Abstract
Alternative splicing (AS) involving NAGNAG tandem acceptors is an evolutionarily widespread class of AS. Recent predictions of alternative acceptor usage reported better results for acceptors separated by larger distances, than for NAGNAGs. To improve the latter, we aimed at the use of Bayesian networks (BN), and extensive experimental validation of the predictions. Using carefully constructed training and test datasets, a balanced sensitivity and specificity of ≥92% was achieved. A BN trained on the combined dataset was then used to make predictions, and 81% (38/47) of the experimentally tested predictions were verified. Using a BN learned on human data on six other genomes, we show that while the performance for the vertebrate genomes matches that achieved on human data, there is a slight drop for Drosophila and worm. Lastly, using the prediction accuracy according to experimental validation, we estimate the number of yet undiscovered alternative NAGNAGs. State of the art classifiers can produce highly accurate prediction of AS at NAGNAGs, indicating that we have identified the major features of the ‘NAGNAG-splicing code’ within the splice site and its immediate neighborhood. Our results suggest that the mechanism behind NAGNAG AS is simple, stochastic, and conserved among vertebrates and beyond.
INTRODUCTION
Alternative splicing (AS) is now well established as a widespread phenomenon in higher eukaryotes and a major contributor to proteome diversity. Over half of the multiexonic human genes are believed to have splice variants (1,2). Large-scale detection of AS usually involves expressed sequence tags (ESTs) or microarray analysis (1,3). However, due to various sampling biases, not all AS events can be detected by these methods; furthermore, exon arrays usually do not probe short distance events. Moreover, nowadays genomic sequence data is being churned out at a much faster rate than transcript data, that is, several genomes have low transcript coverage. Thus, there is a need for independent methods of detecting AS.
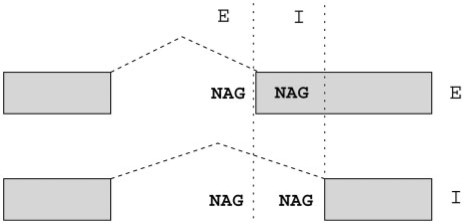
Alternative acceptors are the second most common kind of AS in human, after exon skipping (4). NAGNAG AS, involving tandem acceptors separated by three nucleotides, is a common type of AS, contributing almost half of all cases of conserved alternative acceptor usage (5,6). NAGNAG splicing results in two possible splice variants—splicing after the first AG results in the E (exonic, also known as proximal) isoform, whereas splicing after the second AG results in the I (intronic, also known as distal) isoform (Figure 1)—accordingly, we refer to constitutively spliced NAGNAG acceptors as the E- or I-class, and to usage of both acceptors, or AS, as the EI-class. According to the data present in the Tandem Splice Site DataBase TASSDB (7), 16% (1815 of 10 740) of human NAGNAG acceptors are alternatively spliced. However, 40% (3562) of the remaining NAGNAG acceptors have less than ten ESTs each, thus implying that a subset of these NAGNAGs may simply lack evidence of AS due to insufficient sampling of the transcriptome. An accurate predictive method would give us a meaningful estimate of the number of yet undiscovered alternative NAGNAG acceptors. Previous work on predicting alternative 3′ splicing, while reporting good results overall, had modest results for NAGNAG AS compared to cases involving larger distances (8). This seems to contrast with previous work which reported that a simple model based on splice site strength was enough to explain NAGNAG and other short-distance tandem AS (9).
Figure 1.
NAGNAG alternative splicing. Nomenclature of NAGNAG AS with E and I sites and isoforms.
To improve the prediction of NAGNAG AS, we used Bayesian Networks (BN), which are probabilistic graphical models, and TassDB (7) to carefully construct our training and test datasets. BNs are an increasingly popular machine learning approach to data modeling and classification (10,11). We achieved a high balanced sensitivity and specificity and good results in extensive experimental validation of predictions. We show that the performance on a dataset from literature (8) can be improved by a careful consideration of available transcript evidence to include only strongly supported NAGNAGs as constitutive or alternative. Using a BN learned on human data on six genomes from mouse to worm; we show that the performance is comparable or only slightly inferior to that achieved in human. Our results suggest that the mechanism behind NAGNAG splicing is simple, and maintained in evolution.
MATERIALS AND METHODS
Before describing the materials and methods in detail, we note that an overview of the workflow is provided as Supplementary Data (Supplementary Data File 6).
Feature design and extraction
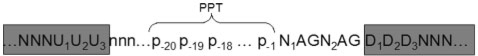
Feature extraction was done using data on NAGNAGs from TassDB (7), using PHP and Perl scripts. The region used for analysis can be seen in Figure 2. Since the composition of the splice site neighborhood influences splicing in general, the base pairs at positions −20 to +3 with respect to the NAGNAG were each used as a single feature, as were the two Ns in the NAGNAG motif. The last three positions of the upstream exon were also included, since they can influence both the process of splicing, as well as reflect any influence of codon usage near the exon boundary. Thus, we had a total of 28 features which each represented a nucleotide, and thus had four possible values (A, C, G, T). A weak polypyrimidine tract (PPT) can contribute to AS, and the number of pyrimidines in the 3′ region of the intron is a measure of PPT strength. Therefore, we designed three features related to the pyrimidine content in the 20-bp region upstream of the NAGNAG: ‘Y-content’, which refers to the number of pyrimidines in this region, ‘MaxY-content’, which is the maximal run of consecutive Ys in this region, and their starting position, ‘MaxY-content position’. Additionally, three more PPT-related features were derived from the 50-bp region upstream of the NAGNAG. Following (12), we measured the maximal number of Ys in a 20-nt window, starting from 50-nt upstream of the NAGNAG. Since U and C are not functionally equivalent, PPTs containing 11 continuous Us are the strongest, and the presence of blocks of purines can be detrimental to splicing (13), we also tested two features called ‘T-strength’ and ‘R-strength’, which measured the longest continuous U (Ts in genomic sequences) and R (A or G) strings, starting from 50-nt upstream of the NAGNAG. Since the architecture of the pre-mRNA plays an important role in constitutive and AS (14), the length of the upstream intron (ending in the NAGNAG motif) as well the length of the upstream and downstream exons were taken as features. Splice site strength, being one of the most important determinants of splicing outcome, was also included as a feature—the strength of the two possible splice sites for each NAGNAG exon, as computed using MAXENTSCAN (15), contributed two more features. Lastly, since GC-content can also play a role in splicing, we measured the GC content of the upstream intron as well as the upstream and downstream exons, leading to three more features. In all, 42 features were used (Table 1).
