Abstract
In Saccharomyces cerevisiae, the Mrt4 protein is a component of the ribosome assembly machinery that shares notable sequence homology to the P0 ribosomal stalk protein. Here, we show that these proteins can not bind simultaneously to ribosomes and moreover, a chimera containing the first 137 amino acids of Mrt4 and the last 190 amino acids from P0 can partially complement the absence of the ribosomal protein in a conditional P0 null mutant. This chimera is associated with ribosomes isolated from this strain when grown under restrictive conditions, although its binding is weaker than that of P0. These ribosomes contain less P1 and P2 proteins, the other ribosomal stalk components. Similarly, the interaction of the L12 protein, a stalk base component, is affected by the presence of the chimera. These results indicate that Mrt4 and P0 bind to the same site in the 25S rRNA. Indeed, molecular dynamics simulations using modelled Mrt4 and P0 complexes provide further evidence that both proteins bind similarly to rRNA, although their interaction with L12 displays notable differences. Together, these data support the participation of the Mrt4 protein in the assembly of the P0 protein into the ribosome and probably, that also of the L12 protein.
INTRODUCTION
The ribosomal P0 protein, the orthologue of bacterial ribosomal L10 protein, is the central component of the eukaryotic ribosomal stalk. This structure is formed by the association of P0 with two heterodimers of the P1 and P2 acidic phosphoproteins, the counterparts of prokaryotic protein L7/L12. Although the stalk seems to essentially fulfill the same role in the translational machinery of all organisms, which is related to the interaction and activity of several soluble factors (1), the eukaryotic P0/(P1–P2)2 complex has some peculiar properties that are not attributed to the prokaryotic structure. These differences have led to the proposal that this complex might modulate ribosomal activity, which does not yet appear to be the case in prokaryotes (2). Among these peculiarities, the eukaryotic domain seems to undergo a process of assembly/disassembly during translation, which can be detected as an exchange between the ribosome bound P1/P2 proteins and the free proteins present in the cytoplasm (3–6). It is not clear when this exchange occurs but it seems most likely to take place during ribosome dissociation, after the termination step. It is possible that as a result of this process, the Saccharomyces cerevisiae ribosome population does not have a homogeneous P1/P2 content. Thus, particles containing different amounts of these acidic proteins, or even totally lacking them, can be detected in the cell (7,8). Since the proposed modulatory role of the ribosome stalk is based on the relative proportion of ribosomes with different P1/P2 content, it is fundamental to understand how this ribosome domain is assembled and how the assembly/disassembly process is controlled.
The stalk must be assembled as part of the ribosome and ribosome assembly in eukaryotes is an extremely complicated process that remains far from being understood (9). A large number of proteins and small rRNA molecules, generically known as trans-acting or assembly factors, participate in the assembly process. Many of these components have been identified in S. cerevisiae, although their specific function is only known in a limited number of cases. In yeast, ribosome biogenesis starts in the nucleolus with the transcription of the pre-rRNAs, which bind to a considerable number of trans-acting factors and ribosomal proteins in order to form the 90S particle (10,11). After three consecutive processing steps, the 90S pre-ribosomal particle, which contains the 35S pre-rRNA, yields a pre-40S subunit (12) and a pre-60S subunit (13). These two particles follow independent pathways in the nucleus that include the formation of a still poorly defined number of intermediates, especially in the case of the pre-60S. Late pre-ribosomal particles are then exported into the cytoplasm where they undergo the final maturation steps.
The information available regarding stalk assembly is still scarce. The P0 protein has been detected in different pre-ribosomal complexes (14–17). However, additional information will be required before a conclusion can be reached in regard to stage in which the protein is assembled. Similarly, there is no direct evidence that the P1/P2 stalk proteins are present in pre-ribosomal particles and it is generally assumed that they are assembled in the cytoplasm.
Significant sequence homology has been found between a number of ribosomal proteins and several assembly factors. Thus, the L24 ribosomal protein (Rpl24) is homologous to Rlp24, as is L7 (Rpl7) to Rlp7 and Rps9 to Imp3 (9,13,18,19). In the case of the Rpl24/Rlp24 pair, it has been proposed that the ribosomal protein might replace the assembly factor in the late pre-ribosome particles as one of the last maturation steps that take place in the cytoplasm (13,20).
Interestingly, the P0 protein and Mrt4 share notable amino acid homology. Mrt4 was initially thought to be involved in mRNA turnover (21) but it was later found in pre-ribosomes, and it is now considered a component of the ribosome assembly machinery (22). The Mrt4 protein has been detected in particles purified using tagged proteins involved in the early and mid-stages of 60S ribosomal subunit assembly (9,15), suggesting that it fulfills a specific role at the beginning of the 60S ribosomal subunit biogenesis pathway. However, Mrt4 has recently been associated with factors involved in the late stages of 60S assembly (23), making its true role far from clear.
