Abstract
Genetic studies have linked many nonsyndromic deafness patients to mutations in genes coding for gap junction proteins. To better understand molecular identities of gap junctions in the cochlea, we investigated the expression of pannexins (Panxs). Western blot and reverse transcription PCR detected the expression of Panx1 and Panx2. Immunolabeling localized Panx1 to the inner and outer sulcus, as well as the Claudius cells. Both Panx1 and Panx2 were expressed in the spiral and Scarpa’s ganglion neurons. These data for the first time showed expressions of Panxs in the cochlea, therefore adding a new family of gap junction proteins to those used to form intercellular transport pathways in the cochlea.
Keywords: Pannexin, gap junction, cochlea, mouse, immunolabeling
Introduction
Gap junctions are intercellular channels directly linking adjacent cells to facilitate exchange of ions, nutrient and signaling molecules. The complete gap junction channels are assembled by close apposition of two gap junction hemichannels. Hemichannels are composed of six compatible connexin (Cx) subunits forming a large size transmembrane pore. Twenty-one and twenty Cx genes have been identified in the human and mouse genome respectively [1]. The essential role of Cxs in normal hearing is demonstrated by genetic studies showing that about half of inherited nonsyndromic deafness cases are caused by mutations in the Cx26 [2], Cx30 [3] genes. Corresponding mouse models of Cx mutant mice are also deaf [4,5]. Protein subunits used to assemble gap junctions in the cochlea are highly heterogeneous [6–9]. The functional roles of the Cxs in the cochlea are currently unclear.
Vertebrate animals express Cxs, while invertebrates utilize innexins as their gap junction proteins. Despite similarities in function and membrane topology, the two protein families share little sequence homology. A new family of gap junction proteins called pannexins (Panxs) has been recently found, which is expressed in both vertebrate and invertebrate animals [10]. In vitro recordings made from Xenopus oocytes demonstrated that Panx1 is able to form hemichannels and whole gap junctions [11], although ability of Panxs to form gap junctions in mammalian cell lines and in vivo has been a matter of debate [12]. The expression pattern of many subtypes of Cxs in the mouse cochlea (e.g., Cxs 26, 29, 30, 31, 43) has been investigated previously [8,9,13,14], and at least some of them (e.g., Cx26 and Cx30) are known to form heteromeric gap junctions [13]. Cxs are currently believed to be the sole class of molecules assembled into the gap junctions and hemichannels in the cochlea. To gain a better understanding of the molecular identity of protein subunits contributing to intercellular coupling in the inner ear, we have analyzed the expression of Panx1 and Panx2 in the mouse cochlea. Our results for the first time showed the cochlear expression of Panxs, suggesting that Panx1 and Panx2 are used as molecular building materials to form the intercellular transport pathways in the cochlea that are known to be essential for normal hearing.
Materials and Methods
Semi-quantitative RT-PCR detection of Panx1 and Panx2
Total RNA was isolated from mouse cochlea with PicoPure™ RNA isolation kit (Arcturus Bioscience, CA). The RNA integrity and concentration were assessed by capillary gel electrophoresis using the Agilent Bioanalyzer 2100 with RNA 6000 PicoChips (Agilent Inc., CA). The density ratio of 28S:18S bands must be >1.8. Total RNA was transcripted to cDNA with the Applied Biosystems’ high capacity cDNA archive kit (Bedford, MA). Relative expression of Panxs was normalized to the expression level of GAPDH. The PCR primers used to amplify Panxs are designed (Vector NTI, Invitrogen, CA) and synthesized (Sigma Biotech, Inc., MO) according to the following table:
| Forward | Reverse | Amplicon Size | |
|---|---|---|---|
| Panx1 | CCCACTTGGCCACGGAGTAT | CACAGTGGGAGGTTTCCAGA | 301 |
| Panx2 | AACCAATTTACTGTTATACTCCGC | CTTCTCAAACAGATTCTGCTCC | 407 |
| GAPDH | CAACTCACTCAAGATTGTC | ACCTGGTCCTCAGTGTAG | 638 |
PCR amplifications were carried out on a GenAmp PCR system 9700 (Applied Biosystems, MA) with the following protocol: 10-min at 95°C denature, 35 cycles of 95°C (30 sec), 55°C (30 sec) and 72°C (30 sec), followed by 10 min at 72°C. PCR products were visualized by electrophoresis on 1% agarose gels stained with ethidium bromide, and they were quantified by a Fluochem™ multipurpose imager (Alpha Innotech Corp., CA).
