Abstract
Cryphonectria hypovirus 1 (CHV1), associated with the picorna-like superfamily, infects the chestnut blight fungus and attenuates the virulence of the host fungus. The genomic RNA of the virus has two continuous open reading frames, A and B, separated by the pentanucleotide UAAUG. We present here evidence suggesting that ORF B is translated from genome-sized virus mRNA by a coupled termination/reinitiation mechanism mediated by the pentamer. In the coupled translation, the overlapping UAA and AUG triplets serve as the stop codon of ORF A and the initiator of ORF B, respectively. This was established by the use of a luciferase assay with a basic construct containing the ORF A sequence and the firefly luciferase gene while retaining the pentamer between the two coding sequences. The proportion of ribosomes reinitiating translation after terminating was determined to be 2.5–4.4% by three independent assay systems in fungal and insect cells. Use of a series of mutant constructs identified two sequence elements, the pentamer and the p40 sequence, that affect the efficiency of coupled translation and virus replication. Together, these results provide the first example of coupled translation facilitated by the pentanucleotide UAAUG in the kingdom Fungi. The mechanism by which the preceding p40-coding sequence promotes reinitiation is discussed.
INTRODUCTION
Eukaryotic mRNAs are generally monocistronic. In concert with other initiation factors, 40S ribosome subunits bind to the cap structure and scan to the first AUG, where in conjunction with 60S subunits they begin translation and continue until they encounter the termination codon (scanning model) (1). However, there are a number of examples of polycistronic mRNAs in which 5′ proximal and downstream open reading frames (ORFs) are translated from single mRNA species (2,3). Such noncanonical translation mechanisms include leaky scanning (4), ribosomal shunt (5), internal ribosome entry site (IRES)-mediated internal initiation (6,7), read-through of termination codons (2), frame-shift (8) and translation coupling (9). The former three mechanisms allow ribosomes access to downstream initiators, while in the latter three a small proportion of ribosomes continue translation of downstream ORFs after or during translation of upstream ORFs. Translation of upstream and downstream ORFs is coordinately regulated and is very likely to be important for their functional roles.
The prototype hypovirus Cryphonectria hypovirus 1-EP713 (CHV1-EP713) (10) is a mycovirus that infects the chestnut blight fungus Cryphonectria parasitica and attenuates the virulence of the fungus in its plant host. This system has attracted attention not only because the virus serves as a biological control agent for chestnut blight (11,12), but also because it is useful for studies on virus/host and virus/virus interactions because of the ease of genetic manipulation of both the virus and its host (12–17). CHV1 is evolutionarily associated with the picorna-like superfamily, which includes the human-infecting poliovirus and the plant-infecting potyvirus (18). The genome of CHV1 is polyadenylated at the 3′ terminus, but whether its 5′ terminus is covalently bound by a viral protein is unknown. CHV1 has two continuous open reading frames (ORFs), A and B, separated by a pentanucleotide, UAAUG (19). The 5′ proximal ORF A encodes the polyprotein p69, which is cleaved cotranslationally into the multifunctional protein p29 and the basic protein p40 by the autocatalytic protease activity of p29 (20). Translation of p69 was shown experimentally to end with the UAA triplet within the pentamer (19). The 3′ proximal ORF B encodes a large polyprotein that also contains a papain-like protease, p48, at the most 5′ proximal region; p48 releases itself from the rest of the ORF B polyprotein (21), which contains replicase-associated domains including RNA-dependent RNA polymerase and RNA helicase (19,22).
Northern blot analysis of ssRNA fractions from virus-infected mycelia with a probe specific for the p29-coding sequence suggested that ORF A is translated from the genome-sized mRNA (23), possibly by an IRES-mediated translation initiation (12,24). However, the mechanism of translation of ORF B has remained enigmatic (12,15,25). ORF B could be translated from subgenomic RNA or polycistronically from full-length transcripts. No extensive search for subgenomic RNAs corresponding to any region of the downstream ORF B has been performed, particularly since their identification would be hampered by the complexity brought about by defective interfering (DI) RNAs smaller than the genome size (26). Shapira et al. (19) predicted coupled translation as a likely mechanism of the translation of ORF B, where the genome-sized plus-sense RNA functions to express both p69 and the ORF B polypeptide. This speculation is supported by the nature of the CHV1 ORF A/B junction sequence, UAAUG. This pentamer is found at the junction of two tandem ORFs in other systems, namely influenza B virus segment 7 (9,27) and the Bombyx mori retrotransposon element SART1 (28), where it is considered to act as a facilitator of coupled translation. Termination/reinitiation has been established for these examples, in which ribosomes translating the upstream ORF stop at the UAA triplet and resume translation of the downstream ORF at the AUG triplet without leaving the template.