Here, we have investigated the functional relationship between P0 and Mrt4, finding that both proteins can interact with the same region in the rRNA but that they are mutually exclusive in the ribosome. These data support the hypothesis that Mrt4 binds first to pre-60S ribosomal particles, occupying the P0 site in the earlier assembly steps, and that it is then later displaced by the ribosomal protein.
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae W303-1B (MAT α, leu2-3,112, ura3-1, trp1-1, his3-11,15, ade2-1, can1-100) was the parental control strain. Saccharomyces cerevisiae W303dM (MAT α, leu2-3,112, ura3-1, trp1-1, his3-11,15, ade2-1, can1-100, MRT4::KanMX4) and W303dMGP0 (MAT α, leu2-3,112, ura3-1, trp1-1, his3-11,15, ade2-1, can1-100, RPP0::URA3-PGAL1-RPP0, MRT4::KanMX4) were derived from S. cerevisiae W303-1B and W303dGP0 (24), respectively, by deleting the MRT4 gene using the KanMX4 module that carries short flanking MRT4 homology regions (25). In the W303dGP0 strain, the genomic copy of the RPP0 gene is under the control of the GAL1 promoter (24).
Isolation and analysis of ribosomes
Cells were grown to an OD600 of ∼0.8 and they were resuspended in ice-cold buffer [10 mM Tris–HCl (pH 7.4), 20 mM KCl, 12.5 mM MgCl2, 5 mM β-mercaptoethanol] containing a protease inhibitor cocktail (aprotinine, leupeptine, pepstatine and PMSF at 10 μg/ml), before they were disrupted by vigorous shaking with glass beads in a FastPrep FP120 (Bio101/Savant) at 4°C. The S30 fraction was obtained by centrifuging the extract at 13 000 r.p.m. for 20 min at 4°C in a Sorvall SS-30 rotor. Ribosomal particles were prepared from the S30 fraction by centrifugation at 4°C in a TL100.3 rotor for 90 min at 90 000 r.p.m. at 4°C. The particles were washed by centrifugation in a 20–40% discontinuous sucrose gradient in 20 mM Tris–HCl (pH 7.4), 500 mM ammonium acetate, 100 mM MgCl2, 5 mM β-mercaptoethanol. The ribosomes were finally stored at –70°C in the same buffer. Ribosomal proteins were analyzed either by SDS–PAGE or by isoelectrofocusing in a pH range of 2.0–5.0 as indicated previously (26). Western blots were performed using Immobilon-P membranes (27), and the stalk proteins and protein L12 (Rpl12) were detected using specific antibodies (28). The antibody to S. cerevisiae ribosomal protein L1 (Rpl1) was a gift from Prof. F. Lacroute (30) and the rabbit antibody to protein Mrt4 was obtained using custom made peptides. When required, ribosomes were washed in increasing NH4Cl concentrations as described previously (31).
Polysome profiles were obtained by 7–50% sucrose gradient centrifugation of total cell extracts as described previously (32).
Enzymes and reagents
Restriction endonucleases were purchased from Roche, MBI Fermentas, New England Biolabs and Amersham, and they were used as recommended by the suppliers. The T4 DNA ligase, calf intestinal alkaline phosphatase and the DNA polymerase I Klenow fragment were obtained from Roche. DNA manipulations were essentially performed as described in ref. (33). PCR reactions were carried out using Pwo DNA polymerase (Roche) and custom made oligonucleotides from Isogen, following the recommendations of Dieffenbach and Dveksler (34).
Plasmids
pFL37-Mrt4/P0
A chimeric MRT4/RPP0 gene was constructed by overlapping PCR using as template DNA fragments corresponding to the coding region of Mrt4 from M1 to I137 and the coding region of P0 from R122 to D312, and including 500 nt from the RPP0 3′ UTR. These fragments were amplified by PCR with the oligonucleotides Mrt4-1/Mrt4-2 and P0-14/P0-Eco (Supplementary Table S1). The MRT4/RPP0 chimera was then digested with the restriction enzymes NdeI and EcoRI, and cloned in the BsP0 plasmid (35) from which the wild-type P0 gene had been removed. The recombinant plasmid, called pBsMrt4/P0, contains the chimera under the control of the S. cerevisiae RPP0 gene flanking regions. Subsequent digestion of this plasmid with EcoRI and XhoI was performed and the resulting fragment containing the chimera was subcloned into pFL37 (35) to obtain the 8.035 Kb pFL37-Mrt4/P0 plasmid.
pTAPC111-Mrt4
A PCR fragment containing the Mrt4 coding region lacking the termination codon was obtained by PCR using the Mrt4-500 and Mrt4BamHIR primers (Supplementary Table S1) and yeast genomic DNA as the template. The fragment digested with BamHI was inserted in pTAPC111 (36) linearized with BamHI and SmaI.
pTAPC111-P0
The P0 coding region was obtained by PCR using the P0-EcoRI and P0-BamHI primers (Supplementary Table S1) and it was inserted into the vector produced by digesting pTAPC111-Mrt4 with BamHI and EcoRI.
