Abstract
Utrophin is the autosomal homolog of dystrophin, the product of the Duchenne’s muscular dystrophy (DMD) locus. Utrophin is of therapeutic interest since its overexpression can compensate dystrophin’s absence. Utrophin is enriched at neuromuscular junctions due to heregulin-mediated utrophin-A promoter activation. We demonstrate that heregulin activated MSK1/2 and phosphorylated histone H3 at serine 10 in cultured C2C12 muscle cells, in an ERK-dependent manner. MSK1/2 inhibition suppressed heregulin-mediated utrophin-A activation. MSK1 over-expression potentiated heregulin-mediated utrophin-A activation and chromatin remodeling at the utrophin-A promoter. These results identify MSK1/2 as key effectors modulating utrophin-A expression as well as identify novel targets for DMD therapy.
Keywords: Utrophin, Heregulin, MSK1/2, Chromatin remodeling, Transcription
Introduction
DMD is the most common fatal X-linked disorder affecting 1 in 3500 boys. The disease is caused by mutations in the DMD gene, leading to defects in dystrophin expression [1,2]. Utrophin is the chromosome 6-encoded autosomal homolog of dystrophin, and bears significant structural and functional similarities to dystrophin [3,4]. Utrophin is driven by two promoters (A & B) and the restricted localization of utrophin-A occurs in part due to selective expression of the utrophin-A gene product in sub-synaptic nuclei [5,6]. Utrophin can functionally substitute for missing dystrophin; upregulation achieved by transgenic means [7,8], viral vectors [9] and pharmacological approaches [10,11] ameliorate the dystrophic phenotype of mdx mice [12].
Concerted efforts have begun to identify the molecular mechanisms regulating utrophin expression. We and others have shown that heregulin (HRG) binding to cell surface erbB/HER receptors leads to ERK-dependant phosphorylation of the ets-related GABPα/β transcription factor complex which in turn leads to enhanced binding of the overlapping ets/N-box site, activation of utrophin-A promoter and increased transcription in cultured muscle [6,13], in a manner similar to that described for acetylcholine receptors [14]. In addition, cognate binding site mapping and activation by other transcription factors (e.g. SP1, NFAT, PGC-1 α) has also been described [12,15,16]. However, transcriptional activation of genes also depends on remodeling of the chromatin structure [17,18]. Nevertheless, the identity of intracellular effectors and chromatin-level changes that ultimately allow increased utrophin-A promoter activation are yet to be described. At periods of intense transcriptional activity phosphorylation at Ser10 and Ser28 of the core histone H3 has been noted [18,19]. Notably, histone H3 Ser10 phoshorylation correlates well with the induction of immediate-early genes as well as with other inducible genes [20]. The well-established targets of ERK- and p38-type MAP kinases, the mitogen- and stress-activated protein kinases-1 and-2 (MSK1/2) are histone H3 Ser10 and Ser28 kinases [20–23], and thereby provide clues for potential downstream effectors for ERK-mediated activation of utrophin-A by heregulin. Here, we identify and characterize MSK1/2 as effectors of heregulin-mediated chromatin remodeling at the utrophin-A promoter.
Results
Heregulin activates MSK
To address the possibility that the major growth factor-stimulated histone H3 kinases MSK1/2 are downstream effectors of heregulin, we determined the levels of the active forms of ERK, p38 and MSK1/2 in the C2C12 muscle cell line after incubation with heregulin at different time points (Fig 1A). Consistent with previous reports, heregulin treatment was found to increase levels of active ERK1/2 and to a minor extent p38 (Fig 1A). Ten minutes of heregulin treatment was sufficient to activate these kinases. Using phospho-specific antibodies recognizing the essential, activating phosphorylation site serine 376 of MSK1/2, we observed that heregulin induced phosphorylation of MSK1/2 with a time course similar to that for activation of ERK (Fig 1A). To independently verify the results we assayed MSK1/2 kinase activity upon heregulin stimulation. As can be seen in Fig 1B, heregulin increased MSK1/2 activity with the peak of activity being observed at 10 minutes and the time course was similar to that noted using phospho-specific antibodies against MSK1/2. To investigate if ERK-dependence contributed to the tight temporal correlation between heregulin-mediated activation of ERK1/2 and MSK1/2, we pretreated C2C12 cells with U0126, a specific inhibitor of MEK, the upstream kinase activating ERK. Heregulin-stimulated activation of MSK1/2 was significantly suppressed in U0126 treated cells (Fig 1C), demonstrating that ERK plays a major role in heregulin-mediated MSK1/2 activation.