Figure 2.
Nomenclature of features used in this study. Nomenclature of sequence features used to analyze NAGNAG splicing. The region used to derive all 42 features is shown, along with the names given to the positional features. Positional features, including the last three nucleotides of the upstream intron, were derived using the database TassDB, which in turn used reference annotations (RefSeq when available, else ENSEMBL).
Table 1.
Features for machine learning used in this study
| Feature subset | Number of features | Motivation |
|---|---|---|
| N1, N2, D1, D2, D3 and positions in the PPT | 25 | NAGNAG splicing is influenced by the NAGNAG motif and its sequence context |
| U1, U2, U3 | 3 | Potential influence on protein context |
| Length of neighboring exons and upstream intron | 3 | The architecture of the pre-mRNA influences AS |
| GC content of neighboring exons and upstream intron | 3 | GC content can influence AS |
| Features related to the pyrimidine content of the PPT | 6 | Composition of the PPT influences splicing |
| Splice site strength of E and I splice sites | 2 | Alternative NAGNAGs tend to have comparable splice site strengths |
Analyses with dataset D1
The dataset D1 used in (8) was provided by Martin Akerman. To derive the features, we used the genomic coordinates to find the NAGNAGs in TassDB (7), since it contains information about all NAGNAGs in the human genome (as of early 2006). In order to use an SVM for comparison, since that is what was used in (8), we used the WEKA package and the SMO implementation of SVMs therein, using a polynomial kernel. To begin with, we used the labels as provided in D1, and then we replaced the labels according to TassDB, and finally we replaced the samples labeled constitutive by samples with ≥10 ESTs (for one variant only) from TassDB. Leave-one-out cross-validation was used, as in (8). For feature selection within WEKA, we used the method ‘CfsSubsetEval’, as well as manual inclusion and exclusion of features. We also repeated the analysis with a Bayesian network to ensure that BNs are a good choice for this task, and found that the BNs did match the performance of the SVM.
Datasets derived from TassDB
The dataset D2 of human NAGNAG acceptors was extracted from TassDB (7) using the criteria: (i) constitutive: ≥10 ESTs supporting either E or I variant, 0 for the other; (ii) alternative: ≥2 ESTs supporting each variant, ≥10% of ESTs supporting minor variant (Supplementary Data File 2). The remaining human NAGNAGs were used for prediction only (Supplementary Data File 3). NAGNAG acceptors from the mouse, rat, chicken, zebrafish, fly and worm genomes were extracted in the same manner. Only NAGNAG acceptors from transcripts with a correct exon–intron structure as well as a correct open reading frame were used.
Validation of splicing class assignments using next-generation sequence data
Published next-generation transcriptome sequence data (Illumina GA II) was retrieved from the Short Read Archive section of the GEO database at NCBI (accession number GSE12946) (16).The dataset comprised 313 million 32-mer readings, obtained from cDNA from nine different human tissues and five breast cell lines. For the analysis we required exact matches of the readings to one of the isoforms. Matches had to overlap at least 6 nt on both sides of the exon–exon junction and were discarded if the same sequence occurred somewhere else in the human transcriptome (RefSeq transcripts or NAGNAG isoform sequences).
Bayesian networks
We used the algorithms for feature selection, model learning and classification as described in (17), and made available via the public webserver BioBayesNet (18). BioBayesNet restricts the structure of the BNs by using the so-called tree-augmented naïve Bayes (TAN) structure (19). In contrast to a naïve Bayes classifier/network, where the attributes are assumed to be independent, a TAN classifier augments the underlying naïve Bayes classifier by allowing at most one additional parent per node. Feature selection was carried out in three stages. First, a ‘discretizer’ applying the algorithm of Fayyad and Irani (20) discards features for which no suitable discriminative intervals are found. Secondly, the sequential feature subset selection (SFFS) algorithm (21) was applied. Thirdly, we enforced inclusion or exclusion of features manually.
Experimental validation and quantification of splice variants
For validation and quantification of splice variants, PCRs were performed using 200 pg cDNA templates from the Human Multiple Tissue cDNA Panels I and II (Clontech, Heidelberg, Germany). For each given gene, a suitable tissue was determined from expression data obtained from the Stanford SOURCE database (21). PCR primers were obtained from Metabion (Supplementary Data File 5), each sense primer labeled with 6-carboxyfluorescein (FAM). Reactions were set up with BioMix Red (Bioline, Luckenwalde, Germany) and 10 pmol primer in 25 µl total volume, according to the manufacturer's instructions. The thermocycle protocol was 2 min initial denaturation at 94°C, followed by 42 cycles of 45 s denaturation at 94°C, 50 s annealing at 56°C, 1 min extension at 72°C, and a final 30 min extension step at 72°C. Each product was diluted 1/40, and 1 µl of the dilution mixed with 10 µl formamide (Roth, Karlsruhe, Germany) and 0.5 µl of GeneScan GS500LIZ (Applied Biosystems, Darmstadt, Germany) were heated to 94°C for 3 min. The mixture was than separated on an ABI 3730 capillary sequencer and analyzed with the GeneMapper 4.0 software (Applied Biosystems). If two peaks with about the expected fragment sizes (with a tolerance of ±3 nt) and distance (3 nt) were visible, the isoform ratios were calculated based on the peak areas.
Information gain
Information gain is defined as the reduction in the entropy of the class variable, given the feature. The formula for information gain is:
IG(Class | Feature) = H(Class) − H(Class | Feature)
where H(Class) is the entropy of the class variable, and H(Class|Feature) is the conditional entropy of the class variable, given the feature. Information gain is a well established measure for feature selection in Machine Learning (22). We used the WEKA package (22) for computing information gain, in order to rank the features according to how informative they were. We also used it for prediction based on SVMs, as implemented in the SMO option, and for prediction using Naïve Bayes classifiers.
The BayNAGNAG webserver
We used WEKA to implement the BNs, and C++ code was written to enable the web browser to interact with WEKA, using the features derived from the user's input along with saved BN models to produce the predicted splicing outcome.
Estimating the number of undiscovered alternative NAGNAGs
To estimate the number of alternative NAGNAGs which lack transcript evidence as of now, we used the accuracy of predictions according to the experimental validation, as follows: We computed the average accuracy of prediction in the three probability intervals f1 = 0.5−0.69, f2 = 0.7−0.89 and f3 = 0.9−1.0, according to the experimental results. If fi is the fraction of experimentally validated predictions in the interval i, and ni is the number of samples in the test dataset which are currently labeled as constitutive, but predicted to be alternative, then the estimated number of yet undiscovered alternative NAGNAGs is
N = n1 * f1 + n2 * f2 + n3 * f3.