Immunolabeling of Panx1 and Panx2 in the mouse cochlea
We used CD1 mice (age range: embryonic day 16 (E16) to postnatal day 45 (P45)) according to the animal use protocol approved by the Institute Animal Care and Use Committee of the Emory University. Mice were anesthetized before they were perfused with 4% formaldehyde. Samples were decalcified in 10% EDTA solution (72 hrs, 4°C), cryo-protected in 20% sucrose solution overnight, and were embedded in the OCT compound. Sections (7 µm) were cut with a cryostat (2800 Frigocut E, Cambridge Instruments, Germany).
The immunolabeling protocol was essentially the same as that previously described [15]. Briefly, cryo-sections were permeabilized and blocked before they were labeled with the Panx1 antibodies (Abs). Two kinds of Panx1 Abs were used and similar results were obtained: (1) polyclonal rabbit anti-human Panx1 Ab, which was characterized previously by Dvoriantchikova et al., 2006 [18]); (2) polyclonal chicken Panx1 Ab (Diatheva, catalog#ANT0027, Fano, Italy). Panx2 Ab was purchased from Invitrogen Corp. (catalog#42–2800). Sections were washed and incubated with secondary antibodies (goat anti-chicken or goat anti-rabbit, Jackson ImmunoResearch Lab, PA). The slides were examined with a conventional fluorescence microscope (Zeiss Axiovert135, Carl Zeiss, PA) equipped with an AxioCam photo documentation system.
Western blotting detection of Panx1 and Panx2 in the cochlea
Cochleae without the bony shell were homogenized in RIPA lysis buffer (catalog#20–188, Upstate, NY) supplemented with protease inhibitor cocktail (Catalog#539134, CalBiochem, CA). Samples were centrifuged at 16,000g for 20 minutes (4°C). Total protein concentrations were measured using the bicinchoninic acid protein assay kit (Pierce, IL). Equal amounts of protein (10 µg) were loaded in 20% SDS-PAGE gels for electrophoresis. Proteins were transferred to Hybond ECL nitrocellulose films (Amersham Pharmacia Biotech). The same Abs used for immunolabeling were used in the Western blot experiments. Results were visualized after processing films with supersignal west femto chemiluminescent substrate (Kodak XAR-5 film, Pierce, IL). Protein abundances were calculated from optical densitometry analysis with the background subtracted. Details were described previously [15].
Results