Figure 1. Heregulin activates MSK1/2 in C2C12 cultured muscle cells.
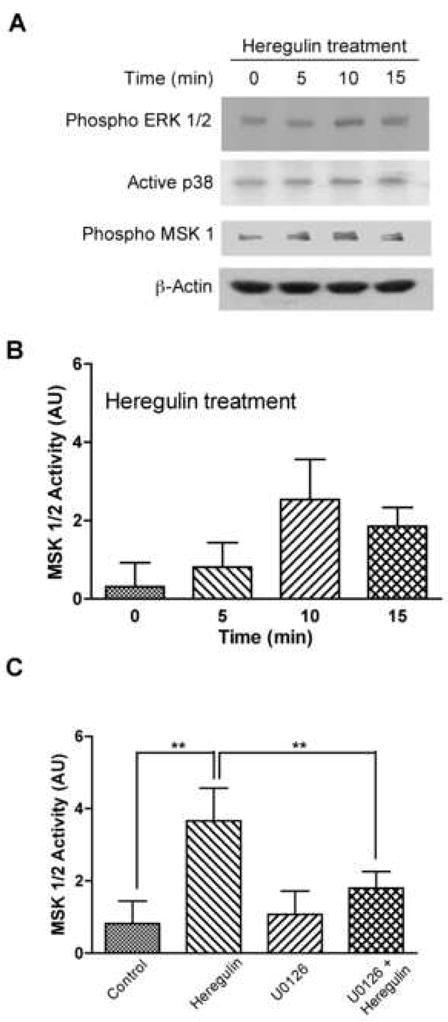
Serum starved C2C12 cells were treated with 2nM heregulin for times indicated and analyzed by western blots. (A) Phospho ERK1/2, active p38 and phospho MSK1/2 antibodies revealing increased phosphorylation of ERK1/2 and MSK1/2 but only slightly increased phosphorylation of p38. β-actin was used as control for equal loading. Total MSK1/2 enzymatic activity (B) measured from cell lysates in parallel experiments shows maximal activation by heregulin at 10′ (Bargraph: 0′. 0.30±0.61AU; 5′. 0.81±0.63AU; 10′. 2.54±1.02AU; 15′. 1.86±0.48AU; n=4). Total MSK1/2 enzymatic activity assays (C) show ERK-dependence. C2C12 cells were serum starved overnight, treated with or without MEK inhibitor, U0126 (10μM) for 15′ and then were incubated with 2nM heregulin for 10′ (control 0.82±0.62AU; HRG 3.67±0.90AU; U0126 1.07±0.65; U0126+HRG 1.8±0.46AU; n=4). **statistical significance p<.01.
Role for MSK in heregulin stimulated utrophin promoter activation
Having demonstrated that heregulin phosphorylates and activates MSK1/2, we asked whether MSK1/2 plays a role in heregulin-stimulated utrophin upregulation. Serum starved C2C12 cells were transfected with a full-length utrophin-A promoter luciferase reporter construct (Fig 2A) in the absence or presence of the MSK1/2 inhibitor GF109203X followed by heregulin-stimulation. As shown in Fig 2B, heregulin-induced utrophin-A promoter activation was suppressed by GF109203X. To independently validate MSK1/2 as an intracellular effector of heregulin-mediated utrophin promoter activation, we co-transfected the utrophin promoter reporter construct along with an expression construct for wild type MSK1 and stimulated with heregulin. While MSK1 overexpression alone did not change the basal level of utrophin promoter activity per se, MSK1 overexpression potentiated the heregulin-mediated activation of the utrophin promoter (Fig 2C). MSK1 overexpression did not show any effect over N-box deleted utrophin promoter (Fig 2E), which is insensitive to heregulin.