We used the validation accuracies for two different thresholds (≥1%, and ≥10%) of abundance of the minor variant, leading to two estimates of the number of yet undiscovered alternative NAGNAGs.
RESULTS
Performance on a dataset from the literature
While an SVM reported in (8) succeeded in predicting AS for alternative acceptors separated by up to a distance of 100 nt, NAGNAG acceptors were shown to be the least predictable (8). To understand the reasons behind that, we obtained the underlying dataset from the authors, called D1 in the following. However, in the following we did not use conservation based features, because we aim at predicting AS using information only from a single genome. Using our own set of 42 features (Table 1), we verified that the reported performance is matched by the BN, as well as by an SVM implementation provided in the WEKA package (22). The predicted NAGNAG class is the one which receives the maximum score or posterior probability from the classifier. We computed the receiver operating curve (ROC), which is a plot of the true positive rate versus the false positive rate, and measured the area under the ROC curve (AUC), which is a standard measure of the quality of a classifier (23). An ideal classifier, which makes no errors, would achieve an AUC of 1. By means of the SMO (Sequential Minimal Optimization) implementation of a support vector machine in WEKA and all our features, the AUC obtained for distinguishing EI and E cases is 0.79, the same as reported (8). Using a subset of features (Table 2) yielded by feature selection improves this to 0.82. Similarly, using all 42 features, the AUC obtained for distinguishing EI and I cases is 0.7, the same as reported (8), and this improves to 0.77 using feature selection.
Table 2.
Performance on the dataset D1, using SVMs
| Classification problem | Original sample labels |
Sample labels according to TassDB |
||
|---|---|---|---|---|
| AUC | Featuresa used | AUC | Features used | |
| E versus EI | 0.82 | N1, N2, MAXENT-E, MAXENT-I, D1, p−1, Y-content, | 0.89 | N1, N2, D1, D3, U1, U2, p−8, p−5, p−2, p−1 |
| I versus EI | 0.77 | N1, N2, MAXENT_E, MAXENT_I, D1, p−2, p−1, GC-intron, | 0.85 | N1, N2, D1, D2, D3, U1, U2, U3, p−19, p−18, p−16, p−13, p−12, p−11, p−10, p−9, p−8, p−6, p−5, p−2, p−4, p−3, p−2, p−1 |
aFor nucleotide nomenclature see Figure 2. Y-content: fraction of the 20-bp upstream of the NAGNAG motif that are pyrimidines, GC_intron: G + C content of the intron ending with the NAGNAG, MAXENT_E, MAXENT_I: MAXENT scores for the É and I splice sites.
To check whether this relatively modest performance was due to the set of constitutive NAGNAGs in D1 being in fact contaminated by alternative NAGNAGs, we searched the Tandem Splice Site DataBase (TassDB) (7) for the NAGNAGs in the D1 dataset, and replaced the labels ‘alternative’ and ‘constitutive’ according to TassDB. Indeed, this revealed that many NAGNAGs in D1 labeled constitutive were in fact alternative according to the transcript evidence in TassDB—119 of 397 (30%) cases assigned to the E-class, and 104 of 177 (58.8%) cases assigned to the I-class, are in fact alternative (EI-class) according to TassDB. Incorporating this information resulted in improved performance—the AUCs achieved were 0.89 for distinguishing EI cases from E cases, and 0.85 for distinguishing EI cases from I cases (Table 2).
However, such relabeling still allows samples which have very low transcript coverage and are thus potentially mislabeled also in TassDB, and it also changes the ratios of the sizes of the various classes, especially for the EI versus I problem. Therefore, we replaced all samples labeled constitutive in D1 by samples from TassDB which had ≥10 ESTs supporting one splice site, and none for the other. Since there are only 331 such samples in the I-class, we randomly chose 331 (of 5032) samples from the E-class. This new mixed dataset yielded significantly improved performance, with AUC values of 0.97 and 0.94 for EI versus E and EI versus I, respectively. Removing all NAGNAGs containing a GAG, as done in (8), did not affect the performance drastically, as we obtained AUC values of 0.96 and 0.92 for EI versus E and EI versus I, respectively. Thus, the use of strict thresholds on EST evidence of constitutive splicing greatly reduces the noise in the dataset, and improves the prediction performance. It must be pointed out that we only used transcript evidence for the human genome, that is, some of the alternative cases might be human-specific.
To further validate the relabeling of samples in D1, we analyzed next-generation transcriptome data (Illumina/Solexa GA II), 313 million sequences, obtained from nine different human tissues and five breast cell lines (16) as an additional source of experimental evidence for NAGNAG isoforms. A total of 7509 NAGNAG cases had sequences specifically matching at least one of the isoforms (total of 363009 sequences). We note that the coverage of the transcriptome by these Solexa data is not exhaustive, so there are likely more examples of AS NAGNAGs than thereby supported. We applied stringent filters on the number of sequences supporting an event—these filters had been previously shown to help in the detection of experimentally reproducible AS (24). To consider a NAGNAG to be alternatively spliced, we required at least two supporting sequences for each isoform, and at least 10% of the total sequences to support the minor isoform. A constitutive NAGNAG had to be supported by at least 10 sequences for one isoform, and 0 for the other. We then computed the intersection of this dataset with D1 (Supplementary Data File 8), and compared the labels of the samples. 203 cases of D1 were found in the filtered Solexa dataset—of 142 cases labeled constitutive in D1, 66 (46%) had evidence for being alternatively spliced. When we repeated the comparison after replacing the labels according to TassDB, there were 74 cases labeled constitutive, of which only 12 (16%) were alternative according to the Solexa data. This underscores the need to use thresholds of transcript support for both constitutive and AS as well as confirms our relabeling.
In-silico performance on a TassDB derived dataset
Having seen that sets of constitutive splice events might in fact be significantly corrupted by (not yet detected) alternative acceptors, we decided to take extra care in our selection of human alternative and constitutive NAGNAGs for training data by considering only NAGNAGs which are strongly supported in TassDB. Thus, a NAGNAG was considered constitutive if it had ≥10 ESTs supporting one splice site, and none for the other. To be considered alternative, there had to be ≥2 ESTs for each splice variant, and ≥10% of the ESTs must support the minor variant. Such filtering of alternative events was not required in D1 as another stringent filter—of conserved AS—had already been applied.