The correct structure of all constructs was confirmed by DNA sequencing.
Tandem affinity purification of protein complexes
Protein complexes were obtained as described previously (37), with the following modifications. Buffer 1 contained 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 1 mM PMFS, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin and 0.1% NP-40. In addition, all absorption and elution buffers were complemented with 1 mM PMFS. IgG Sepharose 6 fast-flow from GE Healthcare and Calmodulin affinity resin from Stratagene were used for purification.
Purification of P0-TAP and Mrt4-TAP particles
Saccharomyces cerevisiae W303dGP0 and W303dM were transformed with the pTAPC111-P0 and pTAPC111-Mrt4 plasmid, respectively. Cells were grown in the appropriate media with glucose as a carbon source and total cell extracts were TAP fractionated as described above. The W303dGP0 strain, which is normally grown in galactose to express the wild-type P0, has to be shifted to glucose to enrich on the P0-TAP fusion protein.
Molecular dynamics
Starting structures
Initial protein models were built with the MODELLER9v4 software. For the Mrt4 and P0 proteins, the PDB 1ZAV chain A structure was used as a template, while the L12 protein was modelled using chain L from PDB 487D. The yeast ribosomal RNA stalk was modelled by mutating the fragment between residues 1145 and 1218 from the PDB 3CC2 chain 0 with the graphic software InsightII (Accelrys Software Inc., San Diego, CA, USA). The L12-RNA-Mrt4 and L12-RNA-P0 complexes were constructed by means of structural alignments using MAMMOTH (38) and with the G, I and 0 chains from PDB 3CC2 as a template. The hydrogen atom positions, standard atomic charges and radii were assigned according to the ff99 force field (39). The two modelled complexes were immersed in cubic boxes of TIP3P water molecules (40) sufficiently large to guarantee that the shortest distance between the solute and the edge of the box was >15 Å. Counter ions were also added to maintain electroneutrality. Three consecutive minimizations were performed: (i) the first involving only hydrogen atoms; (ii) the second only the water molecules and ions; and (iii) the entire system.
Simulation details
The minimized starting structures, prepared as indicated previously, were simulated in the NPT ensemble using Periodic Boundary Conditions and Particle Mesh Ewald method to treat long-range electrostatic interactions. The systems were then heated and equilibrated in two steps: (i) 200 ps of MD heating of the whole system from 100 to 300 K; and (ii), equilibration of the entire system during 1.0 ns at 300 K. The equilibrated structures were the starting points for 25 ns of unconstrained MD simulations at constant temperature (300 K) and pressure (1 atm). The SHAKE algorithm was used to keep bonds involving H atoms at their equilibrium length, allowing a 2 fs time step for the integration of Newton's equations of motion. The ff99 and TIP3P force fields, as implemented in the AMBER 8 package (41), were used to describe the proteins and water molecules, respectively. The last 15 ns of each trajectory were used to sample frames at 1 ps intervals, which were subsequently used for the analysis.
Effective free binding energies were estimated qualitatively using the MM-GBSA approach (42). The MM-GBSA method approaches the free energy of binding as a sum of a molecular mechanics (MM) interaction term, with a contribution of solvation from a generalized Born (GB) model and with a surface area (SA) contribution to account for the non-polar part of desolvation. These calculations were performed for each snapshot of the simulations using the appropriate module within AMBER 8. Averages were taken every 20 snapshots, and a normal distribution was fitted to the measured free energy density.