Figure 2. MSK1 plays a critical role in heregulin-induced utrophin-A promoter activation.
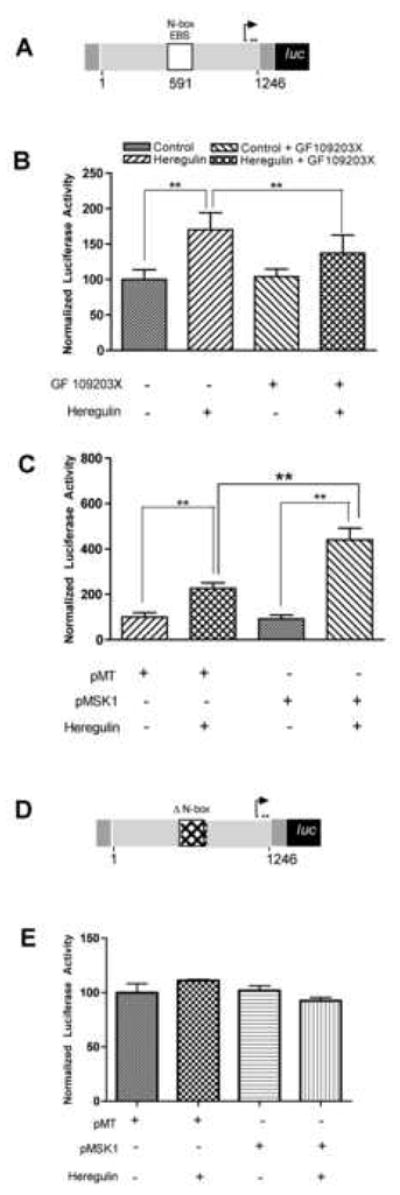
Schematic (A) of the utrophin-A promoter luciferase construct was cotransfected into C2C12 cells along with transfection control pRL-TK. C2C12 cells transfected with utrophin-A promoter-reporter were serum starved and (B) treated with 2nM heregulin and the MSK inhibitor (5μM of GF109203X) for 30′ as indicated and showed significant reduction of heregulin-induced utrophin-A promoter activity in the presence of pharmacological inhibition of MSK1/2. (Bargraph: control 100±13.73; HRG 170.30±24.22; control+GF109203X 104.2±10.65; HRG+GF109203X 137.3±25.47; n=5). These cells were also (C) transfected with either the empty vector pMT or MSK1 expressing constructs (pMSK1) prior to overnight serum starvation and heregulin stimulation and showed potentiation of heregulin-induced utrophin-A promoter activity by MSK1. (Bargraph: pMT 100.0±19.28; pMT+HRG 227.10±23.04; pMSK1 91.71±17.41; pMSK1+HRG 440.50±51.65; n=5). Schematic (D) of the N-box deleted utrophin-A promoter luciferase construct cotransfected into C2C12 cells along with pRL-TK. Cells were also (E) transfected with either empty vector pMT or MSK1 constructs (pMSK1) prior to overnight serum starvation and heregulin stimulation. Neither heregulin nor MSK1 showed upregulation (Bargraph: pMT 100.9±14.58; pMT+HRG 106.5±1.686; pMSK1 102.7±7.617; pMSK1+HRG 93.27±5.339; n=5). Luciferase activity is normalized to pRL-TK-derived luciferase activity (internal control) and expressed as 100% in the control group. ** statistical significance p<.01
The role of MSK1 on utrophin promoter activation was further confirmed by the study of endogenous utrophin level (Fig 3). Taqman Real Time PCR (Fig 3A) and Western blot (Fig 3C, D) showed that MSK1 potentiated the heregulin-stimulated utrophin upregulation, while myostatin, a non-synaptic gene remained insensitive (Fig 3B).