RT-PCR and Western blot detected expressions of Panx1 and Panx2 in the cochlea
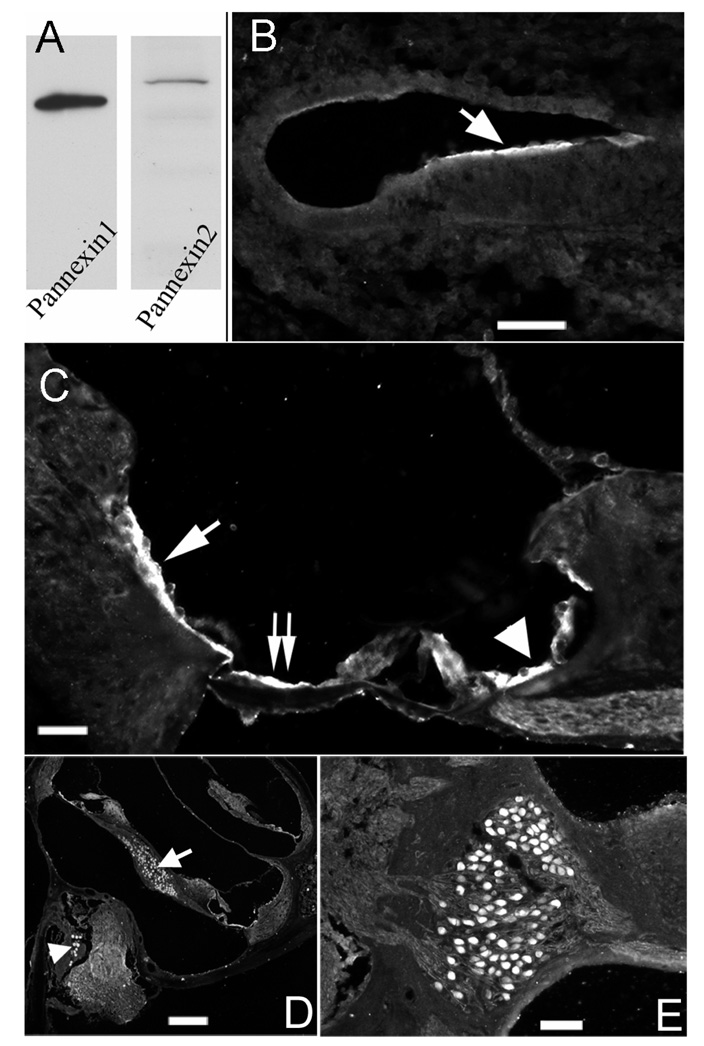
Panx1 and Panx2 transcripts in the cochlea during development were detected by RT-PCR amplifications. mRNAs for both Panxs were found as early as E16.5. PCR band intensity (normalized to GAPDH) of Panx1 was relatively stable at all developmental stages we examined, ranging from E16.5 to P45. In contrast, Panx2 data suggested that its expression level gradually increased as the cochlea matures. Protein expression of Panx1 and Panx2 was assessed by Western blots. Single bands at molecular weight of 48kD and 74kD for Panx1 and Panx2 respectively were detected (Fig. 1A), which are consistent with previously reported data [16,17]. Single band in the Western blots for Panx1 and Panx2 support the specificity of both antibodies used in this study.These data indicate that both Panx1 and Panx2 genes are expressed in the mouse cochlea.
Figure 1.
Western blots of antibodies used in the study and cellular expression patterns of Panx1 detected by immunolabeling. A) Western blot of antibodies against Panx1 and Panx2. B) Panx1 expression pattern at E16.5 in the cochlea. C) Panx1 expression pattern in the organ of Corti of adult cochlea. D) Panx1 immunoreactivity in spiral ganglion (arrow) and Scarpa's ganglion (arrowhead). E) An enlarged picture showing Panx1 immunoreactivity in SG neurons. Scale bars represent 100 µm, 60 µm, 200 µm and 100 µm for B), C), D) and E) respectively.
Immunolabeling pattern of Panx1 and Panx2 in the cochlea
At E16.5, the earliest developmental stage we examined, Panx1 immunoreactivity was detected in the epithelial cells on top of the developing organ of Corti (arrow in Fig. 1B). In the adult cochlea, Panx1 showed a markedly different expression pattern. It was detected in both inner sulcus (arrowhead in Fig. 1C) and outer sulcus (single arrow in Fig. 1C) cells, as well as in the Claudius cells (double arrow in Fig. 2B). Inner phalangeal cells around the inner hair cells were weakly labeled. Panx1 was also detected in the spiral ganglion (arrow in Fig. 1D) and Scarpa’s ganglion (arrowhead in Fig. 1D) neurons. Fig. 1E shows a higher magnification image, indicating that Panx1-specific immunolabeling was uniformly distributed in cell bodies of the neurons.