We first performed high-resolution northern blot analysis to search for a subgenomic transcript covering ORF B, with negative results. We then developed a protocol with the firefly luciferase (Fluc) gene preceded by the CHV1 ORF A sequence to assess the downstream ORF expression in filamentous fungal cells. We present evidence that the downstream Fluc ORF is translated at 2.5–4.4% of the preceding ORF A by a coupled termination/reinitiation mechanism facilitated by the pentamer. Two sequence elements, the pentamer itself and the p40-coding region, were shown to affect the efficiency of coupled translation and virus viability.
MATERIALS AND METHODS
Constructs for transformation, in vitro transcription and transfection
The starting construct ORFA-Fluc for Fluc assay was generated by the PCR-based overlap-extension method (29) with the high-fidelity KOD DNA polymerase (Toyobo). The entire ORF A domain (map positions 496–2364) and the Fluc gene were amplified separately from plasmid pTNR4 with the primer pair P29-3 and P48-4, and from the pGL3 control vector (Promega) with primers Luc-2 and Luc-12 (Table S1). Map positions on the CHV1 genomic RNA are based on Figure 3 of ref. 19. Taking advantage of the overlapping sequences of primers P48-4 and Luc-2, full-length ORFA-Fluc fragments were amplified with a mixture of two partial fragments as template by use of primer pair P29-3/Luc-12 and KOD DNA polymerase. After digestion with EcoRI and XbaI, the fragment was subcloned into the pBluescript SK+ plasmid vector, and subsequently NotI-digested ORFA-Fluc fragments from pBluescript SK+ plasmid were inserted downstream of the C. parasitica glyceraldehyde-3-phosphate dehydrogenase (GPD) gene promoter of pCPXHY2 or pCPXHY3, derivatives of pCPXHY1 (30), to obtain plasmid ORFA-Fluc (Table S2). Mutant variants of ORFA-Fluc were generated by the overlap extension method using ORFA-Fluc as template and mutagenic primers (Table S1). The common forward and reverse primers P29-3 and Luc-12 were used for the mutants in the first and second PCR reactions.
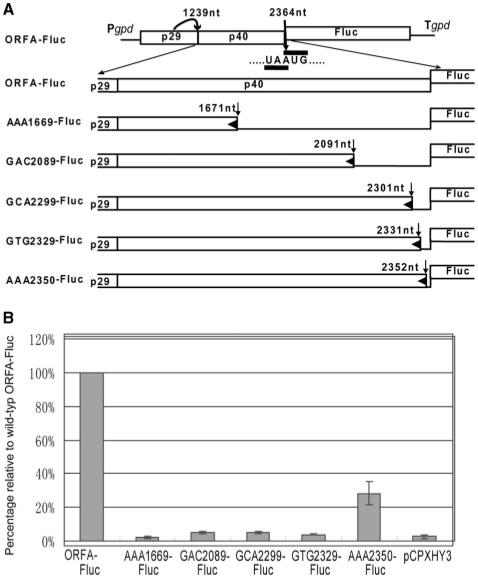
Figure 3.
Effects of insertion of stop codons into the p40-coding domain on levels of Fluc expression from the reporter constructs. (A) Schematic representation of nonsense mutant constructs. A TAA triplet was substituted for AAA (map positions 1669–1671; construct AAA1669-Fluc), GAC (map positions 2089–2091; construct GAC2089-Fluc), GCA (map positions 2299–2301; construct GCA2299-Fluc), GTG (map positions 2329–2331; construct GTG2329-Fluc) and AAA (map positions 2350–2352; construct AAA2350-Fluc) in the context of the basic construct ORFA-Fluc. (B) Levels of Fluc activities in transformants with these constructs were estimated. Mean values and standard deviations were obtained from three data sets. Each set was from 10 independent transformants for each construct.