Figure 3. MSK1 enhances endogenous utrophin expression upon heregulin stimulation.
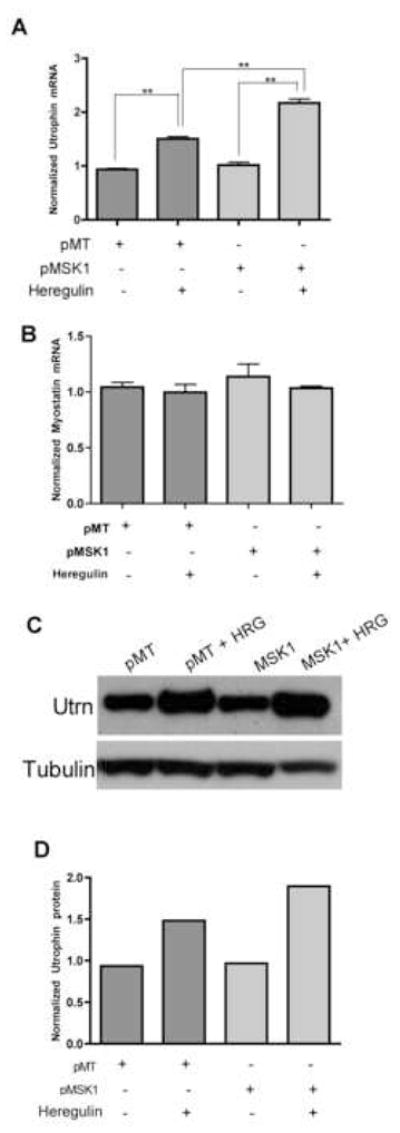
C2C12 cells transfected with either empty vector, pMT or MSK1 construct (pMSK1) were serum starved overnight followed by treatment with 2nM heregulin (HRG) for 30 minutes. MSK1 overexpression showed potentiation of heregulin-mediated utrophin (A) mRNA upregulation (Bargraph: pMT 0.9361±.040; pMT+HRG 1.503±.085; pMSK1 1.018±0.052; pMSK1+HRG 2.168±0.074; n=4). In contrast, heregulin did not show any effect on myostatin (B) mRNA expression (Bargraph: pMT 1.044±0.0.085; pMT+HRG 0.9979±0.139; pMSK1 1.139±0.225; pMSK1+HRG 1.035±0.034; n=4). The effect of MSK1 overexpression on heregulin-mediated utrophin upregulation was translated to protein level as revealed by anti-utrophin western blot (C). Tubulin was used as loading control. The band intensities of Western blot experiment were quantified and the ratio of utrophin and corresponding tubulin bands were plotted (D). ** statistical significance p<.05
Heregulin phosphorylates histone H3 (Ser10) at utrophin promoter
Since MSK1/2 is a known histone H3 kinase, we reasoned that heregulin-mediated activation of MSK1/2 might promote histone H3 phosphorylation. To test this possibility, C2C12 cells were stimulated with heregulin for different times and the extent of total histone H3 phosphorylation was analyzed by western blotting and immunostaining using antibodies that specifically recognize phosphorylation of histone H3 (Ser10). Western blotting showed that heregulin-stimulation led to a small increase in histone H3 phosphorylation within 10 minutes (Fig 4A, 4B). To determine if heregulin-mediated increases in histone H3 (Ser10) phosphorylation occurred at regions of active gene transcription, we first examined nuclei of non-mitotic, heregulin-treated cells by confocal microscopy using antibodies recognizing the phosphohistone H3 (Ser10) residue. Immunostaining revealed increased punctuate speckling representative of localized histone H3 phosphorylation thought to occur at MSK target genes [22] in heregulin-treated muscle cells (Fig 4C). Next we asked whether the heregulin could induce histone H3 phosphorylation at the utrophin-A promoter itself. Using anti-phosphohistone H3 (Ser10) antibodies we performed chromatin immunoprecipitation (ChIP) analysis of the utrophin-A promoter in C2C12 cells that had been transfected with an expression construct for MSK1 and treated with heregulin for 10 minutes or left untreated. Using ChIP we detected ~1.4 fold increased histone H3 (Ser 10) phosphorylation at the utrophin-A promoter after heregulin stimulation (Fig 4D, 4E). These data provide a mechanistic basis for heregulin-mediated increase in utrophin-A promoter activity.
