Abstract
The organization of the eukaryote nucleus into functional compartments arises by self-organization both through specific protein–protein and protein–DNA interactions and non-specific interactions that lead to entropic effects, such as e.g. depletion attraction. While many specific interactions have so far been demonstrated, the contributions of non-specific interactions are still unclear. We used coarse-grained molecular dynamics simulations of previously published models for Arabidopsis thaliana chromatin organization to show that non-specific interactions can explain the in vivo localization of nucleoli and chromocenters. Also, we quantitatively demonstrate that chromatin looping contributes to the formation of chromosome territories. Our results are consistent with the previously published Rosette model for Arabidopsis chromatin organization and suggest that chromocenter-associated loops play a role in suppressing chromocenter clustering.
INTRODUCTION
The eukaryote interphase nucleus is organized into many functionally specialized regions or substructures such as chromosome territories (1), nucleoli, Cajal bodies and speckles (2). The chromosomes are composed of chromatin, a complex of DNA and proteins. The gene-rich euchromatin and repeat-rich heterochromatin, which differ significantly in sequence content, volumetric DNA density, transcriptional activity and epigenetic modifications (3) form distinct substructures that occupy spatially separate nuclear regions. Heterochromatin is mostly localized at the nuclear periphery and around the nucleolus, and euchromatin at the interior of the nucleus (4–6). While this nuclear organization may appear static, the spatial organization of these substructures most likely involves specific as well as non-specific interactions between dynamic constituents (7).
Specific interactions, such as protein–DNA and protein–protein interactions have been well described (8–10). For instance, heterochromatin has been suggested to localize to the nuclear periphery through interactions with lamina proteins in animal nuclei (11). However, in plants, for which no lamina homologues have been described, heterochromatin still localizes peripherally. Apart from such specific interactions, inevitably non-specific interactions also occur. The architecture of the interphase nucleus is thought to arise by self-organization through both types of interactions (12,13). However, the contribution of non-specific interactions to nuclear organization has so far not been well-characterized. Here, we study this by identifying properties of nuclear organization that can be explained through non-specific interactions. Using molecular dynamics simulations of chromatin and comparing the results with microscopy data, we specifically focus on the question where nuclear substructures, especially heterochromatin and nucleoli, will localize due to the effect of non-specific interactions.
Non-specific interactions, and the entropic effects they give rise to, can, based on arguments derived from statistical mechanics, be expected to play a role in this localization. The nucleus is a crowded environment containing up to 0.4 g/ml of macromolecules (14), which can be regarded as a mixture of large and small particles in a dense solution (15). Entropy plays an important role in determining the localization of such particles through depletion attraction (15). This attraction occurs when the translational and rotational degrees of freedom of each particle are limited by all other particles in a crowded environment. Around each particle a zone of excluded volume exists which is inaccessible to the centers of mass of other particles. When less numerous large particles (such as nucleoli and heterochromatic regions) coexist with more numerous small particles, the total entropy gain of the small particles may outweigh the entropy loss of the large particles when the latter aggregate, thus minimizing their excluded volumes. This leads to an apparent force between the large particles, the depletion attraction. Depletion attraction can also occur between large particles and a confinement wall, since a wall too is lined by a large excluded volume. Depletion attraction has in a nuclear context been implicated to be responsible for grouping DNA polymerases together into replication factories (15) and RNA polymerases into transcription factories (16), but can be expected to affect many other structures or functional compartments as well. We here study the effects of depletion interaction on the position of nucleoli and of heterochromatin in interphase nuclei.
Other non-specific interactions arise from the polymer nature of chromosomes. Each chromosome consists of a DNA chain which is compacted by association with proteins to form a chromatin fiber. The first level of compaction occurs through the formation of nucleosomes, consisting of histone proteins, which associate with DNA forming a fiber of approximately 10 nm thickness. In more condensed chromatin, the nucleosomes form a 30 nm diameter fiber, the exact structure of which is still debated (17). Histone 1 and other proteins stabilize this and other higher order chromatin structures. Chromosomes have a high length to thickness ratio and many internal translational and rotational degrees of freedom and therefore are expected to show behavior typical for confined polymers in solution. For instance, the chromatin chains can be expected to resist intermingling and will influence the localization of other functional compartments in the nucleus through exclusion interactions. However, the behavior of confined polymers in general and the effect of the polymer nature of chromatin on nuclear organization in particular are difficult to predict theoretically. Therefore, behavior of confined polymers has been investigated using soft matter computer simulation approaches (18). These methods have already been applied in a biological context to show that the combination of a physically confined genome inside a rod-shaped bacterium and conformational entropy could fully explain the spatial segregation of duplicated circular chromosomes (19).
In interphase nuclei chromatin is not a purely linear chain. Instead, it forms loops through specific interactions, for instance during regulation of gene expression (20), at boundary elements (21) and in transcription factories (22), and non-specific interactions (23) creating in effect a network polymer. The geometry of network polymers influences their localization and mixing properties (19). The geometry of chromatin has been studied initially in human cells where several models have been proposed to describe how the linear 10–30 nm fiber folds into a higher order structure through loop formation. An early approach was the random walk/giant loop (RW/GL) model (24) which proposes a highly flexible backbone to which giant loops, each comprising several Mbp of DNA, are attached. Monte Carlo simulations based on this model, however, showed that it cannot explain the spatial distance distribution between chromatin markers in interphase nuclei. Therefore the multi-loop subcompartment (MLS) model was introduced which does predict the interphase distances rather well (25). Based on the MLS model the spherical chromatin domain (SCD) model (26) was developed which proposes that multiple loops form rosette-like domains of ∼1 Mb in size that are linked by DNA stretches of 120 Kb. Monte Carlo simulations based on the SCD model have been used to show that chromosomes do not have preferred association with any other chromosome. This was done by comparing theoretical association rates between chromosome territories with experimental data on chromosome territory positions in for instance human and Arabidopsis nuclei (27,28). Here, we use a similar approach based on molecular dynamics to determine by simulation the localization of heterochromatin and nucleoli, and the level of chromosome mixing.