RESULTS
The P0 and Mrt4 proteins can not bind simultaneously to the ribosome
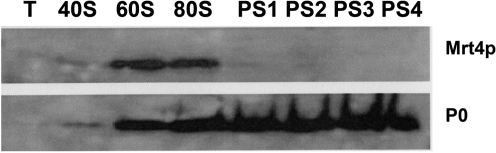
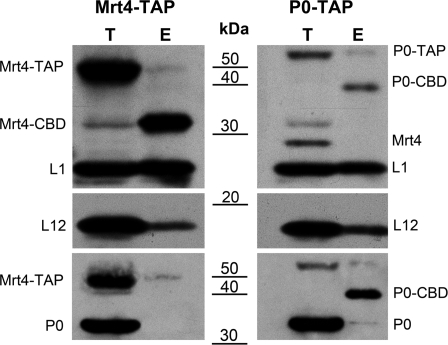
The notable sequence homology between the ribosomal P0 protein and the Mrt4 protein, a putative ribosome assembly factor, suggests they may be functionally related during ribosome assembly. As a first step to explore this hypothesis, the distribution of both proteins was assessed after sucrose gradient centrifugation of a total cell extract from the parental strain (Figure 1). As expected, P0 was present in all the ribosomal particles, except in the 40S subunit, whereas the Mrt4 protein was detected in the 60S and 80S peaks but not in the polysomes. The absence of Mrt4 from the polysomes suggested that it was bound to translation inactive particles, probably pre-ribosomal particles that are found in the gradient overlapping the 60S/80S region. Alternatively, both proteins might be bound to a fraction of the mature ribosomal population that is inactive due to the presence of Mrt4. To resolve this dilemma, particles carrying either tagged Mrt4-TAP or tagged P0-TAP were affinity purified and the presence of the partner proteins was analyzed as described in the Material and methods section. The presence of ribosomal proteins L1 and L12 in the samples eluted from the columns indicate that real protein complexes and not individual proteins were purified (Figure 2). While P0 was absent from Mrt4-TAP purified particles, Mrt4 was not found in P0-TAP ribosomes, indicating that both proteins seem to be unable to bind to the ribosome at the same time.
Figure 1.
Total extracts from S. cerevisiae W303-1B were resolved in a 7–50% sucrose gradient and the fractions corresponding to the gradient top (T), the peaks corresponding to the 40S, 60S and 80S particles, as well as the first four peaks of the polysome region were collected and analyzed by SDS–PAGE. The Mrt4 and P0 proteins were detected in immunoblots using specific antibodies.
Figure 2.
Total extracts from strain W303dGP0 expressing P0-TAP or W303dM expressing Mrt4-TAP were affinity purified using the TAP procedure. Aliquots from the starting extracts (T) and from the last purification step (E) were analyzed by SDS–PAGE and the proteins were detected with specific antibodies. The positions of the untagged proteins are marked, as well as that of the proteins tagged with either TAP, or the fragment remaining after treatment with the protease in the second purification step (CBD). Antibodies against Mrt4, the L1 ribosomal protein and the CBD fragment of TAP were used in the upper panel, that against the ribosomal protein L12 was used in the middle panel, and that against the P0 protein was used in the lower panel. In this last panel, the tagged Mrt4 protein was also detected due to the reaction of the secondary antibody used with the protein A fragment in the TAP tag.
The rRNA binding site of the P0 ribosomal protein can be substituted for the equivalent Mrt4 domain
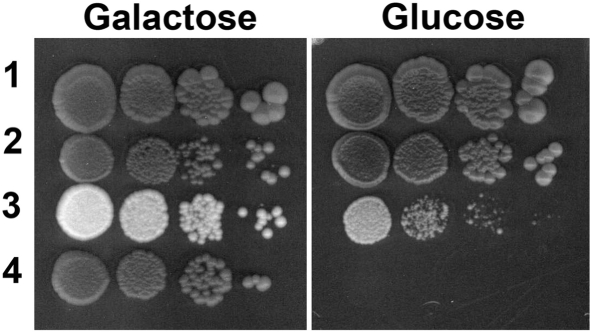
The S. cerevisiae ribosomal P0 protein rRNA binding domain corresponds to the first 121 amino acid residues of the N-terminal domain (43) and it is homologous to the equivalent region of the Mrt4 protein (Supplementary Figure S3). This homology suggests that both proteins might interact with the same region in the 25S rRNA, the GTPase Associated Region (GAR). To test this possibility, a protein chimera was constructed by replacing the first 121 amino acids of P0 with the equivalent region from Mrt4, which contains 16 additional residues at the amino end. The chimeric gene was cloned into the centromeric plasmid pFL37 under the control of the P0 promoter region, yielding the pFL37-Mrt4/P0 plasmid, which was used to transform the S. cerevisiae strain W303dMGP0. This strain is a conditional null P0 mutant that carries the genomic copy of gene RPP0 under the control of the GAL1 promoter and since P0 is an essential protein, the strain does not grow in the presence of glucose (24). As expected, the parental W303dMGP0 was unable to grow in the presence of glucose while transformation with pFL37-Mrt4/P0 permitted growth in this carbon source, albeit at a lower rate than that of the control (Figure 3), indicating that the Mrt4/P0 chimera can partially complement the lack of P0. Indeed, in a YEPD liquid medium the transformed strain expressing the chimera had a doubling time around four-fold higher than the parental S. cerevisiae W303. In these experiments the W303 and W303dM strains were used as a positive control.
Figure 3.
Growth test of S. cerevisiae strain expressing the Mrt4/P0 chimera. Serial dilution of S. cerevisiae W303 (1), W303dM (2), W303dMGP0-Mrt4/P0 (3) and W303dMGP0 (4) plated in rich medium containing either galactose or glucose as a carbon source, and incubated at 30°C for 5 days. Controls (1, 2 and 4) were transformed with an empty plasmid.