Figure 4. Heregulin-induced chromatin remodeling at the utrophin-A promoter.
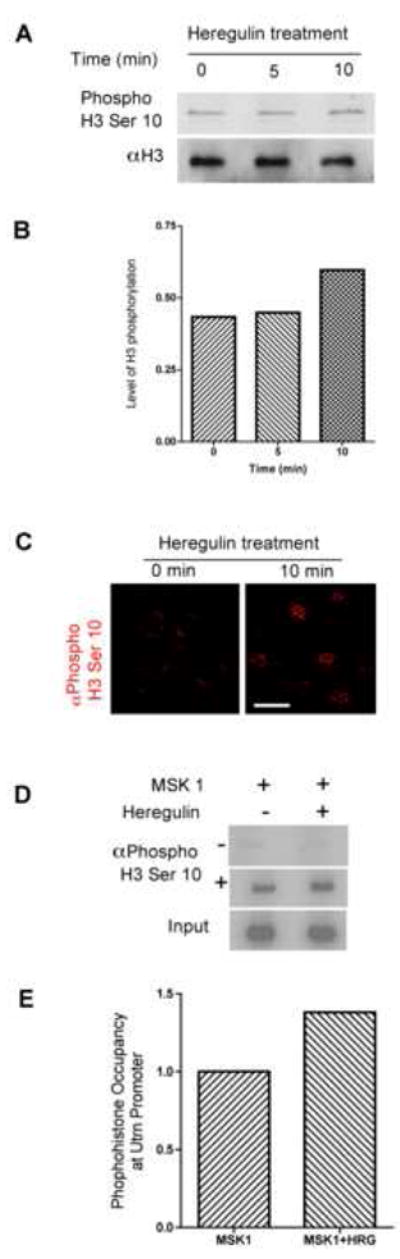
Serum starved C2C12 cells were treated with 2nM heregulin at time points indicated and analyzed by western blots (A) using phosphohistone H3(Ser10) antibody reveals increased phosphorylation. Anti-H3 antibody was used as control for equal loading. The ratio of band intensities of phosphohistone and corresponding histone showed extent of phosphorylation (B). Immunofluorescence (C) using these antibodies demonstrated increases in global phosphorylation of histone H3 and increased punctate, speckling indicative of activation of promoters. Scale bar = 25μm. ChIP (D) was performed on serum starved C2C12 cells that were transfected with MSK1 followed by incubation with 2nM heregulin. Lysates were analyzed using no antibody (top lane) or phosphohistone H3(Ser10) antibody (middle lane). Radioactive PCR using primers from the utrophin-A promoter region revealed that heregulin-induces chromatin remodeling at the utrophin-A promoter. Aliquots of inputs were used as control (lower lane). The band intensities were quantified and the ratio of phosphohistone and corresponding input band as the measure of occupancy of phosphohistone H3 at utrophin promoter showed ~1.4 fold increase upon heregulin treatment (3E).