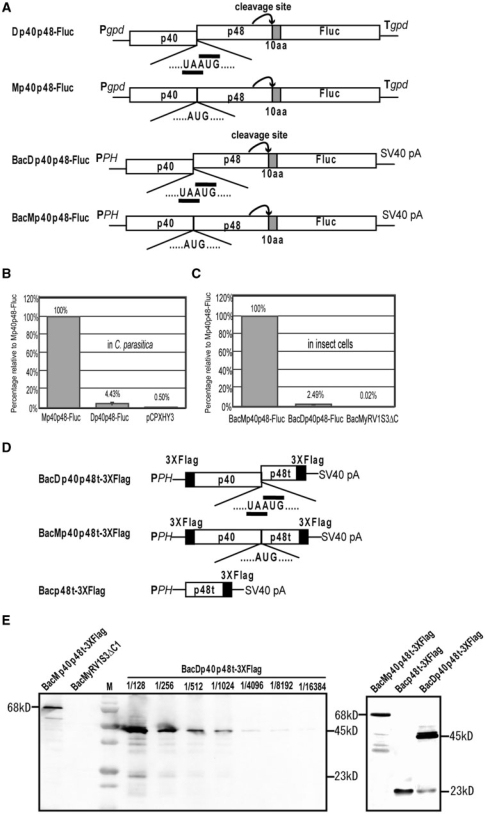
For determination of reinitiation efficiency, three sets of constructs were prepared, each containing a dicistronic and a monocistronic version. The dicistronic construct Dp40p48-Fluc harbors the p40 and p48-coding domain (map positions 1240–3647, ref. 19) and Fluc. After the viral and Fluc components had been amplified separately, they were combined by the overlap PCR method and cloned into the pBluescript SK + plasmid vector. The fragment was subcloned into pCPXHY2 (Dp40p48-Fluc) and the baculovirus transfer vector pFastBacDual (Invitrogen) (BacDp40p48-Fluc). Mp40p48-Fluc and BacMp40p48-Fluc are the monocistronic variants carrying a UA deletion at the pentamer UAAUG, which were made by overlap extension PCR with the dicistronic construct (Dp40p48-Fluc) as template. The third set of constructs is BacDp40p48t-3×Flag and BacMp40p48t-3×Flag. The sequence encoding 3× Flag epitope tag (DYKDHD-G-DYKDHD-I-DYKDHD-DDDK) was added at each end of the p40/truncated-p48 fragment by three progressive PCR reactions using three pairs of overlapping primers. The first and second PCR reactions were performed to amplify p40-truncated p48 fragment (map positions 1240–2909, ref. 19) with the primer pairs Flag5′-1 and Flag3′-1, and Flag5′-2 and Flag3′-2 (Table S1), on plasmid pRFL4. To avoid self-annealing of primers, two partially overlapping fragments (map positions 1240–2753 and 2372–2909, ref. 19) each having 3× Flag at the 5′ or 3′ terminus were generated separately in the third round of PCR with the second PCR products as templates and primer pairs Flag5′-3 and NS23, and Flag3′-3 and p48-17, respectively. These two fragments were ligated taking advantage of the SalI site (map position 2636, ref. 19), and cloned into pGEM-T easy vector (Promega) to obtain Dp40p48t-3×Flag. The insert with the dual tags was subcloned into the baculovirus transfer vector pFastBacDual to obtain construct BacDp40p48t-3×Flag. To prepare monocistronic BacMp40p48t-3×Flag, the 5′ and 3′ terminal regions of Dp40p48t-3×Flag and the middle portion of Mp40p48-Fluc were amplified by PCR with the primer pairs M13F and P40-2, M13F and P40-2 and P40-1 and P48-19. The resulting three fragments were mixed and used as template in overlap extension PCR with the primer set M13F and M13R. The amplified fragment was cloned into the baculovirus transfer vector pFastBacDual to obtain construct BacMp40p48t-3×Flag. Because the p40-coding domain lacks the initiation codon, an AUG triplet was added at the 5′ terminus of each construct.
Full-length and near full-length virus cDNAs were constructed essentially as described by Suzuki et al. (31) and Suzuki and Nuss (32) using the intermediate plasmid pTNR4 that contains the 5′ terminal region of CHV1 cDNA (map positions 1–3704, ref. 19) downstream of the T7 RNA polymerase promoter. Pentamer mutations were introduced into pTNR4 by the overlap extension method to prepare CHV1-UAACG, CHV1-UAAGC, CHV1-UGAUG, CHV1-1A, CHV1-AUG (see Table S2 for sequence information). For NS31, a region spanning map positions 2730–3279 was amplified, digested with BamHI and ApaI, and used to replace the BamHI–ApaI domain (map positions 562–3147, ref. 19) of pTNR4. The 3′-terminal 8.9-kb NheI–SpeI fragment of pRFL4 (covering map positions 3705–12734, ref. 19) was inserted into the intermediate plasmid with modifications.
Sequences and detailed DNA operations for the constructs used in this study (Table S2) are available upon request. All constructs were examined for mis-incorporation that might occur during PCR.