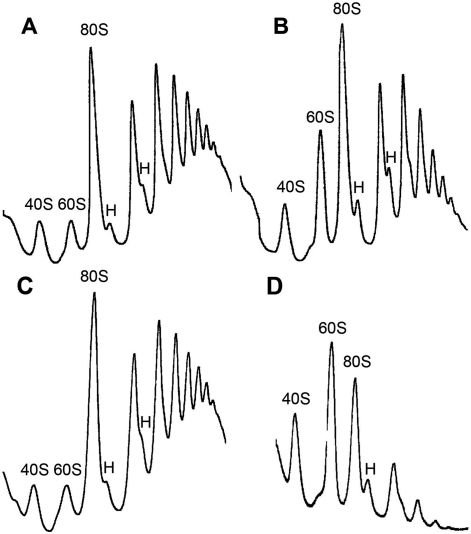
To assess the translational capacity of the chimera, the polysome profile was analysed in sucrose gradients (Figure 4). When compared with extracts from control cells grown in galactose, fewer polysomes were present in extracts from the transformed strain grown in glucose to express the chimera, in agreement with their slow growth rate. In addition, there was an increase of the halfmers as well as of the free 60S peak relative to the 80S peak. In contrast, polysomes were practically absent in extracts from W303dMGP0 transformed with an empty plasmid and maintained in a glucose medium for 48 h, which accumulate free subunits and pre-ribosomal particles, in agreement with previous data from similar conditional P0 null mutants grown in restrictive conditions (24).
Figure 4.
Polysome profiles from the W303dMGP0-Mrt4/P0 strain grown in the presence of galactose (A) or glucose (B), as well as strain W303dMGP0 transformed with an empty plasmid and grown either in galactose (C) or shifted to a medium containing glucose for 48 h (D). Total cell extracts were resolved on 7–50% sucrose gradients in the conditions indicated in the Materials and Methods section. Sedimentation is from left to right. The peaks of free 40S and 60S subunits, 80S free couples/monosomes are indicated. Half-mers are labelled by an H.
These results suggest that the 60S ribosomal subunits carrying the Mrt4/P0 chimera can be engaged in protein synthesis but that the association with the small subunit is probably weaker. Thus, these subunits are probably released from the polysomes during centrifugation, yielding halfmers and accumulating free large subunits.
Deletion of MRT4 leads to a 60S ribosomal subunits shortage and the formation of halfmer polysomes (22), therefore the possibility that some of the observed halfmers are due to a partial complementing capacity of the Mtr4 function in 60S ribosomal subunit biogenesis by the Mrt4/P0 chimera can not be excluded.
Analysis of ribosomes carrying the Mrt4/P0 chimera
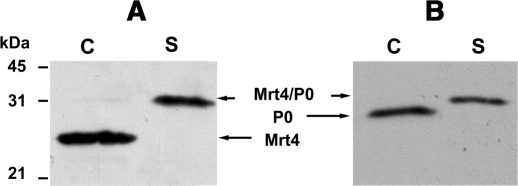
Ribosomes from the transformed W303dMGP0 and parental W303 grown in glucose were resolved by SDS–PAGE, and the presence of P0 and Mrt4 was assessed with the monoclonal antibody 3BH5 specific to the P0 C-terminal domain and a rabbit anti-Mrt4 serum (Figure 5). In the strain expressing the chimera, a protein band was detected in the ribosomes that cross-reacted with both antibodies and that was absent in the particles from the wild-type strain, wherein separate bands corresponding to P0 and Mrt4 were identified.
Figure 5.
Analysis of ribosomes from cells expressing the Mrt4/P0 chimera. Purified ribosomal particles (80 μg) from W303dMGP0-Mrt4/P0 (S) and W303 (C) grown in the presence of glucose were resolved by SDS–PAGE. The proteins were detected by immunobloting using antibodies specific to Mrt4 (A) and P0 (B). Position of the different proteins in the gels and the molecular weight markers are indicated.
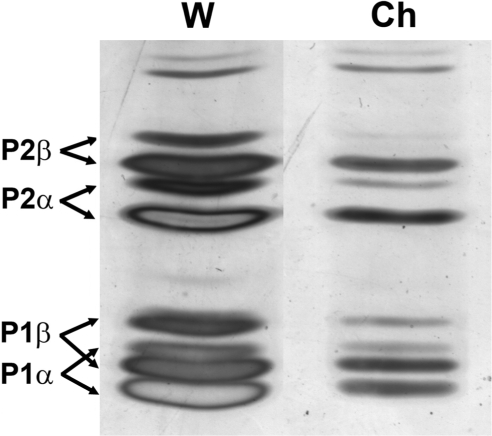
The ribosomes were also resolved by isoelectrofocusing to test whether the other ribosomal stalk components were affected by the presence of Mrt4/P0. A severe reduction in the four, P1α, P1β, P2α and P2β, proteins was evident in ribosomes carrying the chimera (Figure 6).