Discussion
Our data provide a model (Fig 5) for heregulin-mediated upregulation of utrophin promoter-A in muscle. We propose that binding of the neurite-associated growth factor heregulin to cell surface erbB/HER receptors at the NMJ leads to receptor activation by phosphorylation and activation of a variety of intracellular signaling pathways such as ERK. This in turn leads to ERK-dependent phosphorylation and activation of MSK1/2 as well as phosphorylation of transcription factors such as ets-related GABPα/β complex and SP1. Phosphorylation of these and other transcription factors (e.g. NFAT, PGC-1α) in turn leads to recognition and enhanced binding of these transcription factors to their respective binding sites on the utrophin-A promoter, in some cases with synergistic cooperability [12,16]. The recognition and recruitment of the transcription factors to the promoter is accompanied by MSK-mediated chromatin remodeling of the utrophin-A promoter which in turn, facilitates transcription of the utrophin gene.
Figure 5. Transcriptional model of the utrophin-A promoter by heregulin stimulation.
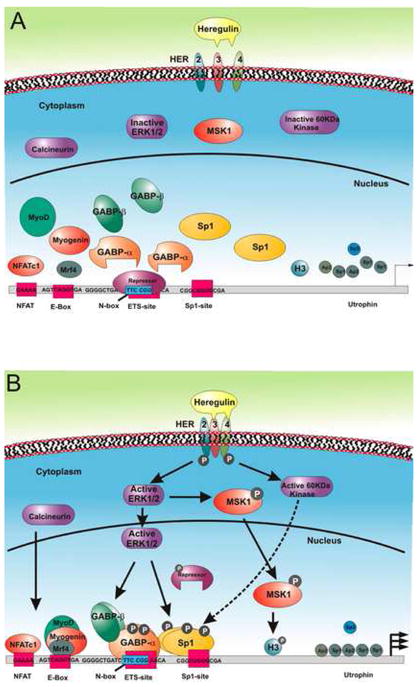
Multiple arrows represent increased transcription; P; phosphorylated protein; dotted and full arrowed lines represent potential and defined signaling cascades respectively. See text for details.
In conclusion, we have identified a role for MSK1/2 as an intracellular effector of heregulin-mediated chromatin remodeling and activation of the utrophin-A promoter in muscle cells. Modulating MSK1/2 or related histone kinases as well as histone H3 phosphorylation and/or acetylation at the utrophin enhanceosome may provide a novel pharmacological strategy to achieve utrophin upregulation in DMD patients. In the hemoglobinopathies, some degree of success has already been achieved using drugs such as sodium butyrate to activate fetal hemoglobin promoter in order to functionally substitute for mutated adult haemoglobin [24]. Similar strategies may be able to achieve therapeutic utrophin upregulation in DMD skeletal muscle, however this hypothesis needs to be tested in vivo.
Material and methods
Note: Details provided as Supplementary data (S1).
C2C12 muscle cell cultures
The C2C12 mouse muscle cell lines were cultured as suggested by provider (ATCC, Rockville, MD, USA). Five ×104 cells/35mm were plated 16h prior to transfection and serum-starved overnight prior to incubation with 2nM heregulin (R&D Systems, Minneapolis, MN, USA) or 5μM Bisindolylmaleimide I-GF109203X, (Calbiochem, San Diego, CA, USA).
Plasmid constructs and transfection
The pPUBF utrophin-A promoter luciferase reporter contains the human utrophin-A promoter cloned into pGL2 (Promega, Madison, WI, USA) and was kindly provided by Dr. Kay Davies (Oxford University, Oxford, UK). Renilla luciferase (pRL-TK) construct was used to control for transfection efficiency. The pMSK1 construct contains wild type human MSK1 cloned into pMT2 [25]. Plasmids were transfected using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA).