Baculovirus expression of reporter sequences and western blotting
Reporter constructs for determination of ORF B translation efficiency were cloned into the baculovirus transfer vector pFastBacDual (Invitrogen). Reporter sequences on pFastBacDual were moved to baculovirus DNA retained as bacmid in DH10Bac Escherichia coli cells and then transfected into Spodoptera frugiperda cells (Sf9) according to the manufacturer's protocol (Bac-to-Bac Baculovirus Expression System, Invitrogen). Cell culturing and preparation of cell lysates were performed according to Matsuura et al. (33) and Suzuki et al. (34).
Proteins were fractionated on polyacrylamide gel containing SDS in Laemmli's buffer system (35) and electro-transferred onto nitrocellulose membrane. Antigen/antibody reactions were detected as described by Suzuki et al. (34).
In vitro transcription and northern blot analysis
Transcripts were synthesized in vitro as described by Suzuki et al. (31) using cDNA of wild-type CHV1 and two mutant deletion variants. The full-length virus cDNA clone pRFL4 and a p69 deletion mutant, Δp69b, were reported earlier by Suzuki et al. (31) and Suzuki and Nuss (32). An additional mutant virus termed ‘NS31’ contains a further deletion from map positions 567 to 2729 (19). SC57 covers a fragment of cDNA of the CHV1 genome from map positions 2161 to 3709. cDNAs of the pentamer mutants CHV1-UAACG, CHV1-UAAGC, CHV1-UGAUG, CHV1-1A (UAAAUG) and CHV1-AUG are described above. These cDNA plasmid clones were digested with Spe I or Nhe I, and subjected to in vitro transcription with T7 RNA polymerase (Promega). Sizes of transcripts were predicted to be ∼12.7 kb for pRFL4, 10.9 kb for Δp69b and 10.6 kb for NS31.
Total RNA was extracted from CHV1-infected fungal mycelia grown in potato dextrose broth (PDB, Difco) by the method of Suzuki et al. (23). Single-stranded RNA was enriched by two rounds of LiCl precipitation. In vitro synthesized and ssRNA fractions from mycelia were electrophoresed in 0.7% agarose under denaturing conditions for 90 min at 135 V constant voltage in a Mupid mini-gel apparatus (Advance) and transferred to Hybond-N+ nylon membrane (Amersham Biosciences). The blot was pre-hybridized and hybridized with a probe specific for the ORF B sequence at map positions 2364–3619 (19) as described by Suzuki et al. (23).
Transformation and transfection
Spheroplasts of C. parasitica EP155 (ATCC 38755) prepared by the methods of Churchill et al. (36) were transformed with reporter constructs cloned downstream of the GPD promoter in the fungal expression vectors pCPXHY2 or pCPXHY3, derivatives of pCPXHY1 (30). Surviving spheroplasts were regenerated on media containing osmotic stabilizer. Monokaryotic fungal strains were obtained by single conidial isolation.
Transfection was performed by the protocol described by Chen et al. (37). In vitro transcription of viral cDNA was performed as described above. Synthetic transcripts (5 or 15 μg) were electroporated into 5 × 105 spheroplasts using a BioRad Gene Pulser at settings of 1.5 kV, 200 Ω and 25 μF.
Luciferase assay
Fungal strains were cultured on PDA (Difco) overlaid with cellophane (PDA-cellophane) for 3–4 days at 24–26°C. Mycelia were harvested with sterilized toothpicks from PDA-cellophane cultures. Known amounts (10–30 mg) of mycelia were placed into a Röhren tube (Sarstedt) and homogenized with a pestle in the presence of 100 μl of Fluc substrate solution (PicaGene, Toyo Ink) containing a cocktail of proteinase inhibitors (Nacalai Tesque). After 1 min of homogenization, Fluc activity was measured in a Minilumat LB 9506 luminometer (Berthold) over 50 s. For maintenance, strains were cultured on regeneration plates (36) and stored at 4°C in a refrigerator until use.
Spodoptera frugiperda cells were infected with recombinant baculovirus carrying the Fluc gene and were harvested 3 days post-infection by centrifugation at 8000 r.p.m. for 5 min. After being washed with phosphate-buffered saline (PBS), pH 7.2, the resulting pellet was resuspended in 200 μl of lysis buffer (160 μl PBS, 40 μl cell lytic agent; PicaGene) containing the proteinase inhibitor cocktail. The lysate was incubated at room temperature for 1 min. Samples were subjected to centrifugation at 12 000 r.p.m. for 30 s. To assess Fluc activity with the luminometer, 100 μl of substrate solution (PicaGene) containing the proteinase inhibitors was added to 20 μl of the resulting supernatant.