Most previous modeling studies have simulated human nuclei or chromosomes. However, the large size of the human genome and the dispersed localization of its heterochromatin make it less suitable for whole-genome simulations. Our study is based on the model plant Arabidopsis thaliana, which has a small completely sequenced genome of ∼150 Mb (29). The five chromosome pairs occur in territories that are distributed randomly except for chromosomes 2 and 4 which bear the nucleolar organizing regions (NOR) and therefore associate more frequently with each other and the nucleolus (28). On each of these chromosomes the NOR is located close to the telomere of the short chromosome arm. Fifteen percent of the Arabidopsis genome consists of heterochromatin which is concentrated around the centromeres and at the NORs. The chromosome arms are predominantly euchromatic. In the interphase nucleus, the heterochromatic (peri)centromere and NOR regions localize to 6 to 10 chromocenters, which are preferentially located at the periphery of the nucleus (4,30). A model has been proposed in which the euchromatic arms form loops consisting of 0.1–1.5 Mb protruding out of the heterochromatic chromocenter, resulting in a rosette-like structure (30).
We have implemented this chromatin organization model (and for comparison other models in which loops are not associated with the chromocenter) using self-avoiding polymer chains and performed computer simulations based on Molecular Dynamics on these models. Parameter settings such as chromosome lengths and nuclear size, density and heterochromatin content were based on experimental data. We then compared the predictions derived from our simulations regarding the stability of chromosome territories, the localization of heterochromatic chromocenters and the nucleolus with microscopy data of Arabidopsis leaf mesophyll nuclei and found that entropic forces can by themselves explain the localization of the nucleolus and heterochromatin in vivo.
METHODS
Simulation design
Molecular dynamics setup
Chromatin was modelled as a polymer chain consisting of particles and simulations were performed using the ESPResSo (31) software package as described in e.g. in Arnold and Jun (32). Particles are modelled as point centers of a repulsive force, with an interaction range between particles establishing an excluded volume around each particle. The radius of this excluded volume is hereafter treated as the particle radius. For each combination of particle types a Weeks–Chandler–Andersen potential [a Lennard–Jones potential that is shifted and truncated to only include the repulsive part of the potential (33), formula 1] was used to define non-bonded particle-particle interactions. The basic length scale in our simulation is the effective distance between the centers of two of the smallest particles in the simulation, set by the parameter σ. The energy scale ϵ is chosen such that at room temperature (T = 298 K) KBT/ϵ = 1. Analogous to (32) the time unit is τ = σ√(m/ϵ) = 1, where m is the (irrelevant) particle mass. The attractive part of the potential was cut off at 1.12‱246σ by setting shift to 0.25ϵ and setting the remainder of the potential to 0. Bonded interactions were defined by a harmonic spring potential between two particles with distance r according to formula 2. R was set to the sum of the radii of the particles involved, K was used as a configurable parameter to control the elasticity of the bonds. To prevent bond extensions that would allow chains to pass through each other, Kharmonic was set to 50.0.
Formula 1
Formula 2
Starting configurations for simulations were obtained in several ways. For models involving only linear chains, ESPResSo's pseudo self avoiding walk algorithm was used to create the chains. For models with internal loops, geometrical arrangements were used as starting configurations. Similarly, the initial spatial distribution of chains through the nucleus was either determined randomly or followed a geometrical arrangement.
The simulation was performed by successive velocity-Verlet MD integrations with a time step set to 0.01 in the natural units determined by the (arbitrary) mass of the particles, with the temperature controlled by a Langevin thermostat. The simulations took place within a spherical confinement defined by an LJ potential similar to formula 1 between the virtual perimeter of the confining sphere and all other particle. Initially, the radius of the confining sphere was between 3 and 10 times larger than the eventual radius depending on the spatial size of the initial configuration, which had to fit within the confinement. In all cases, simulations started with a equilibration phase in which the initial configuration was allowed to relax while the LJ potential was ‘capped’ in order to avoid excessive repulsive potentials due to high degrees of particle overlap in the original configuration. During the equilibration phase the LJ cap was slowly increased, until the cap exceeded all LJ interactions in the simulation and the cap was removed. Simultaneously, the confining sphere was slowly shrunk until it reached its eventual size. After the confining sphere reached its final size and the LJ cap was removed, all simulations were further equilibrated until parameters such as total energy, end-to-end distance and contour length, and mixing stabilized (Supplementary data Figure 1).
Number of monomers
The maximum computationally feasible monomer resolution for a single simulation was ∼10 kb/monomer (30 000 monomers in total in the simulation) but at this resolution each experiment takes 6 weeks to complete (on 4 AMD Opteron type 248 processors). Since large series of parameter settings had to be evaluated, a lower monomer resolution of ∼75 kb/monomer (4000 monomers in total) was selected at which simulations (each of which lasted at least 107 integration time steps after equilibration) completed within 1–7 days. We note that if one assumes that the chromosomal DNA is well modelled by a yeast-like 30 nm fiber, which has an estimated Kuhn length of about 400 nm (34), the most appropriate monomer size would be estimated as 400 nm/(10 µm Mbp−1) ∼ 40 kb. Unfortunately, there is not enough data available on chromatin structure in Arabidopsis to more accurately determine the proper coarse-graining length. However, polymer physics suggests that the global phenomena discussed in this work, i.e. the relative positioning of the different chromosomes are relatively insensitive to the actual number of monomers (35).The number of monomers in the simulation did prove to have some effect on one of our observables, the mixing score (defined below). In an unmixed state, longer polymers have a higher ratio of internal monomers (which only interact with other monomers belonging to the same chain) to external monomers (interacting with monomers belonging to other chains), resulting in a lower score. However, since scores are normalized through determining the extreme scores of no mixing and perfect mixing, the mixing parameter is corrected for the influence of monomer numbers. To make sure no unexpected monomer number dependence occurs in specific chromatin conformations, we only compared simulations in which the number of monomers was equal (4000 monomers). At a typical monomer volume fraction of 15% and assuming a nucleus 5 μm in diameter, the resulting monomer diameter is 169 nm.