Western Blotting
C2C12 cells were lysed, resolved on 10% SDS-PAGE gels (for utrophin, NuPage 3–8% Tris Acetate gel was used) and electrotransferred. Membranes were probed with Anti-Active ERK, anti-Active P38 (1:1000; Promega, Madison, WI, USA), Anti-phospho-MSK1 (Ser376) (1:2000), Anti-β actin (1:4000; Cell Signaling, Danvers, MA, USA), anti-phosphohistone H3, the mitosis marker (1: 1000; Upstate, Lake Placid, NY, USA.), Anti-utrophin and Anti-Tubulin (1: 5000, Sigma, USA) overnight at 4°C. Blots were washed, incubated with appropriate secondary antibodies and visualized.
MSK1/2 Assay
The MSK1/2 activity in the whole cell lysate was assayed using a QTL Lightspeed MSK1/2 Assay Kit (QTL Biosystems, Santa Fe, NM, USA). C2C12 cells were lysed and 7.5μg protein containing lysate was used in the MSK1/2 Assay according to the manufacturer’s protocol.
RNA isolation and Taqman Real time PCR
RNA was isolated using RNAeasy kit (Qiagen Sciences, Maryland, USA) followed reverse transcription and cDNA corresponding to 25ng of RNA was used for Taqman Real Time PCR assayes (Applied Biosystems) for utrophin (Mm01168866_m1), myostatin (Mm00440328_m1) and β actin (4352933E) according to the manufacturer’s protocol. The expression of utrophin and myostatin were quantified by ΔΔCt method using β-actin as endogenous control.
Immunohistochemistry
Cells were fixed with cold methanol and processed for immunohistochemistry as previously described [11]. Cells were incubated with rabbit anti-phosphohistone H3 (Ser 10) antibody (Upstate) and visualized using AlexaFluor 546 conjugated goat anti-rabbit secondary antibody using confocal imaging using a Nikon TE300 microscope
Chromatin Immunoprecipitation Assay
ChIP was performed with mitosis marker, phosphohistone H3 (Ser10) rabbit polyclonal antibody according to the manufacturer protocol (Upstate). Presence of utrophin-A promoter was detected by PCR using [α-32P]dCTP with primers 5′CCCAAACTCAACAACCTCAGTAAAC3′ and 5′CAAATTGTCCGAAAATGTGTG TCA3′ designed to amplify 151bp of utrophin A promoter (NCBI Accession No X95524). Products were resolved on gels and imaged using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA).
Image Quantification
Band intensities were quantified by Image Quant-5.2.
Statistical analysis
Student’s t test was used throughout this study to calculate p values for determination of statistical significance. All results are shown as mean ± SD.
Supplementary Material
Acknowledgments
We thank L. Sørensen, B. Kofoed and L. Jensen for technical assistance and to K. Davies, for gift of reagents. We thank S. Bogdanovich, T. Choudhuri, E. Pierce, N. Lukinova and reviewers for helpful suggestions. This work was supported by grants from the Danish Cancer Society (MGH), University of Copenhagen (TOBK), and by grants from the AFM (France), DPP (The Netherlands), MDA (USA) and the NIH (USA) to TSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angus LM, Chakkalakal JV, Mejat A, Eibl JK, Belanger G, Megeney LA, Chin ERRN, Jasmin BJ. Calcineurin-NFAT signaling, together with GABP and peroxisome PGC-1{alpha}, drives utrophin gene expression at the neuromuscular junction. Am J Physiol Cell Physiol. 2005;289:C908–917. doi: 10.1152/ajpcell.00196.2005. [DOI] [PubMed] [Google Scholar]
- Burton EA, Tinsley JM, Holzfeind PJ, Rodrigues NR, Davies KE. A second promoter provides an alternative target for therapeutic up-regulation of utrophin in Duchenne muscular dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14025–14030. doi: 10.1073/pnas.96.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Negri T, Cozzi F, Colpo R, Andreetta F, Croci D, Davies KE, Cornelio F, Pozza O, Karpati G, Gilbert R, Mora M. Dystrophic phenotype of canine X-linked muscular dystrophy is mitigated by adenovirus-mediated utrophin gene transfer. Gene Therapy. 2003;10:750–757. doi: 10.1038/sj.gt.3301941. [DOI] [PubMed] [Google Scholar]