This TassDB dataset (called D2 in the following) consists of 5363 constitutive (5032 E, 331 I) and 902 alternative NAGNAGs. We also repeated the comparison with the filtered Solexa dataset (Supplementary Data File 9) as in the previous section—2890 cases of D2 were found in the filtered Solexa data, and of the 2466 cases labeled constitutive, only 37 (1.5%) had evidence of AS in the Solexa data. The much lower number of mislabeled constitutive samples in D2 when compared to the original D1, further justified the choice of stringent filters.
D2 was partitioned into two equal parts, and then, in turn, we used half of the data to train the BNs, and the remaining half was used for testing. The test set remained untouched while the training set was used for discretization, feature selection and learning the BN. Finally, the BN which had been learned on the training set was used to classify the samples in the test set. This procedure was carried out twice, using each half for training and testing in turn, and the average of the two runs was taken as the final performance.
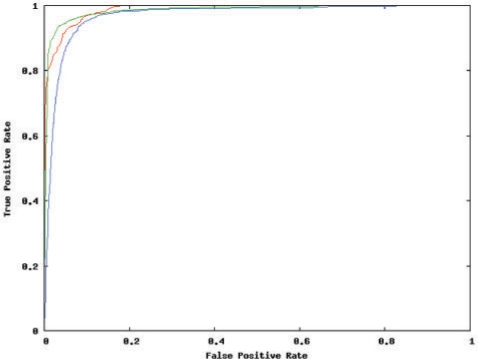
We classified each candidate as belonging to one of the three classes (EI/E/I). The BN achieved AUC of 0.96, 0.97 and 0.98 respectively for identifying EI, E and I variants, as seen in the ROC plot (Figure 3). The balanced sensitivity and specificity obtained was 92%, 95% and 93% (EI/E/I). We would like to note that in contrast to (8), which divided this classification into two sub-tasks, namely predicting EI versus E, and EI versus I, we treat it as a 3-class problem, thus covering all three possible splicing outcomes at the same time.
Figure 3.
In-silico performance of the Bayesian network. ROC plot showing the performance achieved on the 3-class [I-class (red), E-class (green), and EI-class (blue)] classification problem. The I-class is relatively the easiest to predict, whereas the EI-class, or AS, is the hardest.
Another noteworthy difference is that while (8) reported worse performance for distinguishing between EI and I cases, compared to distinguishing between EI and E cases, in the 3-class problem, the highest performance is achieved in predicting the I-class, that is, constitutive usage of the downstream acceptor. This is intuitively easy to grasp, since the scanning mechanism (24) implies that the upstream acceptor is preferentially used, so that constitutive usage of the downstream acceptor is only likely when the upstream splice site is quite weak, for example, when we have a GAGHAG pattern (H = A, C or T). Previous experimental work on 3′ splicing (25), as well in-silico analyses of NAGNAG splicing (26,27) have shown that the nucleotide preceding the AG can influence the choice of 3′ splice site, with the following order of preference: CAG > TAG > AAG > GAG. Consistent with this, 227 of 331 I cases (68.6%) in the D2 dataset have a GAGHAG pattern. The order of preference is also reflected in the sequence logos (28,29) constructed using the D2 data (Supplementary Data File 1).
Removing all NAGNAGs containing a GAG from the training and test sets results in AUC of 0.90, 0.94 and 0.90 respectively for identifying EI, E and I variants. Removing such NAGNAGs can be considered, as GAGs are believed to rarely serve as functional splice sites (8,30), and therefore such NAGNAGs are considered ‘implausible’ for the purposes of AS (31). However, since TassDB contains 182 alternative NAGNAGs of this kind (of which 59 have ≥2 ESTs supporting each variant), we decided to include them. The BN achieves AUC of 0.83, 0.98 and 0.99 respectively for identifying EI, E and I variants on the subset of GAG-containing NAGNAG motifs and predicts 6% of the EST-supported ones to be alternatively spliced. On the other hand, among the currently known constitutively spliced GAG-containing NAGNAG acceptors eight (1.2%) are being predicted to be alternative.
Experimental validation
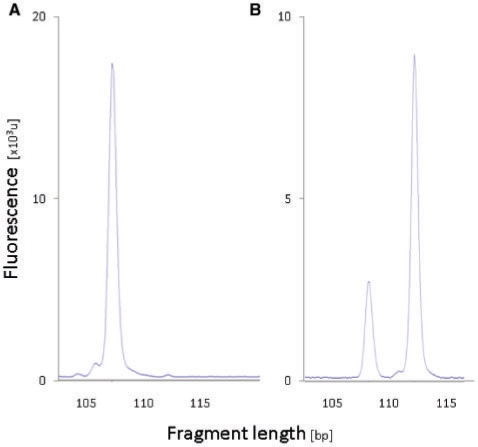
Having established that highly accurate predictions of NAGNAG splicing are possible in-silico, we decided to perform extensive experimental validation of predictions. Experimental validation was performed using RT–PCR followed by capillary electrophoresis with laser-induced fluorescence detection. NAGNAG AS appears in our experimental readout as two fluorescence peaks separated by three nucleotides (Figure 4). To avoid false positive results due to noise, a threshold has to be defined above which the intensity of the minor peak is considered as a robust signal of AS. Accordingly, we measured the accuracy of predictions against the threshold of the isoform ratio, that is, the abundance of the minor transcript (lower peak).
Figure 4.
Experimental validation of predictions using RT–PCR and quantification by capillary electrophoresis. Experimental results indicating (A) constitutive NAGNAG splicing of VPS13D exon 27 and (B) alternative NAGNAG splicing of INPP5E exon 6, minor isoform abundance 24%.
Candidates for experimental work were chosen from both, the entire D2 dataset described in the previous subsection (termed ‘training data’), and the remaining 4475 (913 EI, 3206 E and 356 I) human NAGNAGs in TassDB (termed ‘test data’). The BN learned on D2 uses 14 features (Figure 5) and was applied to classify both training as well as test data (Supplementary Data Files 2 and 3, respectively), and candidates were chosen based on the classification results. Besides the prediction of AS for exons with low EST coverage, we decided to select candidates of several different types, as explained in the following, where P(EI) refers to the probability of being alternative.