Implementing chromatin chains, chromocenters and loops
Linear chains (LC) and linear chains with chromocenters (LCC) models were implemented by linear chains of monomers with relative lengths as in Arabidopsis (29), with the LCC model including a chromocenter at the genomically appropriate position. Chromocenters were implemented as single monomers with size as measured in Arabidopsis leaf mesophyll cells (see below). The rosette model was implemented by dividing the monomers belonging to the euchromatic chromosome arms equally over the desired number of loops. In the standard situations, 15 loops per chromosome were included because this resulted in loops of 1–2 Mb. This loop size corresponds to the upper range of sizes suggested in (30). In smaller loops, the monomer resolution becomes a limiting factor. In loop size variation experiments, the distribution of monomers over loops was changed but the total number of monomers stayed the same.
Each loop consists of a linear stretch of regular polymer chain, of which both ends are joined to the chromocenter by harmonic springs of which the length has been adjusted to reflect the center–center distance between a chromocenter and a euchromatic monomer. Upon these bonds no rotational constraints are placed.
The loops and chromocenter (LAC) model was implemented by adding a bond between two monomers that are one loop size away in sequence. This creates a loop since the monomers between these two monomers no longer participate in the main polymer chain. All loops were of the same size as the bonds in the rosette model with 15 loops per chromocenter, for comparison.
In the LAC 100% model, bonds were added in all positions that qualify according to the loop size criterion, creating a very short main chain bristling with loops. In the LAC 50% model, there was a 50% chance that a bond was actually created at each of these locations, leading to loops being on average of equal size as the linear stretches in between the loops, and in the LAC 10% model only a 10% chance.
For both Rosette and LAC models, geometrical arrangements were employed as starting configurations. No difference was observed after equilibration between starting configurations in which these geometrical arrangements were randomly distributed through space and configurations in which these models were oriented on a spiral as described in the section on starting configurations below.
The influence of initial starting configurations on mixing parameter
Two types of starting configurations were employed. In the first, linear chains were introduced at random positions in the confining sphere before compression, and the monomers of each chain were positioned by the pseudo random walk algorithm of the ESPResSo software. The other starting configuration was designed to achieve initial spatial separation of all chains and consisted of a geometrical arrangement in which the positions of the linear chains’ middle monomers were distributed over a sphere surface as described by the following equations in polar coordinates: rcenter = 2/3 rconfining sphere, ϕ = 6π ichain/nchain, θ = −9/20 π + π ichain/nchain, with ichain the i-th chain and nchain the total number of chains. The other monomers were positioned on a cylindrical spiral with a periodicity of 12 monomers, a dislocation of one monomer diameter per winding, and a radius of 12/π times the monomer radius. No significant difference was observed in the value of the mixing parameter after 10 000 time units between simulations with these two starting configurations, or by visual inspection of the configurations.
Sampling of ensemble averages of parameters from simulations
Sample configurations were stored every 103 integration steps (1.6 MD time units) after equilibration was achieved. These stored configurations were then used for calculation of ensemble averages of all measured parameters. Since all simulations were performed for at least 107 cycles after equilibration, at least 104 configurations were used to calculate ensemble averages of the parameters, except for mixing, which for computational reasons was, in all but one simulation, calculated only once every 104 integration steps (for the simulation on which Supplementary data Figure 1 is based, mixing was calculated every 103 integration steps).
Derivation of model/simulation parameters from biological data
Calculation of chromatin density in Arabidopsis nuclei
The volume density of all chromatin (the DNA and all attached proteins/nucleic acids) in Arabidopsis nuclei is unknown. However, a lower and upper limit for this density can be estimated. An upper limit derives from the observation that the nucleoplasm is a molecularly crowded solution containing 0.1–0.4 g/ml of macromolecules (7). This would roughly translate to a 10–40% volume density of macromolecules, providing an upper limit to chromatin density. However, not all macromolecules in the nucleus are bound to chromatin.
A lower limit can be derived from estimations of known components of chromatin. DNA itself can be modelled as a cylinder of 300 million base pairs long, 0.33 nm per nucleotide high and 2.4 nm wide. This results in a total volume of 1.8 μm3, or 2.7% of total nuclear volume assuming a spherical nucleus with a diameter of 5 μm (volume 65 μm3). In Arabidopsis chromatin, one nucleosome is associated with ∼200 bp of DNA. The molecular weight of each nucleotide is ∼340 Da, and therefore 200 bp of DNA weighs roughly 135 kDa. These are associated with ∼120 kDa of core histones. Assuming that the specific weight of DNA and proteins is similar, the DNA–histone complex roughly has a double volume compared to naked DNA. Therefore chromatin occupies at least 5% of the nuclear volume. This lower estimate ignores many components of chromatin, such as other chromatin proteins and attached RNA and small molecules. As a compromise between the lower and upper limits, we use a density of 15 percent of the nuclear volume for all simulations.
Measurements of chromocenter size
Using a Zeiss LSM 510 confocal microscope z-stack image series were produced of 45 Arabidopsis thaliana ecotype Colombia leaf mesophyll nuclei expressing H2B-YFP fusion protein (36). Of these, 24 were of sufficient quality to be used for chromocenter size analysis. The nuclear volume occupied by euchromatin and heterochromatin fractions were measured based on a semi-automated thresholding algorithm. Pixels belonging to the nucleus were separated from background pixels by thresholding based on a smoothed background mask. The threshold levels used to distinguish between background, euchromatin and heterochromatin were set by a human operator. The relative volume of the heterochromatic fraction was determined by adding up the volumes represented by each pixel above the threshold and dividing by the total nuclear volume. To calculate the radius of chromocenters relative to total nuclear radius for use in simulations, this value was divided by 10 (chromocenter number in simulations) and subsequently the cubic root was taken.
Calculation of nucleolus position and size
In the same dataset as used for the measurement of chromocenter size, all images containing more than one nucleolus were discarded, leaving 39 images for analysis. Pixels belonging to the nucleus were separated from background pixels by thresholding based on a smoothed background mask. The threshold levels were determined by a human operator. The center of volume of the nucleus was determined as the intersection point of the medial planes of the nucleus in x, y and z dimensions. Thresholding methods were unable to consistently determine the nucleolar radius and center position because noise dominated the signal. Therefore, the center and diameter of the nucleolus were determined by a human operator using the measure function of the ImageJ software package in the z-stack image containing the median nucleolar section. The nucleolar radial position was measured as relative distance of the nuclear center to the nucleolar center compared to the distance from the nuclear center to the nuclear periphery along the line running through both centers. The resulting relative nuclear eccentricity of the nucleolus was binned into 10 subsequent spherical shells of equal volume (taking into account the peripheral excluded volume).