- Chaubourt E, Fossier P, Baux G, Leprince C, Israel M, De La Porte S. Nitric oxide and l-arginine cause an accumulation of utrophin at the sarcolemma: a possible compensation for dystrophin loss in Duchenne muscular dystrophy. Neurobiol Dis. 1999;6:499–507. doi: 10.1006/nbdi.1999.0256. [DOI] [PubMed] [Google Scholar]
- Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- Davie JR. MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Sci STKE. 2003;2003:PE33. doi: 10.1126/stke.2003.195.pe33. [DOI] [PubMed] [Google Scholar]
- Davie JR, Spencer VA. Control of histone modifications. Journal of Cellular Biochemistry. 1999;Suppl 32–33:141–148. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Potter AC, Tinsley JM, Wood SJ, Vater R, Young C, Metzinger L, Vincent A, Slater CR, Davies KE. Postsynaptic abnormalities at the neuromuscular junctions of utrophin-deficient mice. Journal of Cell Biology. 1997;136:883–894. doi: 10.1083/jcb.136.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M, Jensen CJ, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO Journal. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramolini AO, Angus LM, Schaeffer L, Burton EA, Tinsley JM, Davies KE, Changeux JP, Jasmin BJ. Induction of utrophin gene expression by heregulin in skeletal muscle cells: role of the N-box motif and GA binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3223–3227. doi: 10.1073/pnas.96.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Davies KE. Pharmacological strategies for muscular dystrophy. Nature Reviews. Drug Discovery. 2003;2:379–390. doi: 10.1038/nrd1085. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Hoffman EP, Kunkel LM. Identification of a chromosome 6-encoded dystrophin-related protein. Journal of Biological Chemistry. 1990;265:16717–16720. [PubMed] [Google Scholar]
- Khurana TS, Rosmarin AG, Shang J, Krag TO, Das S, Gammeltoft S. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein alpha/beta. Molecular Biology of the Cell. 1999;10:2075–2086. doi: 10.1091/mbc.10.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana TS, Watkins SC, Chafey P, Chelly J, Tome FM, Fardeau M, Kaplan JC, Kunkel LM. Immunolocalization and developmental expression of dystrophin related protein in skeletal muscle. Neuromuscular Disorders. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Krag TO, Bogdanovich S, Jensen CJ, Fischer MD, Hansen-Schwartz J, Javazon EH, Flake AW, Edvinsson L, Khurana TS. Heregulin ameliorates the dystrophic phenotype in mdx mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13856–13860. doi: 10.1073/pnas.0405972101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DR, Hill DF, Dickson G, Spurr NK, Byth BC, Marsden RF, Walsh FS, Edwards YH, Davies KE. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- Mejat A, Ravel-Chapuis A, Vandromme M, Schaeffer L. Synapse-specific gene expression at the neuromuscular junction. Ann N Y Acad Sci. 2003;998:53–65. doi: 10.1196/annals.1254.008. [DOI] [PubMed] [Google Scholar]
- Miura P, Jasmin BJ. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we? Trends Mol Med. 2006;12:122–129. doi: 10.1016/j.molmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Olivieri NF, Weatherall DJ. The therapeutic reactivation of fetal haemoglobin. Human Molecular Genetics. 1998;7:1655–1658. doi: 10.1093/hmg/7.10.1655. [DOI] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen-and stress-induced phosphorylation of histone H3 and HMG-14. Embo J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Mahadevan LC, Clayton AL. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction.[see comment] Seminars in Cell & Developmental Biology. 1999;10:205–214. doi: 10.1006/scdb.1999.0302. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene.[see comment] Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Chromatin remodeling and transcription. Current Opinion in Genetics & Development. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