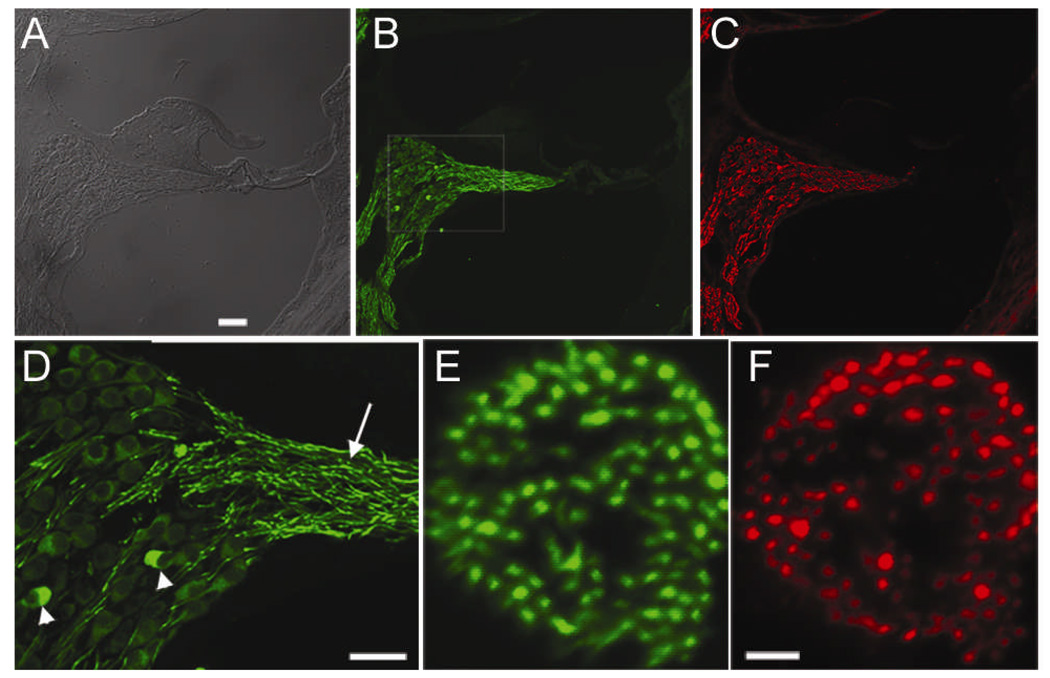
Figure 2.
Panx2 expression in the cochlea detected by immunolabeling. Cochlear sections double labeled with Panx2 (B) and neurofilament (C) antibodies. Panel A) is the differential interference contrast (DIC) image of the same section. D): Boxed area in B) is enlarged to show the details. Arrow and arrowheads point to fiber and soma of spiral ganglion neurons, respectively. E) & F) are cross sections of auditory nerve bundle double immunolabeled with Panx2 (E) and neurofilament (F) antibodies. Scale bars represent 60 µm (A-C), 60 µm (D) and 10 µm (E & F).
Double immunostaining of cochlear sections (Fig. 2A) with antibodies against Panx2 (Fig. 2B) and neurofilament (Fig. 2C) showed that spiral ganglion neurons were the only cells labeled by Panx2 antibody in the cochlea (Fig. 2B), since the general labeling patterns of Panx2 and neurofilament totally overlapped (comparing Fig. 2B&C). While the expression of Panx1 was mostly observed in the soma of neurons (Fig. 1E), the intensity of the Panx2 labeling was noticeably stronger in the fibers of the SG neurons (Fig. 2D). Only a few cell bodies of SG neurons were labeled at an intensity comparable to that in the nerve fibers (arrowheads in Fig. 2D). Double immunostaining for Panx2 (Fig. 2E) and neurofilament (Fig. 2F) on the cross sections of the auditory nerve bundles showed a remarkable overlap in the labeling by the two antibodies, further supporting that Panx2 is expressed by the SG neurons. Double immunolabeling of Panx2 (Fig. 3B) and neurofilament (Fig. 3C) further localized Panx2 expression to both cell bodies (arrowhead in Fig. 3B) and fibers (arrow in Fig. 3B) of the Scarpa’s ganglion neurons.
Figure 3.
Immunoreactivity of Panx2 in Scarpa’s neurons. Panel A) shows a DIC image of the section, which was double immunolabeled with an antibody against Panx2 (B) and neurofilament (C). Arrow and arrowheads point to fiber and soma of the Scarpa’s ganglion neurons, respectively. The scale bar represents approximately 50 µm.
Discussion
Gap junctions play vital roles in normal hearing because null and point mutations in Cxs (e.g., Cxs26, 30, 31, 32) are well known genetic causes of deafness in both human patients [2,3,18] and animal models [4,5]. Identities of the molecular components used to build gap junctions in the cochlea are therefore essential for understanding the normal cochlear physiology and mechanisms of deafness. Cxs are currently considered the sole molecules assembled into gap junctions and hemichannels in the cochlea. Our data showed expressions of Panx1 and Panx2, which belong to the Panx family of gap junction protein subunits, in the mammalian cochlea. These data suggest the Panx family of gap junction proteins are candidates of molecular building material suitable for assembling gap junction-mediated intercellular and/or hemichannel communication pathways in the cochlea.