Analysis of simulations
Calculation of chromocenter clustering
Distances were measured between each combination of two chromocenters. A randomly chosen initial chromocenter was assigned to the first cluster and all other chromocenters close (distances < 2.2 times the chromocenter radius) to the first chromocenter were assigned to its cluster. This analysis was repeated for each new chromocenter in the cluster. When no more chromocenters could be assigned to the first cluster, a so far unclustered chromocenter was picked and assigned to the next cluster, after which clustering proceeded in the same way as for the first cluster until all chromocenters were assigned to a cluster.
Mixing parameter calculation
To quantify the amount of mixing in any single simulation state a mixing score was assigned to each monomer. The 10 closest neighbours to the monomer were determined and the mixing score was equal to the amount of unique/different chains these 10 neighbor monomers belonged to. A mixing value was obtained by averaging all monomer scores for the configuration, and to obtain the score for a model, sample configurations were drawn from a simulation at fixed MD time intervals after equilibration and their mixing scores averaged. Error bars show the standard deviation of the averaged mixing scores. To determine the range of MP values that can be encountered in simulation states, the mixing algorithm was applied to two situations representing the extremes of perfect mixing and no mixing. To simulate perfect mixing, all repulsive Lennard–Jones potentials between monomers were switched off, leaving only bond interactions, which in effect meant monomers belonging to different polymer chains did not interact at all. This resulted in an upper limit on the mixing parameter. Minimal mixing was achieved by assigning a 10 times stronger repulsive potential between monomers of different chains than between monomers in the same chain (but with the same interaction range cut-off), enforcing the formation of (stable) chromosome territories. This resulted in a lower limit for the mixing value. Based on these values all scores are linearly re-normalized to a 0 to 1 range. In this way, the mixing parameter becomes independent of monomer number. For alternative ways to quantify mixing, we refer to Supplementary Material.
RESULTS
Computer simulations of interphase chromosomes
To determine the contribution of non-specific interactions on the localization of heterochromatic regions in Arabidopsis, chromatin was modelled as polymer chains and implemented in simulations using the ESPResSo software package (31). The chromosomes were modelled as 10 self-avoiding chains of monomers, with the relative lengths (in base pairs) of the five Arabidopsis chromosomes, and inserted into a confining sphere representing the nuclear envelope. Monomers were scaled to occupy 15% of the confinement volume based on a chromatin density estimation of 15 volume percent (see Methods section). As a first approximation, the chromosomes were composed of identical monomers. As a compromise between resolution and performance, all simulations were performed at a resolution of 75 kb per monomer. Since 75 kb is larger than the experimentally observed persistence length [3–20 kb (37)] of a 30 nm chromatin fiber there is no correlation of chain orientation over stretches of 75 kb. We therefore connected adjacent monomers by simple harmonic springs without rotational constraints (Figure 1, LC model). For a more detailed description of the simulation system and the way simulations were carried out, we refer to Methods section.
Figure 1.
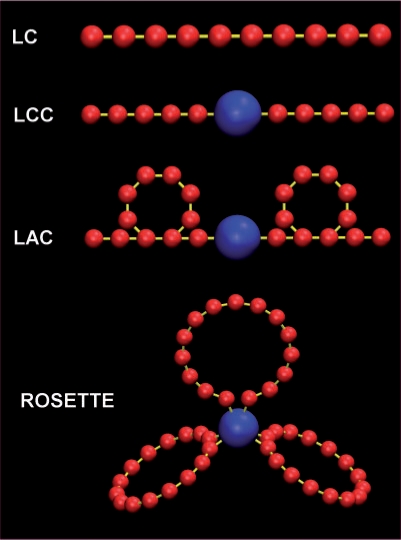
Graphical depiction of the various models. The most simple model is the linear chain (LC) model, in which chromosomes are modelled as consisting of identical monomers (red) arranged in linear chains with harmonic spring potentials (yellow) connecting the monomers. The linear chains with chromocenters (LCC) model is almost identical to the LC model, but models the centromeric area of the chromocome as a large chromocenter (blue). An expansion of the LCC model is the looped arms with chromocenters model (LAC), in which the chains contain loops. In the Rosette model (after Fransz et al. 2002) the chromosome arms loop out from a chromocenter in several loops. Chromocenters, monomers and bonds are not drawn according to scale.
Most of the heterochromatin of an Arabidopsis chromosome is present in the centromeric and flanking pericentromeric regions and form a compact chromocenter. The chromocenters were modelled as a single spherical monomer and positioned in the linear chain at the position of the centromere, resulting in the linear chromosome chromocenter (LCC) model (Figure 1). Since chromocenters in Arabidopsis nuclei vary in shape from near spherical to elongated shapes, we measured their total volume relative to the total nuclear volume in intact leaf mesophyll cells using H2B-YFP expressing seedlings (see Methods section). In these nuclei, chromocenters occupy 6.4% (± 0.02%, n = 24) of the nuclear volume. Dividing this volume equally over 10 spherical chromocenters results in a chromocenter radius of 19% of the nuclear radius (calculation in Methods section). The total amount of monomers was kept at 4000, implying that the euchromatin resolution slightly increased to 65 kb/monomer).
We expanded the LCC model by including chromatin loops in two different ways (Figure 1), based on previously described models of interphase chromatin organization. We thereby assume that loops are formed by a so far unknown mechanism that favors intrachromosomal loops over interchromosomal loops. In Arabidopsis, FISH data suggest that chromosomes contain euchromatic loops of ∼0.1–1.5 Mb emanating from chromocenters (30), which we implemented in simulations (the Rosette model). In our model, we initially set the loop size to ∼1.5–2 Mb (in order to restrict the number of monomers needed to provide sufficient resolution in the loops) and included 15 loops per chromosome (Figure 1). The terminal monomers of each loop are attached to the center of the chromocenter by a harmonic spring potential, allowing these loop attachment points to slide over the chromocenter surface freely.