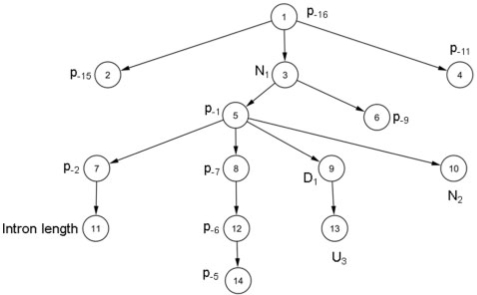
Figure 5.
Bayesian network to predict NAGNAG alternative splicing. The 14-feature Bayesian network learned on the D2 dataset. Note that the class node, which has an edge to all other nodes, is omitted for ease of visualization. Thus, this is just the augmenting tree in the TAN classifier.
Class 1: NAGNAGs from the training data, labeled constitutive, but given a P(EI) ≥0.9 by the BN
As control, constitutive training NAGNAGs with a P(EI) ≤0.1 were chosen. With these candidates, we wanted to test whether the BN can find alternative acceptors even within the ones which had strong transcript support in favor of being constitutive. At a minor variant abundance threshold of 4%, the validation rate is 100% for both cases (6/6; Table 3), and controls (2/2). These results indicate that even strong transcript support (EST coverage ≥10) can miss alternative splice events and cannot ‘prove’ that an exon is indeed constitutive. The highest number of ESTs among these six was 36, for SNTA1 (NM_003098, exon 8) for which we detected 4% usage of the acceptor that was unsupported by ESTs. The highest observed splice ratio of 43% was obtained for C3ORF34 (NM_032898 exon 2) which was originally covered by 21 ESTs confirming the E acceptor exclusively.
Table 3.
Accuracy of prediction against threshold of the minor splice variant
| Threshold of the minor splice variant (%) | Experimentally confirmed predictions of AS |
|
|---|---|---|
| Class 1a (%) | Class 3b (%) | |
| 10 | 50 | 60 |
| 8 | 50 | 90 |
| 6 | 67 | 90 |
| 4 | 100 | 100 |
aSix predictions with P(EI) ≥ 0.9.
bTen predictions with P(EI) ≥ 0.9.
Class 2: NAGNAGs from the training data, labeled alternative, but given a P(EI) ≤0.1 by the BN
As control, alternative training NAGNAGs with a high P(EI) were also chosen. With this class, we wished to identify constitutive NAGNAGs which had erroneous transcript evidence of being alternative. At a minor variant abundance threshold of 4%, the validation rate is 80% (4/5) for cases being constitutive and 100% (6/6) for controls being alternative.
Class 3: NAGNAGs from the test data, labeled constitutive, but given a P(EI) ≥0.5 by the BN
Candidates were chosen from each interval of 0.1 between 0.5 and 1. As controls, two test acceptors, labeled constitutive and with P(EI) ≤0.1 where chosen. The underlying consideration was to test not only the ability of the BN to identify alternative NAGNAGs among the acceptors with low transcript coverage, but also to test whether a higher P(EI) corresponded to a higher accuracy of prediction. The results of the experiments on candidates from this class demonstrates that for a given threshold of isoform ratios, higher posterior probabilities result in more reliable predictions (Table 4). The validation rate for the controls at a minor abundance of 4% is 50% (1/2).
Table 4.
Accuracy of predictions against posterior probability
| P(EI)a | Accuracy of predictions |
|---|---|
| 0.9–1 | 100% (10/10) |
| 0.7–0.89 | 80% (8/10) |
| 0.5–0.69 | 50% (5/10) |
aAbundance of the minor splice variant ≥ 4%.
Class 4: NAGNAGs from the test data, labeled alternative, but given a P(EI) ≤0.1 by the BN
As control, alternative NAGNAGs with a P(EI) ≥0.9 were also chosen. Note that the difference when compared to Class 2 is that these are alternative NAGNAGs with relatively weaker transcript support. For Class 4, the validation rates at a minor abundance of 4% are 83% (5/6) for cases being constitutive, and 50% (3/6) for controls being alternative.
In all, 63 NAGNAGs were investigated (Supplementary Data File 4), and the experiments confirmed that the BN can accurately predict NAGNAG-splicing outcome in 81% (38/47) of candidates. Surprisingly, the validation rate for controls was lower with 75% (12/16).
Most informative features
Next we asked which the most informative features for our classification problem are. By measuring the information gain, we identified the two Ns in the NAGNAG motif, and the splice site scores, to be by far the most informative features (Table 5), which is also in agreement with the literature (8,9,25–27). The downstream N (N2, Figure 2) is the most informative feature, followed by the splice site score of the I acceptor. The next two most informative features are the upstream N (N1, Figure 2), and the splice site score of the E acceptor. The nucleotides immediately upstream and downstream of the NAGNAG acceptor (positions −1, +1, −2 and −3) are the next four informative features, and the nucleotide at position +3 is ranked 10, reflecting the highly localized nature of NAGNAG splicing. The feature ranked 9 is the GC-content of the downstream exon—NAGNAGs whose downstream exon has a higher GC-content are enriched in usage of the I acceptor and correspondingly in alternative NAGNAG splicing.
Table 5.
Top 10 features according to the information gain
| Featurea | Information gain |
|---|---|
| N2 | 0.492 |
| MaxEntScan I | 0.448 |
| MaxEntScan E | 0.252 |
| N1 | 0.199 |
| p−1 | 0.040 |
| D1 | 0.020 |
| p−2 | 0.014 |
| p−3 | 0.005 |
| G/C content of 3′ exon | 0.005 |
| D3 | 0.004 |
aFor nomenclature see Figure 2.
We note that while the splice site scores are very informative, they are not present in the 14 feature BN learned on D2 (Figure 5)—this is because the relevant information is already captured by N1, N2 and the immediate neighborhood. The splice site scores are based on information that also uses positions which are relatively distant from the NAGNAG, and likely not strongly influential on the splicing outcome. Moreover, using the splice site scores introduces a systematic bias against the downstream acceptor, since the ‘PPT’ (polypyrimidine tract) now contains an AG dinucleotide.