Similar to previous reports [19], our results showed that Panx1 and Panx2 have generally similar cellular distribution but differ in the developmental expressions levels. Both Panx1 and Panx2 are expressed in spiral and Scarpa’s ganglion neurons, which are consistent with reports showing that neurons are the predominant cell type expressing Panx1 and Panx2 in the CNS [11,19]. We also found Panx1 was expressed in inner and outer sulcus cells, as well as the Claudius cells. Functional counterparts of these cochlear cells are glial cells in the CNS. At least some reports support the suggestion that Panx1 is expressed in the glial cells [11,12].
What functional roles do Panxs play in the cochlea? Bruzzone [11] showed that Panx1 forms homomeric gap junctions. In contrast, Panx2 can only form functional gap junctions when co-assembled with Panx1. Unlike immunolabeling patterns observed for Cx26 and Cx30, which showed distinctive plaques in the cell membrane [15], Panx1 and Panx2 immunolabeling patterns appeared to be diffused intracellularly (Fig. 1–Fig. 3). This pattern of protein localization argues against the hypothesis that Panxs form gap junctions in the cochlea. This notion is consistent with the results reported showing that Panx1 reconstituted in mammalian cells does not transfer fluorescent dyes and no electrophysiological coupling is detected among cell pairs after transfections [12].
Supporting cells in the organ of Corti are the generator of spontaneous electrical activities in auditory nerves before onset of hearing. Supporting-cell-derived ATP excites both inner hair cells and spiral ganglion neurons. The firing activities synchronized with Ca++ waves are believed to contribute to the refinement of tonotopic organization in the auditory pathway [20]. However, the molecular conduit for releasing intracellular ATP into extracellular space in the cochlea remains unclear. Although Cx hemichannels remain candidates to provide such a passage, physiological properties of Panx hemichannels seem to be better suited for ATP signaling in cochlea. Cx hemichannels are closed at physiological level (1–2 mM) of extracellular Ca++. Panx hemichannels, in contrast, are known to have large unitary conductance (~500 pA) and capable of releasing intracellular ATP into extracellular space when intracellular Ca++ concentration is raised [21,22]. In good agreement, recent studies have revealed that Panx1 hemichannels, rather than connexin ones, mediate the intracellular Ca++–dependent ATP secretion from taste receptor cells [23]. Further experiments are required to test this novel hypothesis for Panxs’ involvement in Ca++-wave-triggered ATP release in the cochlea.
Conclusion
We presented Western blot, reverse transcription PCR and immunolabeling data to show the expression of Panx1 and Panx2 in the neuronal cells in the cochlea. Panx1 was also found in the cochlear supporting cells. These results suggested a new family of gap junction proteins suitable for assembling into gap junctions and hemichannels in the intercellular transport pathways in the cochlea.
Acknowledgments
This study was supported by grants to XL and WT from NIDCD (RO1-DC006483 and R21-DC008353, R21 DC008672) and from the Deafness Research Foundation. VIS received supports from NIH grants R01-EY14232 and P30 EY014801. We thank Mrs. Lian Li for her contribution in technical aspects of the experiments.