The simulations of the Rosette model were compared with simulations of a model in which loops exist, but do not attach to chromocenters. Loops were introduced in the arms of chromosomes by adding additional harmonic spring potentials between monomers not in sequence, resulting in the loops and chromocenter (LAC) model (Figure 1, Methods section). Several variations have been tested: a model where every monomer is in a loop (LAC 100%), one where 50% of the monomers are incorporated into loops, and one in which only 10% of the monomers are incorporated into loops (see Methods section). The loop size was again set to 1.5–2 Mb.
Time traces of the total energy, end-to end distance, contour length and mixing in a LC model simulation show that equilibration takes about 1.6 × 104 MD time units (107 integration cycles, Supplementary data Figure 1). The bell-shaped curves of fluctuations around mean values of these parameters after equilibration (Supplementary Figure 1) and autocorrelation curves of these fluctuations (Supplementary Figure 1) show that the simulations are properly equilibrated and that correlations in the fluctuations decay over a time span on the order of 102 MD time units, short compared to total simulation time (4.2 × 104 MD time units), thus allowing sufficient sampling for equilibration statistics.
We first examined how the various models behaved with respect to the formation of stable chromosome territories. Therefore, we quantified mixing from a sample of equilibrated simulation states from each model by calculating a measure for mixing (mixing parameter, MP) from the frequency of interaction between monomers of different chains (see Methods section), which are normalized on a scale from 0 (no mixing) to 1 (full mixing, Supplementary data Figure 2). In a simulation with linear chromosomes (LC model) at a 15% volume density, mixing occurs to a MP value of 0.7. Because of uncertainty in the actual in vivo chromatin density (Methods section), simulations at different densities were also performed. These show that there is a positive relationship between chromatin density and mixing (Supplementary Figure 3). Mixing already occurs to a MP value of 0.35 at 5% density, which represents an absolute minimal value for chromatin density in vivo (calculation in Methods section) and MP reaches 0.9 at a density of 30%. Simulations of the LCC model showed that the addition of chromocenters has no significant effect on the mixing behaviour of the polymers (at 15% density MP values are 0.67 and 0.68, respectively). However, the level of mixing of looped chromosomes is theoretically expected to be reduced (26), because polymers that contain internal branches or loops are expected to mix less than linear polymers (19). In our simulations, the introduction of loops reduced mixing dramatically: in simulations of the Rosette model (15 loops) the chromosomes mix to a MP value of only 0.07. Variation of the number of loops in the Rosette model showed that the level of mixing is dependent on the number of loops, but that even in simulations in which chromosomes had just 3 loops, the level of mixing was already reduced from a MP value of 0.7 to a value of 0.35. Simulations of the LAC model lead to similar results: an increasing percentage of monomers in loops leads to progressive decrease in the amount of mixing. The Rosette and LAC 100% models, which have identical loop sizes and loop numbers, both almost completely prevent mixing (Figure 2 first column, Supplementary Figure 4).
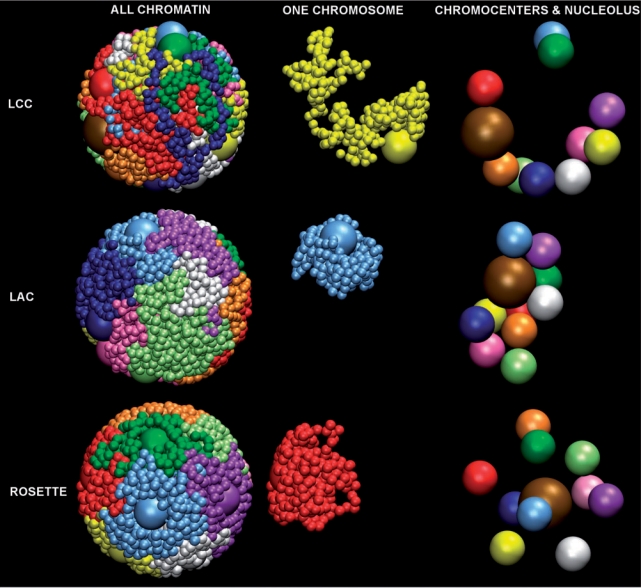
Figure 2.
Overview showing orthographically rendered snapshots of single configurations from LCC (top row), LAC (middle row) and Rosette (bottom row) model simulations containing 10 chromosomes, with chromocenters, in different colors. Of each configuration, three images are provided: left column shows all monomers in the simulation, middle column shows one chromosome, and the right column shows the localization of chromocenters and nucleoli (brown) only.
Chromocenter positions in simulations of the various models
In Arabidopsis, chromocenters preferentially are found within the nuclear periphery, and 2 or more chromocenters can be fused as between 6 and 10 spatially separated chromocenters are usually observed (4,30,38). The localization and fusion of chromocenters was analyzed in simulations of models of the previous section (LCC, LAC, Rosette models). Average radial positions of chromocenters were determined for each model, and distances between chromocenters were measured to determine the frequency of fusion (Methods section).
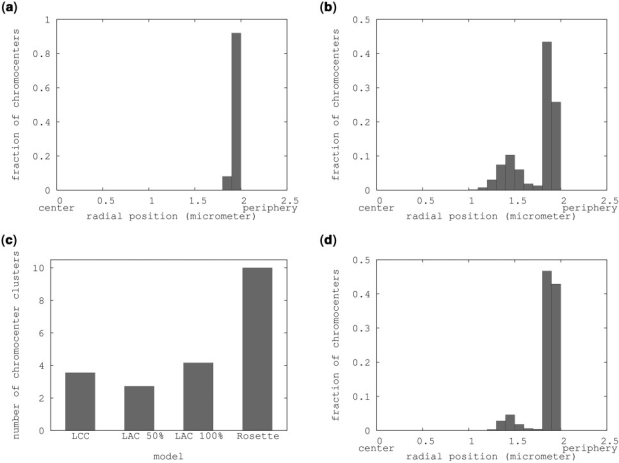
In all three models, the chromocenters preferentially localize to the periphery. However, the models differ in chromocenter distribution over the periphery. For the LCC model, this results in peripheral localization of all chromocenters (Figure 3a). Clustering analysis (Supplementary Figure 5) shows that most chromocenters cluster together in large groups of 3–6 chromocenters. On average about four clusters are present (Figure 3c).