Prediction on the mouse, rat, chicken, zebrafish and fly genomes
To test how the BN trained on human performs on data from other species, we first extracted mouse NAGNAG data from TassDB (7), using the same EST-based filtering criteria as for the D2 data above. The performance on the mouse NAGNAG data was nearly identical to that on human (Table 6, Supplementary Data File 10). Encouraged by this, we used the same EST-based filtering in rat, chicken, zebrafish and fly, and predicted NAGNAG splicing using the BN. The performance achieved for the three vertebrates was very similar to that on human and mouse, whereas the performance on fly data, while not as high as that on the others, was still quite good (Table 6). Investigating the cause behind the reduction in performance on the Drosophila genome, we found that excluding positions not in the immediate neighborhood of the NAGNAG—in particular, excluding all features except the two Ns in the NAGNAG, and the two nucleotides immediately upstream, lead to a slight improvement on Drosophila data. This simplified BN trained on human D2 data with just four features, also almost matched the performance of the previous BN with 14 features (Figure 5) on the other five genomes, as well as when evaluated by the above outlined experimental results (data not shown).
Table 6.
Area under the ROC curve for the three classes and six organisms
| Organism | AUC |
||
|---|---|---|---|
| EI | E | I | |
| Human | 0.967 | 0.985 | 0.989 |
| Mouse | 0.966 | 0.982 | 0.989 |
| Rat | 0.967 | 0.985 | 0.991 |
| Chicken | 0.972 | 0.983 | 0.986 |
| Zebrafish | 0.967 | 0.983 | 0.992 |
| Fruitfly | 0.924 | 0.971 | 0.952 |
Prediction on the worm genome
TassDB also contains data from the worm genome, however, there are no examples of constitutive I variants with 10 or more ESTs. We used the 3-class BN with four features (the two Ns in the NAGNAG, and the two nucleotides immediately upstream) trained on human D2 data to predict NAGNAG-splicing outcomes for Caenarhabditis elegans, and obtained AUC values of 0.93 for predicting the EI and E-classes. Only one sample was predicted to belong to the I-class. A 2-class BN trained on human D2 data from only the E and EI-classes produced the same AUC values. A closer look at the data revealed that none of the 391 NAGNAGs (369 E, 22 EI) had G as the upstream N (N1, Figure 2; which is most often the case for constitutive I variants) in the NAGNAG. Thus, it appears that the splice site sequence context is different in NAGNAG splicing in C. elegans, compared to vertebrates. This is in agreement with previous studies that identified an extended 3′ splice site consensus in C. elegans (28).
Performance using a minimal set of features
Since reducing the number of features lead to an improvement in prediction of NAGNAG AS in Drosophila and worm, we asked how many features we could omit without a significant drop in performance on the human D2 dataset. We found that using only the two Ns in the NAGNAG motif, or only the splice site scores (computed by MAXENTSCAN) led to only slightly worse performance. We also found that using a naïve Bayes classifier instead of a BN (with the same features), led to only a minor drop in performance.In order to compare the impact of leaving out features, we compared the error rates of classification using different feature subsets under a 10-fold cross-validation setting with D2. The results show that the error rate is lowest (5.9%) when using only N1, N2, p1, p2 and D1, that is the two Ns in the NAGNAG, and the immediate two upstream and one downstream positions. The error rate using only the MAXENTSCAN scores (7.4%), is higher than that obtained using all features (7.1%), only N1 and N2 (6.7%), or the 14-feature BN we used for the experimental validation (6.3%). We would also like to point out that there is practically no difference in the computational cost of using the various models—the cost of extra features in training the models is not much, and more importantly, once trained, the various models take near-identical time to classify new data.
Webserver and performance on examples from the literature
To further validate our classifier, we tested it on examples of experimentally studied NAGNAGs from the literature (30), which includes interesting examples of tissue-specific variations of the isoform ratio. As shown in Table 7, the results were promising—13/17 (76%) of the cases were predicted to be alternative. An additional 5/7 cases from (29) were also correctly predicted (data not shown). Thus, the performance on these cases from the literature further underscores the usefulness of our classifier. To enable others to do similar experiments as well as reproduce our results and/or predict NAGNAG AS in candidate acceptors of their interest, we developed a webserver—BayNAGNAG, available at: http://www.tassdb.info/baynagnag/
Table 7.
Predictions of the 14-feature BN on experimentally studied cases from the literature (30)
| Gene | Isoform ratios (E:I) in different tissues (30) | P(EI) | P(E) | P(I) |
|---|---|---|---|---|
| DRPLA | 8:2–9:1 | 0.76 | 0.22 | 0.02 |
| GHRHR | 2:8 | 0.92 | 0.04 | 0.05 |
| BAIAP2 | 1:9–0:10 | 0.88 | 0.04 | 0.07 |
| PTMA | 0:10–1:9 | 0.14 | 0.33 | 0.53 |
| IGF1R | 7:3–8:2 | 0.56 | 0.43 | 0 |
| PAX3 | 0:10-10:0 | 0.72 | 0.03 | 0.25 |
| PAX7 | 0:10–9:1 | 0.69 | 0.13 | 0.18 |
| LEP | 1:9–10:0 | 0.61 | 0.38 | 0.02 |
| DNMT1 (Mouse) | 4:6–6:4 | 0.58 | 0.07 | 0.35 |
| CAST | 9:1–10:0 | 0.90 | 0.08 | 0.03 |
| MAN2B1 | 0:10–3:7 | 0.23 | 0.67 | 0.10 |
| PSEN2 | 7:3 | 0.45 | 0.55 | 0 |
| LAP1B | 0:10–10:0 | 0.84 | 0.15 | 0.01 |
| NOXO1 | 0:10–9:1 | 0.08 | 0.91 | 0.01 |
| CCL20 | 4:6–9:1 | 0.80 | 0.18 | 0.02 |
| SGNE1 | 4:6–8:2 | 0.48 | 0.41 | 0.11 |
| TGFA | 5:5–9:1 | 0.93 | 0.04 | 0.03 |
A user can provide a NAGNAG motif along with the upstream and downstream sequence context, the intron length and the last base of the upstream exon. These are then used to predict the class, and the posterior probabilities of all three classes are provided as output. Predictions using two different BNs are provided—one which uses 14 features (Figure 5) and was used in the experimental validation, and the other trained on MAXENTSCAN (15) scores (of the E and I) splice sites only. Furthermore, we also provide an additional file (Supplementary Data File 7) with the required information for all 10 740 human NAGNAGs used in our study.
Estimating the number of undiscovered alternative NAGNAGs
Using the accuracy of predictions according to the experimental validation, we estimate the number of yet undiscovered alternative NAGNAGs in the human genome (10 740 NAGNAGs, 8925 constitutive, 1815 alternative) to be 258–515. The corresponding estimates for mouse (8735, 7386, 1349), fly (1589, 1411, 178) and worm (4697, 4661, 34) genomes are 214–417, 106–214 and 101–185, respectively.
