo C, Petryszak R, Selengut JD, Sigrist CJ, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–8. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis PP, Omer A. Small non-coding RNAs in archaea. Current Opinion in Microbiology. 2005;8:685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Michalak P. RNA world - the dark matter of evolutionary genomics. Journal of Evolutionary Biology. 2006;19:1768–1774. doi: 10.1111/j.1420-9101.2006.01141.x. [DOI] [PubMed] [Google Scholar]
- 13.Clore GM, Gronenborn AM, Bax A. A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. Journal of Magnetic Resonance. 1998;133:216–221. doi: 10.1006/jmre.1998.1419. [DOI] [PubMed] [Google Scholar]
- 14.Varner SJ, Vold RL, Hoatson GL. An efficient method for calculating powder patterns. Journal of Magnetic Resonance Series A. 1996;123:72–80. doi: 10.1006/jmra.1996.0215. [DOI] [PubMed] [Google Scholar]
- 15.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Nuclear Magnetic Resonance of Biological Macromolecules, Pt B. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 16.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. Journal of Magnetic Resonance. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 17.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. Journal of Magnetic Resonance. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Valafar H, Tian F, Prestegard JH. Rapid Classification of Protein Fold Families Using a Statistical Analysis of Dipolar Couplings. In: Valafar F, editor. Mathematics and Engineering Techniques in Medicine and Biological Sciences (METMBS) Las Vegas: 2001. pp. 146–151. [Google Scholar]
- 19.Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK, Prestegard JH. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. Journal of Magnetic Resonance. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 20.Warren JJ, Moore PB. A maximum likelihood method for determining D-a(PQ) and R for sets of dipolar coupling data. Journal of Magnetic Resonance. 2001;149:271–275. doi: 10.1006/jmre.2001.2307. [DOI] [PubMed] [Google Scholar]
- 21.Gershenfeld NA. The nature of mathematical modeling. Cambridge University Press; Cambridge, U.K.; New York: 1999. [Google Scholar]
- 22.Fukunaga K. Introduction to statistical pattern recognition. Academic Press; Boston: 1990. [Google Scholar]
- 23.Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chemical Reviews. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 24.Prestegard JH, Mayer KL, Valafar H, Benison GC. Determination of protein backbone structures from residual dipolar couplings. Methods Enzymol. 2005;394:175–209. doi: 10.1016/S0076-6879(05)94007-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valafar H, Mayer KL, Bougault CM, LeBlond PD, Jenney FE, Jr, Brereton PS, Adams MW, Prestegard JH. Backbone solution structures of proteins using residual dipolar couplings: application to a novel structural genomics target. J Struct Funct Genomics. 2004;5:241–54. doi: 10.1007/s10969-005-4899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Science. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackledge M. Recent progress in the study of biomolecular structure and dynamics in solution from residual dipolar couplings. Progress in Nuclear Magnetic Resonance Spectroscopy. 2005;46:23–61. [Google Scholar]
- 28.Tolman JR. Dipolar couplings as a probe of molecular dynamics and structure in solution. Current Opinion in Structural Biology. 2001;11:532–539. doi: 10.1016/s0959-440x(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 29.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. Journal of the American Chemical Society. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 30.Prestegard JH, Kishore AI. Partial alignment of biomolecules: an aid to NMR characterization. Current Opinion in Chemical Biology. 2001;5:584–590. doi: 10.1016/s1367-5931(00)00247-7. [DOI] [PubMed] [Google Scholar]
- 31.Poole FL, 2nd, Gerwe BA, Hopkins RC, Schut GJ, Weinberg MV, Jenney FE, Jr, Adams MW. Defining genes in the genome of the hyperthermophilic archaeon Pyrococcus furiosus: implications for all microbial genomes. J Bacteriol. 2005;187:7325–32. doi: 10.1128/JB.187.21.7325-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams MWW, Dailey HA, Delucas LJ, Luo M, Prestegard JH, Rose JP, Wang BC. The Southeast Collaboratory for Structural Genomics: A high-throughput gene to structure factory. Accounts of Chemical Research. 2003;36:191–198. doi: 10.1021/ar0101382. [DOI] [PubMed] [Google Scholar]
- 33.Sugar FJ, Jenney FE, Jr, Poole FL, 2nd, Brereton PS, Izumi M, Shah C, Adams MW. Comparison of small- and large-scale expression of selected Pyrococcus furiosus genes as an aid to high-throughput protein production. J Struct Funct Genomics. 2005;6:149–58. doi: 10.1007/s10969-005-3341-3. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
- 35.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Rna-Ligand Interactions Pt A. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 36.Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. Journal of the American Chemical Society. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- 37.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. Journal of Magnetic Resonance. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 38.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Grishaev A, Steren CA, Wu B, Pineda-Lucena A, Arrowsmith C, Llinas M. ABACUS, a direct method for protein NMR structure computation via assembly of fragments. Proteins-Structure Function and Bioinformatics. 2005;61:36–43. doi: 10.1002/prot.20457. [DOI] [PubMed] [Google Scholar]
- 40.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 41.Moseley HNB, Sahota G, Montelione GT. Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. Journal of Biomolecular Nmr. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]