Figure 3.
Chromocenter localization and clustering. (a) Histogram showing the fraction of chromocenters in each radial position bin in a LCC model simulation. (b) Distribution of chromocenter radial positions in the Rosette model. (c) Clustering analysis on the simulations of (a and b) showing the average amount of chromocenter clusters in each simulation. (d) Distribution of chromocenter radial positions in the rosette model including a 1.5 μm nucleolus.
A similar localization and clustering is observed in simulations of the LAC models. Chromocenters in the LAC model mostly localize into one or two big peripheral clusters (Figure 3c, Supplementary Figure 5).
In simulations of the Rosette model, the radial chromocenter distribution is bimodal, with one large peak representing peripheral chromocenters and the other smaller peak an inner shell of chromocenters (Figure 3b). The preferred localization of chromocenters is on the periphery of the nucleus and chromocenters only localize more internally when the outer shell is filled with chromocenters and their associated loops (Supplementary Table 1). Clustering analysis of simulations of the Rosette models revealed that chromocenters with 5 or more loops do not associate with other chromocenters (Figure 3c), but chromocenters with 3 loops can occasionally form clusters of 2 chromocenters (data not shown).
While all models predict the radial localization of chromocenters correctly, the LCC and LAC models predict association of many or even all chromocenters (Figure 2), thus reducing the number of chromocenter clusters to 4 per nucleus or even less. This is lower than the 6 to 10 chromocenters that are normally observed in Arabidopsis nuclei. The Rosette model never shows any clustering (Figure 2), while in most Arabidopsis nuclei some clustering occurs. Furthermore, the inner shell of chromocenters that was found in simulations of the Rosette model does not occur in Arabidopsis nuclei. So additional parameters must affect the behavior of chromocenters. Since the nucleolus forms a large excluded volume in nuclei and therefore could affect the positioning of chromocenters, we included it in the simulations.
Effect of nucleolus on chromocenter positions
The nucleolus is the most conspicuous sub-nuclear structure. Due to its size, the nucleolus represents a significant excluded volume within the nucleus. Therefore it is to be expected that the nucleolar position is determined by non-specific interactions and that the presence of a nucleolus influences the localization of chromocenters. To test this, we measured nucleolar positions and sizes in Arabidopsis nuclei and compared the results with those obtained in simulations.
To visualize the nucleolus in vivo, Arabidopsis plants expressing a H2B-YFP construct were used (36). Whole, living seedlings were observed using a confocal microscope. In z-stacks made of nuclei of leaf mesophyl cells the nucleolus can readily be observed as a spherical region of low fluorescence within the nucleus. Nuclei with more than one nucleolus (which occurred in ∼10–20% of nuclei) were omitted from the data analysis.
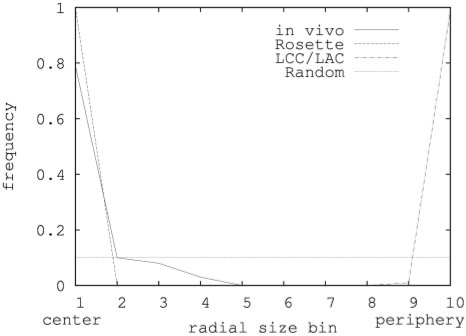
To determine the nuclear/nucleolar radius and nucleolar radial position, a threshold-based automated approach was adopted (see Methods section). This analysis revealed that in Arabidopsis leaf mesophyl nuclei the nucleolus/nucleus radius is 0.30 (±0.05, n = 39). The nucleolus was found to be localized in most cases at or near the centre of the nucleus (Figure 4).
Figure 4.
Nucleolus positions in Arabidopsis. On the horizontal axis nucleolus position bins are shown each representing a shell of 0.1 times the nuclear volume available to the nucleolus. On the vertical axis, the fraction of nucleoli in each bin is shown. The solid line shows measured positions in A. thaliana mesophyl nuclei, the dashed line shows the prediction derived from the Rosette model, the line with alternating dashes and dots shows predictions from the LAC model, and the dotted line shows random localization (assuming the nucleolus to have an equal chance to localize to every available position in the nucleus).
Models were designed to simulate the behaviour of the nucleolus in silico. A nucleolus was added to the Rosette (15 loops) model as a sphere of 0.30 times the nuclear diameter. As a first approach the nucleolus was not attached to the chromatin and this resulted in a central position of the nucleolus in the nucleus (Figure 2). In addition, in the presence of a nucleolus more chromocenters localize to the periphery compared to simulations of the Rosette model without a nucleolus (Figure 3b and d). The small amount of internally localized chromocenters in Figure 3d is due to chromocenters that remain there only for short periods of time during the simulation. Clustering of chromocenters does not take place.
In the same way as for the Rosette model, a nucleolus was introduced into the LCC and LAC 100% models. In these simulations the nucleolus localizes to the nuclear periphery (Figure 2, Figure 4). So only the Rosette model is consistent with the central nucleolar position observed in vivo.
In Arabidopsis, the nucleolus organizing regions (NORs) are located at the ends of the short arms of chromosomes 2 and 4. Since usually 2–3 of the NORs are associated with the nucleolus (30), we tested the effect of attaching the monomers at the ends of all NOR containing arms to the nucleolus on the localization of nucleolus and chromocenters in the Rosette model. This did neither influence the localization of chromocenters nor that of the nucleolus (data not shown).
In FISH studies, telomeres were found exclusively localized around the Arabidopsis nucleolus (30). In the LCC/LAC and Rosette models, the terminal monomers do not have a preferred localization (data not shown). Hypothesizing that the in vivo localization is caused by physical interaction between telomeres and the nucleolus, we attached all chromosome ends to the nucleolus in the Rosette model. This again did neither affect the localization of the nucleolus nor the chromocenters.