References
- 1.Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, et al. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- 2.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 3.Grifa A, Wagner CA, D'Ambrosio L, Melchionda S, Bernardi F, Lopez-Bigas N, et al. Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nature genetics. 1999;23:16–18. doi: 10.1038/12612. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Salmon M, Ott T, Michel V, Hardelin JP, Perfettini I, Eybalin M, et al. Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr Biol. 2002;12:1106–1111. doi: 10.1016/s0960-9822(02)00904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teubner B, Michel V, Pesch J, Lautermann J, Cohen-Salmon M, Sohl G, et al. Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Human molecular genetics. 2003;12:13–21. doi: 10.1093/hmg/ddg001. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad S, Chen S, Sun J, Lin X. Connexins 26 and 30 are co-assembled to form gap junctions in the cochlea of mice. Biochem Biophys Res Commun. 2003;307:362–368. doi: 10.1016/s0006-291x(03)01166-5. [DOI] [PubMed] [Google Scholar]
- 7.Forge A, Becker D, Casalotti S, Edwards J, Marziano N, Nevill G. Gap junctions in the inner ear: comparison of distribution patterns in different vertebrates and assessement of connexin composition in mammals. J Comp Neurol. 2003;467:207–231. doi: 10.1002/cne.10916. [DOI] [PubMed] [Google Scholar]
- 8.Cohen-Salmon M, Maxeiner S, Kruger O, Theis M, Willecke K, Petit C. Expression of the connexin43- and connexin45-encoding genes in the developing and mature mouse inner ear. Cell Tissue Res. 2004;316:15–22. doi: 10.1007/s00441-004-0861-2. [DOI] [PubMed] [Google Scholar]
- 9.Tang WX, Zhang YP, Chang Q, Ahmad S, Dahlke I, Yi H, et al. Connexin29 is highly expressed in cochlear Schwann cells and it is required for the normal development and function of the auditory nerve of mice. J Neurosci. 2006;26:1991–1999. doi: 10.1523/JNEUROSCI.5055-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–R474. doi: 10.1016/s0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- 11.Bruzzone R, Hormuzdi SG, Barbe MT, Herb A, Monyer H. Pannexins, a family of gap junction proteins expressed in brain. Proc Natl Acad Sci U S A. 2003;100:13644–13649. doi: 10.1073/pnas.2233464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Grinspan JB, Abrams CK, Scherer SS. Pannexin1 is expressed by neurons and glia but does not form functional gap junctions. Glia. 2007;55:46–56. doi: 10.1002/glia.20435. [DOI] [PubMed] [Google Scholar]
- 13.Ahmad S, Chen S, Sun J, Lin X. Connexins 26 and 30 are co-assembled to form gap junctions in the cochlea of mice. Biochem Biophys Res Commun. 2003;307:362–368. doi: 10.1016/s0006-291x(03)01166-5. [DOI] [PubMed] [Google Scholar]
- 14.Forge A, Becker D, Casalotti S, Edwards J, Marziano N, Nevill G. Gap junctions in the inner ear: comparison of distribution patterns in different vertebrates and assessement of connexin composition in mammals. J Comp Neurol. 2003;467:207–231. doi: 10.1002/cne.10916. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Ahmad S, Chen S, Tang W, Zhang Y, Chen P, et al. Cochlear gap junctions coassembled from Cx26 and 30 show faster intercellular Ca2+ signaling than homomeric counterparts. Am J Physiol Cell Physiol. 2005;288:C613–C623. doi: 10.1152/ajpcell.00341.2004. [DOI] [PubMed] [Google Scholar]
- 16.Dvoriantchikova G, Ivanov D, Panchin Y, Shestopalov VI. Expression of pannexin family of proteins in the retina. FEBS Lett. 2006;580:2178–2182. doi: 10.1016/j.febslet.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Zappala A, Cicero D, Serapide MF, Paz C, Catania MV, Falchi M, et al. Expression of pannexin1 in the CNS of adult mouse: cellular localization and effect of 4-aminopyridine-induced seizures. Neuroscience. 2006;141:167–178. doi: 10.1016/j.neuroscience.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 18.Xia JH, Liu CY, Tang BS, Pan Q, Huang L, Dai HP, et al. Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nature genetics. 1998;20:370–373. doi: 10.1038/3845. [DOI] [PubMed] [Google Scholar]
- 19.Vogt A, Hormuzdi SG, Monyer H. Pannexin1 and Pannexin2 expression in the developing and mature rat brain. Brain Res Mol Brain Res. 2005;141:113–120. doi: 10.1016/j.molbrainres.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE. The origin of spontaneous activity in the developing auditory system. Nature. 2007;450:50–55. doi: 10.1038/nature06233. [DOI] [PubMed] [Google Scholar]
- 21.Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–68. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Braet K, Aspeslagh S, Vandamme W, Willecke K, Martin PE, Evans WH, et al. Pharmacological sensitivity of ATP release triggered by photoliberation of inositol-1,4,5-trisphosphate and zero extracellular calcium in brain endothelial cells. Journal of cellular physiology. 2003;197:205–213. doi: 10.1002/jcp.10365. [DOI] [PubMed] [Google Scholar]
- 23.Huang YJ, Maruyama Y, Dvoryanchikov G, Pereira E, Chaudhari N, Roper SD. The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6436–6441. doi: 10.1073/pnas.0611280104. [DOI] [PMC free article] [PubMed] [Google Scholar]