DISCUSSION
We have demonstrated that BNs can produce highly reliable predictions of NAGNAG-splicing outcomes. Once transcript evidence had been carefully considered to create a training dataset, the BN achieved high performance, not only in-silico with a balanced sensitivity and specificity of ≥92%., but also according to extensive experimental validation. Altogether, we investigated the AS of 63 NAGNAGs in one to two tissues and confirmed our predictions in 81% of cases and 75% of controls (4% threshold for the minor isoform). The surprisingly low confirmation rate of controls is primarily due to the 50% (3/6) success rate for low expressed genes (Class 4). Likely, some of these failures are false negatives as AS may take place in other cell types than those tested. In turn, this implies that also some non-confirmed case predictions of AS are false negatives within our experimental setup. Summing up cases and controls with P(EI) ≥0.9, the confirmation rate is 89% (25/28) despite that the just discussed problematic Class 4 controls are included. It is natural to ask why ESTs failed to detect the predicted AS in Class 1 candidates, which was successfully validated by our experiments. In our opinion, some of these cases are easily explained by the low minor abundance, which implies that it is not surprising if a relatively low number of ESTs fails to detect AS. For instance, the NAGNAG belonging to the gene NF1 in Class I has a minor abundance of 0.05, so one would expect to see, on average, 1 EST out of 20 supporting the minor variant. However, since this NAGNAG is only covered by 10 ESTs, it is not surprising that AS is not detected.
To the best of our knowledge, this is the first instance of such extensive validation of in-silico predictions of NAGNAG splicing, and is also among the most extensive experimental validations of non-EST based methods of predicting AS published so far.
The single biggest factor contributing to the performance of the BN was the preparation of the training dataset. As we showed by prediction on the dataset D1 from literature (8), judicious use of transcript evidence, especially a threshold on the number of transcripts required to label an exon as constitutive, makes a big difference. A strict threshold on the EST evidence required to label a splice site as constitutive or alternative is required to minimize the noise inherent in EST databases, and the performance of a classifier can only be as good as the quality of the data that it is trained with.
The most informative features (Table 5) are the two Ns in the NAGNAG motif, and the splice site scores. To some extent, the scores for the upstream and downstream splice sites, and the upstream and downstream Ns can be substituted by each other. The nucleotides immediately neighboring the NAGNAG are the next most important, while other features make only small contributions to the prediction performance. Thus it is evident that most of the information required for prediction is encoded in the immediate splice site neighborhood.
A BN trained on human data achieved near-identical performance on the mouse, rat, chicken and zebrafish genomes, indicating that the determinants of NAGNAG splicing outcome are conserved among vertebrates. Furthermore, the fact that the most informative features were the two Ns in the NAGNAG motif, and its immediate neighboring nucleotides, suggests that the mechanism is simple in nature and maintained in evolution. Given the relatively low transcriptome coverage in rat, chicken and zebrafish, one might ask whether the subset of NAGNAG acceptors we studied for these genomes represent the highly expressed subset of genes and thus likely enriched in conserved alternative events. However, this would not appear to be the case, as we obtain nearly identical results for mouse, which has much higher transcriptome coverage. Thus, our BN should be useful to annotate NAGNAG splicing in animal genomes that currently lack extensive transcript data.
The BN trained on human data was also able to predict NAGNAG AS in the Drosophila genome, though with a drop in performance. However, training using data from Drosophila itself did not improve the performance, indicating that the mechanism may well be conserved between vertebrates and Drosophila. Furthermore, using only four features (the two Ns in the NAGNAG, and the two nucleotides immediately upstream), a BN trained on human data achieved good performance on the worm genome, which contains no instances of the I-class with strong EST support.
This suggests that perhaps what is different in NAGNAG splicing in C. elegans, compared to vertebrates is not the mechanism but rather the evolutionary constraints on the splice site sequence context.
Simpler approaches like using only the two Ns in the NAGNAG motif, or only the splice site scores (computed by MAXENTSCAN), or using a naïve Bayes classifier, led to only slightly worse performance, indicating that the other features and the corresponding dependencies learned by the BN are weak in their discriminative power, and in generalization to other datasets. All this points to a simple and stochastic mechanism, at least in as much as predicting the class (EI/E/I) of NAGNAG splicing is concerned. This is in agreement with (9), who proposed a model based on the sequence context from −6 to +6 at the intron-exon boundary, that is, from −3 to +6 with respect to the NAGNAG, or 15 positions in all. We have shown that the class (EI/E/I) of NAGNAG splicing can be predicted in the vast majority of cases with even fewer positions, that is, −2 to +1 with respect to the NAGNAG, or 9 positions in total. However, the prediction of splice ratios and their tissue and/or developmental stage dependent changes has to involve additional cis and/or trans features and can not be based on a simple stochastic mechanistic assumption. We note that the possibility of such a mechanism does not preclude regulation or a biological function (5,32). Stochastic splice site selection might in fact help production of constant splicing ratios, which have been observed in some NAGNAG sites with clear functional implications (5). At a qualitative level, the stronger splice site seems to correspond to the more abundant variant in most cases, thus supporting a model in which the two splice sites compete for binding to the spliceosome. However, quantitative prediction of the precise abundance is much more challenging. Since NAGNAG AS is frame-preserving (and thus not subject to NMD), save for the ∼2% of the cases which introduce an in-frame stop codon (25), the vast majority of cases should lead to different proteins Studies so far have found evidence of both cases where such proteins have variations in function, as well as those in which there is no noticeable difference, and thus the AS is apparently just ‘tolerated’ by the cell [(5) and the references therein].
We also estimated that there are up to several hundred undiscovered alternative NAGNAGs in the human, mouse, fruitfly and worm genomes. We note that these numbers could be an underestimate, since we only consider predictions with P(EI) ≥ 0.5. Given the current level of annotations of the rat, chicken and zebrafish genomes, genomic information about a substantial fraction of NAGNAG acceptors is likely lacking, therefore such estimation would not be meaningful.
Despite the experimentally validated accuracy achieved in predicting the outcome of NAGNAG splicing at the ‘ternary level’ (EI, E or I), the ‘NAGNAG-splicing code’ is not completely solved. Open questions are the isoform ratios and their tissue specificity observed for several NAGNAGs (25,30,33). Here, sequence features may contribute to the isoform ratio although we consider them uninformative for discrimination at the class level, constitutive versus alternative. Prediction of isoform ratios should also address the influence of the sequence context in the intron and in particular of the branch point on the isoform ratios (27). This is a particularly hard task since computational identification of the branch point is an unsolved issue in the splicing field. Finally, the current limitation in studying isoform ratios is that the available transcript data reflect the natural situations with low resolution. In the future a considerably higher amount of transcript data provided by next-generation sequencing technologies might allow an accurate approximation of isoform ratios and ultimately to decipher the splicing code completely.