DISCUSSION
By implementing models for nuclear organization in molecular dynamics simulations, we show that non-specific interactions are sufficient to explain the peripheral localization of heterochromatic chromocenters and central localization of nucleoli in interphase Arabidopsis nuclei. It is therefore not necessary to explain this localization through specific interactions such as between heterochromatin proteins and the nuclear envelope or lamina (39,40) and the involvement of nucleolar proteins in a nuclear matrix (41). Interestingly, chromocenters localize peripherally regardless of the way euchromatin arms are represented: as linear chains attached to the chromocenter (LCC model), chains containing loops (LAC model), or as loops emanating from the chromocenter (Rosette model). This implies that the peripheral chromocenter localization of other species than Arabidopsis could be explained through depletion interactions even if their chromosomes do not form rosette structures. Also, even in species or cell types where heterochromatin does not form chromocenters, heterochromatin is still more condensed and less dynamic than euchromatin. Such a difference in structure could lead to depletion attraction between the less dynamic heterochromatin and the nuclear periphery or the nucleolus, leading to spatial separation of eu- and heterochromatin such as observed in for instance human nuclei.
In animals, many interactions between lamina proteins and heterochromatin proteins have been discovered (40). However, the existence of these interactions does not imply that they are the direct or the sole cause of peripheral heterochromatin localization. Here, we show that peripheral heterochromatin localization is expected to occur even without the presence of any lamina proteins. Interactions between lamina proteins and heterochromatin may further stabilize this localization in animals and may be important to keep heterochromatin away from nucleopores, where active transcription occurs (42).
Predictions of the nucleolar localization in simulations based on the Rosette model fit more closely to those observed in vivo than in simulations of the LAC model. So an Arabidopsis model of chromatin organization in which euchromatic loops extend from heterochromatic chromocenters (Rosette model) has sufficient self organizing potential, whereas an organization in which loops do not attach to chromocenters (LAC) would require specific interactions for nucleolus positioning.
However, neither model can fully explain the observed clustering of chromocenters. Clustering of chromocenters could be expected based on depletion interactions, because it would result in an entropic gain. In the LAC model, that is exactly what happens. However, in the Rosette model clustering of chromocenters does not occur because the emanating loops prevent depletion interactions between chromocenters. The in vivo situation, in which chromocenters cluster to a limited extent and form approximately 6 to 10 spatially separate structures, is somewhere in between the extremes predicted by our models. Limited clustering is observed in Rosette models that include few chromocenter-emanating loops (Supplementary Figure 2). While the loop sizes in these simulations (∼5 Mb) are larger than observed in vivo [1.5 Mb (30)], DNA is more flexible than our simulated polymers and therefore might permit some limited association between chromocenters surrounded by loops of sizes as observed in vivo.
Alternatively, the in vivo clustering could be explained by hypothesizing that limited chromocenter clustering occurs when the Rosette structure is not fully present, for instance during chromosome decondensation after mitosis and during major developmental cell fate changes. Chromocenter clustering in many species is a dynamic process associated with cell differentiation and other major developmental cell fate changes (43). In Arabidopsis, chromocenters partially decondense during floral transition (44) and upon dedifferentiation after protoplastation (45). Presumably, Rosette loops also dissociate when chromocenters decondense. Our results on the clustering of Rosette structures with few loops show that shielding becomes less efficient when fewer loops are present and that in the case of three loops per chromocenter, chromocenters can cluster. Therefore rosette structure disruption (loop release) during major cell fate switches might allow chromocenters to cluster.
Here, we show that chromatin loops also have a major effect on formation of separated chromosome territories, as was predicted in ref. (25). We have provided quantitative proof that in the absence of chromatin loops, chromosomes mix. The introduction of loops in the megabase size range proved to be sufficient for chromosome territory stability in our simulations. Two parameters determine the amount of mixing in looped chromatin: average loop size and the percentage of monomers in loops. Both the Rosette and LAC models achieve similar MP values when loop amounts and sizes are the same (15 loops per chromosome/loop size ∼1.5–2 Mb), indicating that the MP value is not dependent on a specific loop model. This indicates that preventing mixing by loop formation may be a universal mechanism irrespective of how loops are formed in a certain species. Recent work has provided computational evidence that the inability of human chromosomes (represented as a 30 nm fiber) to fully mix is caused by the slow relaxation kinetics of very long entangled polymers involved (46). This would imply that chromosome territory formation could in fact also be explained by kinetic arguments, rather than being a consequence of equilibrium statistics as we argue is the case here. In that respect it should be pointed out that not only are the Arabidopsis chromosomes significantly shorter than human ones, they also have, due to the presence of the chromocenters, a markedly different topology. In this case, one expects the relaxation time to be dominated by the size of the longest loop present, which is at least an order of magnitude smaller than the total chromosome length. Due to the very strong length dependence of the relaxation time (∼ L3) this would already imply a reduction in timescales of three orders of magnitude. Moreover, our chromosomes are confined to a nuclear volume with a radius comparable in size to the radii of gyration of the chromosome arms/loops involved. There is strong evidence that under these conditions polymer dynamics is significantly enhanced with respect to bulk behavior (47), which was not taken into account in ref. (25).
Some recent data indicates that territory segregation is not complete (48). Our models predict different amounts of mixing depending on factors such as density and loop architecture, and quantitative data on the level of mixing in vivo could be used to test them. However, unfortunately such data are not yet available for Arabidopsis. Still, our simulation methods provide a new way to test models of large-scale chromatin organization against biological data and lead to new insights about the effects of large-scale chromatin looping. For instance, it has to our knowledge not been suggested before that chromocenter-emanating loops act as barriers between chromocenters, preventing their coagulation. Our methods can be applied to the chromatin organization of other species, although resolution problems may occur with genomes of larger size than Arabidopsis. To overcome this limitation and to improve the speed of simulations of Arabidopsis nuclei, a more coarse-grained potential to represent larger chromatin chain subdomains could be developed (49).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre for BioSystems Genomics (CBSG); the Netherlands, Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO). The work of B.M. is part of the research program of the Stichting voor Fundamenteel Onderzoek der Materie (FOM), which is supported by the Nederlandse organisatie voor Wetenschappelijk Onderzoek (NWO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Willemse for providing H2B-YFP expressing Arabidopsis plants and A. Arnold for help with simulation design using the ESPResSo software package.