Figure 6.
Estimation of stalk P1 and P2 proteins in ribosomes containing Mrt4/P0. Ribosomes (200 μg) from W303 (W) and W303dMGP0-Mrt4/P0 (Ch) grown in the presence of glucose were resolved by isoelectrofocusing in a 2.5–5.0 pH range. Unidentified bands in the upper part of the gels, which can be used as input control, indicate that a comparable amount of ribosomes was analyzed in both samples.
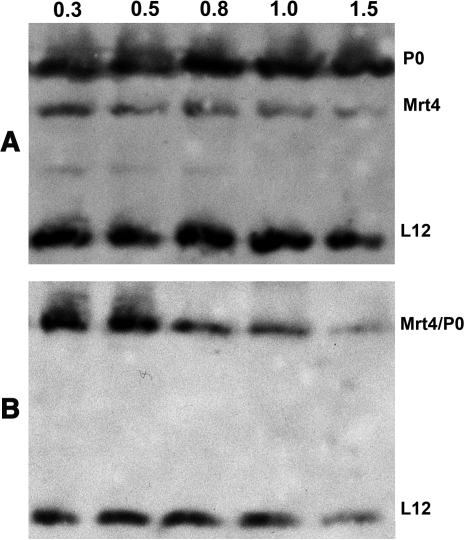
The P0 protein binds tightly to rRNA and in contrast to its prokaryotic counterpart, the L10 protein, it is only weakly displaced from the ribosomes by washing with high salt buffers. To test the affinity of the Mrt4/P0 protein, the ribosomes containing the chimera were treated with increasing concentrations of NH4Cl and ribosomes from the parental W303 strain were used as a control. The amount of P0, Mrt4, Mrt4/P0 and ribosomal protein L12 in the treated ribosomes was estimated in immunoblots (Figure 7), while P0 is practically unaffected by the treatment in the control strain, the chimera began to be removed from the ribosomes at a concentration of 0.8 M NH4Cl. The association of the L12 protein was only mildly affected at 1.5 M NH4Cl in the control, while in the ribosomes containing the chimera it is first released from the particles at lower salt concentrations. Moreover, the Mrt4 in the pre-ribosomal particles present in the control ribosomal preparation was also partially released at high NH4Cl.
Figure 7.
Effect of high salt treatment on ribosomal stalk stability. Ribosomes from W303 (A) and W303dMGP0-Mrt4/P0 (B) grown in the presence of glucose were resolved after being treated with increasing NH4Cl concentrations and the amount of P0, L12 and Mrt4 was estimated in immunoblots probed with specific antibodies.
Structure of the P0 and Mrt4 rRNA binding site
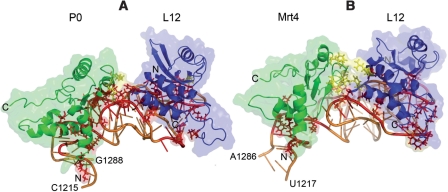
All the previous biochemical data strongly suggest that P0 and Mrt4 bind to the same site in the 25S rRNA ribosomal GAR, as supported by their significant protein sequence homology, predicting that the RNA binding site in both proteins is also likely to be well conserved. Unfortunately, the crystal structure of these proteins has not yet been resolved, information that would confirm this prediction. However, the high resolution structure of the complex formed by the 23S rRNA GAR domain, protein L11 and the NTD of L10E in Haloarcula marismortui is available (44). Given the close homology between these components and their S. cerevisiae orthologues, namely the 25S rRNA GAR domain and the ribosomal proteins L12 and P0, respectively, we have modelled the structure of the equivalent yeast complex and analyzed this through molecular dynamics simulations (Figure 8A). Likewise, a similar complex containing the Mrt4 NTD instead of P0 NTD was also modelled (Figure 8B). A summary of the protein–RNA interactions detected are shown in Supplementary Tables S2 and S3. Although both these complexes were similar, some interesting differences were also detected. The P0 and Mrt4 proteins both interact with the 25S RNA GAR, mainly through the sugar-phosphate backbone and principally at four points around nucleotides G1218, G1229, G1258 and G1281, the equivalent residues to those found in the H. marismortui structure (44). However, in the complex containing Mrt4 there are fewer contacts. From an energetic point of view, and considering that only the relative numbers are really significant, the MM-GBSA analysis of both models indicated that P0 binds a little tighter than Mrt4. Indeed, the predicted binding affinity for the 25S rRNA GAR-L12 complex was of −152.64 (SD 9.53) and −124.93 (SD 9.20) kcal/mol, respectively (Supplementary Figure S2A). However, the most interesting differences were found in the interaction of the P0 NTD and the Mrt4 NTD with the L12 protein in their respective complexes (Supplementary Figure S3). Our model predicts that the L12 protein contacts P0 through a F109L12–K174P0 hydrophobic interaction, as well as a R111L12–R173P0 ionic interaction mediated by the U1257 phosphate group, both at positions equivalent to those reported for the L10E–L11 interaction in H. marismortui (44). In contrast, L12 interacted with Mrt4 at three sites, two of which were similar to those found in the P0 interaction, while a third R117L12–E107Mrt4 ionic contact took place. This stronger interaction seems to pull the three L12 β strands closer to Mrt4, in this way filling the gap between both proteins in the P0 complex and inducing a significant conformational change in L12. Indeed, the predicted affinity of L12 binding was −271.59 (SD 12.8) kcal/mol in the P0 complex as opposed to −175.39 (SD 16.5) kcal/mol in the case of Mrt4 complex (Supplementary Figure S2B).