CONCLUSIONS
BNs can produce highly reliable predictions of NAGNAG-splicing outcomes once transcript evidence had been carefully considered to create training dataset. This indicates that we have identified, on a qualitative level, the most important features of the ‘NAGNAG-splicing code’. As a BN trained on human data achieved near-identical performance on other genomes from mouse to zebrafish and most of the information needed for prediction is encoded in the immediate splice site neighborhood, we conclude that the mechanism is simple in nature and maintained in evolution, as well as that our BN should be useful to annotate NAGNAG splicing in animal genomes that currently lack extensive transcript data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (SFB604) and the German Ministry of Education and Research (0313652D, 01GS0426, 01GR0504). Funding for open access charge: University of Freiburg.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We thank Martin Akerman for providing the dataset D1 and Nicole Ulbricht for skilful technical assistance.
REFERENCES
- 1.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason Pl, Albrecht M, Hegyi H, Giorgetti A, et al. The implications of alternative splicing in the ENCODE protein complement. PNAS. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 4.de la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiller M, Platzer M. Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Gene. 2008;24:246–255. doi: 10.1016/j.tig.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Sugnet CW, Kent WJ, Ares M, Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pacific Symposium on Biocomputing. 2004;9:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 7.Hiller M, Nikolajewa S, Huse K, Szafranski K, Rosenstiel P, Schuster S, Backofen R, Platzer M. TassDB: a database of alternative tandem splice sites. Nucleic Acids Res. 2007;35:D188–D192. doi: 10.1093/nar/gkl762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akerman M, Mandel-Gutfreund Y. Does distance matter? Variations in alternative 3′ splicing regulation. Nucleic Acids Res. 2007;35:5487–5498. doi: 10.1093/nar/gkm603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chern T-M, van Nimwegen E, Kai C, Kawai J, Carninci P, Hayashizaki Y, Zavolan M. A simple physical model predicts small exon length variations. PLoS Genetics. 2006;2:e45. doi: 10.1371/journal.pgen.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian networks. Nat. Biotech. 2006;24:51–53. doi: 10.1038/nbt0106-51. [DOI] [PubMed] [Google Scholar]
- 11.Beaumont MA, Rannala B. The Bayesian revolution in genetics. Nat. Rev. Genet. 2004;5:251–261. doi: 10.1038/nrg1318. [DOI] [PubMed] [Google Scholar]
- 12.Zhang MQ. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 13.Coolidge CJ, Seely RJ, Patton JG. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox-Walsh KL, Dou Y, Lam BJ, Hung S-p, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl Acad. Sci. USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 16.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pudimat R, Schukat-Talamazzini E-G, Backofen R. A multiple-feature framework for modelling and predicting transcription factor binding sites. Bioinformatics. 2005;21:3082–3088. doi: 10.1093/bioinformatics/bti477. [DOI] [PubMed] [Google Scholar]
- 18.Nikolajewa S, Pudimat R, Hiller M, Platzer M, Backofen R. BioBayesNet: a web server for feature extraction and Bayesian network modeling of biological sequence data. Nucleic Acids Res. 2007;35:W688–W693. doi: 10.1093/nar/gkm292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Machine Learning. 1997;29:131–163. [Google Scholar]
- 20.Fayyad UM, Irani KB. Multi-interval discretization of continuousvalued attributes for classification learning. IJCAI. 1993;2:1022–1027. [Google Scholar]
- 21.Pudil P, Novovicova J, Kittler J. Floating search methods in feature selection. Patt. Recognition Lett. 1994;15:1119–1125. [Google Scholar]
- 22.Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques. 2nd. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 23.Ling C, Huang J, Zhang H. Canadian Artificial Intelligence Conference. Heidelberg: Springer Berlin; 2003. pp. 329–341. [Google Scholar]
- 24.Szafranski K, Schindler S, Taudien S, Hiller M, Huse K, Jahn N, Schreiber S, Backofen R, Platzer M. Violating the splicing rules: TG dinucleotides function as alternative 3′ splice sites in U2-dependent introns. Genome Biol. 2007;8:R154. doi: 10.1186/gb-2007-8-8-r154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- 26.Akerman M, Mandel-Gutfreund Y. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res. 2006;34:23–31. doi: 10.1093/nar/gkj408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai K-W, Tarn W-Y, Lin W-c. Wobble splicing reveals the role of the branch point sequence-to-NAGNAG region in 3′ tandem splice site selection. Mol. Cell Biol. 2007;27:5835–5848. doi: 10.1128/MCB.00363-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollins C, Zorio DAR, Macmorris M, Blumenthal T. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA. 2005;11:248–253. doi: 10.1261/rna.7221605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai K-W, Lin Quantitative analysis of wobble splicing indicates that it is not tissue specific. Genomics. 2006;88:855–864. doi: 10.1016/j.ygeno.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Tadokoro K, Yamazaki-Inoue M, Tachibana M, Fujishiro M, Nagao K, Toyoda M, Ozaki M, Ono M, Miki N, Miyashita T, et al. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: the case of Gln in DRPLA affects subcellular localization of the products. J. Hum. Genet. 2005;50:382–394. doi: 10.1007/s10038-005-0261-9. [DOI] [PubMed] [Google Scholar]
- 31.Tsai K-W, Tseng H-C, Lin W-c. Two wobble-splicing events affect ING4 protein subnuclear localization and degradation. Exp. Cell Res. 2008;314:3130–3141. doi: 10.1016/j.yexcr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Atkinson TP, Dai Y. Activation-induced changes in alternate splice acceptor site usage. Biochem. Biophys. Res. Commun. 2007;358:590–595. doi: 10.1016/j.bbrc.2007.04.158. [DOI] [PubMed] [Google Scholar]
- 33.Schindler S, Szafranski K, Hiller M, Ali G, Palusa S, Backofen R, Platzer M, Reddy A. Alternative splicing at NAGNAG acceptors in Arabidopsis thaliana SR and SR-related protein-coding genes. BMC Genomics. 2008;9:159. doi: 10.1186/1471-2164-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.