REFERENCES
- 1.Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum. Genet. 1982;62:201–209. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- 2.Handwerger K, Gall J. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Histone modifications and nuclear architecture: a review. J. Histochem. Cytochem. 2008;56:711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Y, Spector D. Centromere positioning and dynamics in living Arabidopsis plants. Mol. Biol. Cell. 2005;16:5710–5718. doi: 10.1091/mbc.E05-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye Q, Callebaut I, Pezhman A, Courvalin J, Worman H. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 6.Solovei I, Grandi N, Knoth R, Volk B, Cremer T. Positional changes of pericentromeric heterochromatin and nucleoli in postmitotic Purkinje cells during murine cerebellum development. Cytogenet. Genome Res. 2004;105:302–310. [Google Scholar]
- 7.Chubb J, Bickmore W. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell. 2003;112:403–406. doi: 10.1016/s0092-8674(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 8.Lorson C, Hahnen E, Androphy E, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1998;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishov A, Sotnikov A, Negorev D, Vladimirova O, Neff N, Kamitani T, Yeh E, Strauss J, Maul G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert M, Matera A. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim. Biophys. Acta. 2008 doi: 10.1016/j.bbamcr.2008.07.018. [Epub ahead of print; 10.1016/j.bbamcr.2008.07.018; 20 July 2008] [DOI] [PubMed] [Google Scholar]
- 12.Misteli T. The concept of self-organization in cellular architecture. J. Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock R. Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model. Biol. Cell. 2004;96:595–601. doi: 10.1016/j.biolcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Bohrmann B, Haider M, Kellenberger E. Concentration evaluation of chromatin in unstained resin-embedded sections by means of low-dose ratio-contrast imaging in STEM. Ultramicroscopy. 1993;49:235–251. doi: 10.1016/0304-3991(93)90230-u. [DOI] [PubMed] [Google Scholar]
- 15.Marenduzzo D, Micheletti C, Cook P. Entropy-driven genome organization. Biophys. J. 2006;90:3712–3721. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marenduzzo D, Finan K, Cook P. The depletion attraction: an underappreciated force driving cellular organization. J. Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tremethick D. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Cacciuto A, Luijten E. Self-avoiding flexible polymers under spherical confinement. Nano. Lett. 2006;6:901–905. doi: 10.1021/nl052351n. [DOI] [PubMed] [Google Scholar]
- 19.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl Acad. Sci. USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel J, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 21.Wallace J, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson D, Hassan A, Errington R, Cook P. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toan N, Marenduzzo D, Cook P, Micheletti C. Depletion effects and loop formation in self-avoiding polymers. Phys. Rev. Lett. 2006;97:178302. doi: 10.1103/PhysRevLett.97.178302. [DOI] [PubMed] [Google Scholar]
- 24.Sachs R, van den Engh G, Trask B, Yokota H, Hearst J. A random-walk/giant-loop model for interphase chromosomes. Proc. Natl Acad. Sci. USA. 1995;92:2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Münkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 26.Cremer T, Kreth G, Koester H, Fink R, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- 27.Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- 28.Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak M, Fuchs J, Schubert I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113:258–269. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- 29.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2001;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 30.Fransz P, De Jong J, Lysak M, Castiglione M, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl Acad. Sci. USA. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limbach H, Arnold H, Mann B, Holm C. ESPResSo - An Extensible Simulation Package for Research on Soft Matter Systems. Comput. Phys. Commun. 2006;174:704–727. [Google Scholar]
- 32.Arnold A, Jun S. Time scale of entropic segregation of flexible polymers in confinement: implications for chromosome segregation in filamentous bacteria. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:031901. doi: 10.1103/PhysRevE.76.031901. [DOI] [PubMed] [Google Scholar]
- 33.Weeks J, Chandler D, Andersen H. Role of repulsive forces in determining the equilibrium structure of simple liquids. J. Chem. Phys. 1971;54:5237–5247. [Google Scholar]
- 34.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser S. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. USA. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Gennes P. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell University Press; 1979. [Google Scholar]
- 36.Willemse J. A Microscopic Analysis of Arabidopsis Chromatin. the Netherlands: Thesis Wageningen University; 2006. Histone 2B exchange in arabidopsis; pp. 45–60. [Google Scholar]
- 37.Langowski J. Polymer chain models of DNA and chromatin. Eur. Phys. J. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- 38.Berr A, Schubert I. Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics. 2007;176:853–863. doi: 10.1534/genetics.107.073270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baricheva E, Berrios M, Bogachev S, Borisevich I, Lapik E, Sharakhov I, Stuurman N, Fisher P. DNA from Drosophila melanogaster beta-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene. 1996;171:171–176. doi: 10.1016/0378-1119(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 40.Taddei A, Hediger F, Neumann F, Gasser S. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 41.Calikowski T, Meulia T, Meier I. A proteomic study of the arabidopsis nuclear matrix. J. Cell Biochem. 2003;90:361–378. doi: 10.1002/jcb.10624. [DOI] [PubMed] [Google Scholar]
- 42.Taddei A. Active genes at the nuclear pore complex. Curr. Opin. Cell Biol. 2007;19:305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Ceccarelli M, Morosi L, Cionini P. Chromocenter association in plant cell nuclei: determinants, functional significance, and evolutionary implications. Genome. 1997;41:96–103. [Google Scholar]
- 44.Tessadori F, Schulkes R, van Driel R, Fransz P. Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J. 2007;50:848–857. doi: 10.1111/j.1365-313X.2007.03093.x. [DOI] [PubMed] [Google Scholar]
- 45.Tessadori F, Chupeau M, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 2007;120:1200–1208. doi: 10.1242/jcs.000026. [DOI] [PubMed] [Google Scholar]
- 46.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Com.p Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin K, Obukhov S, Chen J, Huh J, Hwang Y, Mok S, Dobriyal P, Thiyagarajan P, Russell T. Enhanced mobility of confined polymers. Nat. Mater. 2007;6:961–965. doi: 10.1038/nmat2031. [DOI] [PubMed] [Google Scholar]
- 48.Branco M, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen J, Addison C, Louis A. Polymer solutions: from hard monomers to soft polymers. J. Phys.: Condens. Matter. 2005;17:S3185–S3193. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.