Figure 8.
Predicted structure of the complexes formed by the S. cerevisiae 25S rRNA GAR, the L12 protein and the NTD of either the Mrt4 (B) or P0 (A). Residues involved in protein–RNA and protein–protein interactions are marked in red and yellow, respectively. C and N indicate the carboxy and amino ends of the proteins.
DISCUSSION
In contrast to the ribosomal P0 protein, Mrt4 is exclusively associated with particles moving to the 60S/80S region in sucrose gradients. Moreover, affinity chromatography purification shows that neither P0 is present in Mrt4-TAP purified particles, nor is Mrt4 present in P0-TAP ribosomes. These results are in agreement with the suggestion that Mrt4 acts as a ribosomal assembly factor and they are compatible with the hypothesis that both proteins compete for the same site in such particles. In fact, the strong amino acid homology in both proteins, especially at the NTD where the P0 rRNA binding site is located (43), indicates that their incompatibility in the ribosome might be due to the fact that both proteins bind to the same site in the nucleic acid molecule. Indeed, in the case of P0 the binding site corresponds to the highly conserved GTPase associated region, the so called GAR domain of the 25S rRNA. A P0 protein carrying the NTD region from Mrt4, the Mrt4/P0 chimera, could partially complement the growth deficiency derived from the absence of the stalk component, which is in full agreement with this proposal. Moreover, Mrt4/P0 is found in the ribosomes from the cells expressing the chimera, indicating that the Mrt4 domain in this protein indeed binds to the 25S rRNA GAR. However, this interaction is apparently weaker since the chimera is released from the ribosomes at lower salt concentrations than those required to release P0, although this might be expected since the Mrt4 protein is also washed off pre-ribosomal particles under less stringent conditions.
The presence of Mrt4/P0 also affects the binding of the P1 and P2 acidic proteins that are present in lower amounts in the ribosomes containing the chimera, probably one of the reasons for the slow growth phenotype of this strain. It seems that the binding site for the P1/P2 heterodimers is altered in the chimera, despite the fact it is located between residues 189 and 260 of the CTD, apparently far from the rRNA binding site. However, this effect is in agreement with previous data showing that modifications in the region downstream of the P1/P2 binding site have a negative effect on the binding of these proteins (43). These long range effects are interesting with regards the acidic protein exchange process that takes place during translation in eukaryotes and indeed, they suggest that the stability of the P0–P1/P2 complex is susceptible to conformational changes in the P0 NTD. If so, cyclical changes induced in P0 during translation, perhaps at the termination stage, could facilitate the release of P1/P2.
Together, the biochemical data suggest that P0 and Mrt4 do indeed bind to the same site in the rRNA. Moreover, they provide experimental support for the proposal that Mrt4 binds first to the pre-ribosome and that it is replaced by P0 at a later stage, as previously speculated for the Rlp24 and Rpl24 proteins (13). However, in contrast to the Rpl24/Rlp24 pair, this exchange is likely to take place in the nucleus since P0 has been reported in nuclear pre-ribosomal particles (14,16,17), and in fact, the P0 protein is not found free in the cytoplasm. How this replacement takes place currently remains unclear, although it probably involves the action of other assembly factors. An ongoing analysis of pre-ribosomal particles from cells lacking Mrt4, not an essential protein, should provide useful information in this respect.
The crystal structure of the ribosomal stalk in yeast mature ribosomes and pre-ribosomal particles is unfortunately not available. However, using the high resolution structure of the H. marismortui ribosomal stalk base (44), we modelled the corresponding S. cerevisiae complex formed by the 79 nucleotide long 25S rRNA GAR, the L12 protein and the P0 NTD, as well as an equivalent complex containing Mrt4. A molecular dynamics analysis of both structures identified expected similarities, as well as some interesting differences. The Mrt4 protein and the P0 NTD interact in a similar manner with the other two components, although the estimated energy of interaction indicates that Mrt4 binding to the complex is weaker, probably due to the fewer contact points with the rRNA. This is in agreement with the biochemical data showing that the assembly factor starts to be released at lower salt concentrations. Nevertheless, from a structural point of view, the most interesting difference lies in the interaction of both proteins with the L12 protein. The contact between Mrt4 and L12 is predicted to be stronger and results in a notable conformational change of the ribosomal protein in the complex. Both proteins come closer together, filling up an empty space existing in the P0 complex (Figure 8 and Supplementary Figure S2). In agreement with these structural data, the estimated binding energy for L12 is notably higher in the Mrt4 than in the P0 modelled complex. The biological significance of these differences is currently obscure since very little is known about the functional role of Mrt4, but our data suggest that this protein might also have an important role in the assembly of the protein L12.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación (Spain) (BFU2006-00365 to J.P.G.B., BFU2007-60151 to J. de la C., BFU2007-64280/BMC to M.R., CSD2006-00023); Fundación Ramón Areces (institutional grant to C.B.M.S.O.). Funding for open access charge: Ministerio de Ciencia e Innovación (Spain).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M.C. Fernández Moyano for expert technical assistance. We acknowledge the generous allocation of computer time at the Barcelona Supercomputing Centre.
REFERENCES
- 1.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 2.Ballesta JPG, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Progr. Nucleic Acids Res. Mol. Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 3.Zinker S, Warner JR. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]
- 4.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 5.Scharf K-D, Nover L. Control of ribosome biosynthesis in plant cell cultures under heat shock conditions. II. Ribosomal proteins. Biochim. Biophys. Acta. 1987;909:44–57. doi: 10.1111/j.1432-1033.1986.tb09971.x. [DOI] [PubMed] [Google Scholar]
- 6.Remacha M, Jimenez-Diaz A, Santos C, Zambrano R, Briones E, Rodriguez Gabriel MA, Guarinos E, Ballesta JPG. The proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 1995;73:959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- 7.Guarinos E, Santos C, Sanchez A, Qiu DY, Remacha M, Ballesta JP. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 2003;50:703–712. doi: 10.1046/j.1365-2958.2003.03733.x. [DOI] [PubMed] [Google Scholar]
- 8.Saenz-Robles MT, Remacha M, Vilella MD, Zinker S, Ballesta JPG. The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta. 1990;1050:51–55. doi: 10.1016/0167-4781(90)90140-w. [DOI] [PubMed] [Google Scholar]
- 9.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 10.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunbar DA, Dragon F, Lee SJ, Baserga SJ. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA. 2000;97:13027–13032. doi: 10.1073/pnas.97.24.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 21.Zuk D, Belk JP, Jacobson A. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics. 1999;153:35–47. doi: 10.1093/genetics/153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 23.Lebreton A, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008;36:4988–4999. doi: 10.1093/nar/gkn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos C, Ballesta JP. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994;269:15689–15696. [PubMed] [Google Scholar]
- 25.Wach A, Brachat A, Pölhmann R, Philippsen P. New heterologous modules for classical or PCTR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 26.Zambrano R, Briones E, Remacha M, Ballesta JPG. Phosphorylation of the acidic ribosomal P proteins in Saccharomyces cerevisiae. A reappraisal. Biochemistry. 1997;36:14439–14446. doi: 10.1021/bi971494o. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose filters: procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JPG. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 29.Santos C, Ballesta JPG. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J. Biol. Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 30.Petitjean A, Bonneaud N, Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal proteins. Mol. Cell Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Madrid F, Reyes R, Conde P, Ballesta JPG. Acidic ribosomal proteins from eukaryotic cells. Effect on ribosomal functions. Eur. J. Biochem. 1979;98:409–416. doi: 10.1111/j.1432-1033.1979.tb13200.x. [DOI] [PubMed] [Google Scholar]
- 32.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. edn. [Google Scholar]
- 34.Dieffenbach CW, Dveksler GS. PCR Primer. A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 35.Rodriguez-Gabriel MA, Remacha M, Ballesta JPG. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J. Biol. Chem. 2000;275:2130–2136. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 36.Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2007;27:1207–1221. doi: 10.1128/MCB.01523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 41.Case D, Darden T, Cheatham TE, Simmerling C, Wang J, Duke R, Luo R, Merz K, Wang B, Pearlman D, et al. AMBER8. University of California, San Francisco: 2004. [Google Scholar]
- 42.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. Calculating structures and free energies of complexe molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 43.Santos C, Ballesta JP. Characterization of the 26S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol. Microbiol. 2005;58:217–226. doi: 10.1111/j.1365-2958.2005.04816.x. [DOI] [PubMed] [Google Scholar]
- 44.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.