RESULTS
No apparent subgenomic RNA corresponding to ORF B is synthesized in infected mycelia
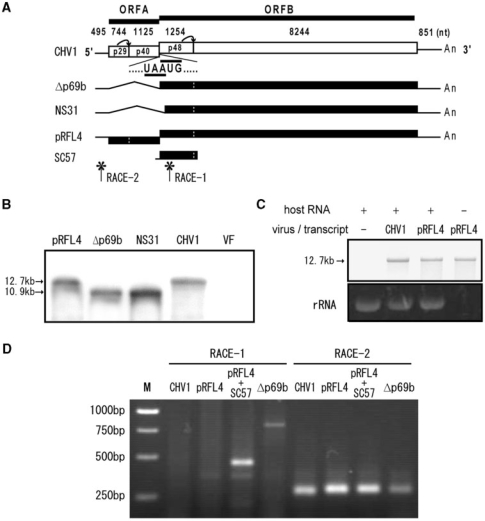
In an attempt to eliminate the possibility that CHV1 ORF B is translated from subgenomic RNA, we performed a northern blot analysis. First, we noted that infection of C. parasitica by CHV1 results in frequent generation of defective interfering (DI)-like RNA smaller than the full-length genome (26). The presence of DI-like RNA would make it difficult to distinguish putative subgenomic mRNA from transcripts from DI-like RNAs. Therefore, we tested over 15 cultures infected with CHV1 for the presence of DI-like RNAs and found that some did not harbor DI-like RNA. The cultures lacking DI-like RNA were subjected to northern analysis. As expected, mRNA corresponding to the entire genome size, with the same migration position as in vitro synthesized full-length viral transcripts (Figure 1A and B, lane pRFL4), was detected specifically in CHV1-infected cultures (Figure 1A and B, lane CHV1), but not in virus-free cultures (Figure 1B, lane VF). We predicted that subgenomic mRNA corresponding to ORF B would be ∼10–11 kb in size, similar to the size markers used in Figure 1B (lanes Δp69b and NS31; 10.9 kb and 10.6 kb, respectively), but no such mRNA was observed in virus-infected cultures.
Figure 1.
Schematic representation of the CHV1 genome, and northern blot and 5′-RACE analyses of CHV1 RNA. (A) The genetic organization of the infectious cDNA clone of CHV1-EP713 (plasmid pLDST) (30) is shown at the top. The plus strand consists of the 5′ noncoding region of 495 nt, the ORF A coding domain of 1869 nt, the ORF B domain of 9498 nt and a 3′ noncoding region of 851 nt, being in total 12 712 nt in length excluding the poly(A) tract (19). ORF A encodes two polypeptides, p29 and p40, which are released from polyprotein p69 by the autocatalytic activity of the papain-like protease domain within p29. ORF B also encodes the papain-like protease p48, responsible for self-cleavage from the N-terminal portion of the encoded polyprotein. The junction between ORF A and ORF B is composed of the pentamer sequence 5′-UAAUG-3′ (map positions 2362–2366, ref. 19), in which the UAA portion is assumed to serve as the termination codon of ORF A and the AUG portion as the 5′-proximal translation initiation codon of ORF B. (B) Detection of CHV1 transcripts by northern blotting. Single-stranded RNA was isolated from CHV1-infected (lane CHV1) and virus-free (lane VF) mycelia (strain EP155), and electrophoresed in 0.7% agarose gel in the TAE buffer system. After being blotted onto nylon membrane, fractionated RNA was probed with DIG-labeled DNA sequence specific for the p48 sequence (map positions 2364–3619, ref. 19) amplified by PCR with p48N1 and p48C2 (Table S1). In vitro synthesized transcripts from viral cDNA clones, pRFL4 (12.7 kb), Δp69b (10.9 kb) and NS31 (10.6 kb), were electrophoresed in parallel and used as size markers. (C) Estimation of levels of CHV1 transcript produced in infected mycelia. Lane CHV1+ was loaded with 1 μg of ssRNA obtained from a deletion mutant of Dicer-like gene 2 (Δdcl-2) (53) infected with wild-type CHV1. DCL2 is required for production for DI RNAs (53), which would make it difficult to identify presumable subgenomic RNA. CHV1 full-length transcripts derived from pRFL4 (60 ng) alone (pRF4–) or mixed with ssRNA (940 ng) from virus-free Δdcl-2 (pRF4+) were electrophoresed in 1.0% agarose gel. Northern blotting was carried out as stated above. (D) Agarose gel electrophoresis of 5′-RACE products. Classic 5′ RACE was performed as described by Hillman et al. (54). Two 5′-RACE reactions (RACE-1and RACE-2) were set with different primer pairs for each of four RNA preparations: ssRNA fractions (1 μg) from CHV1-infected Δdcl2 (lane CHV1) and Δp69b-infected Δdcl2 (lane Δp69b), and in vitro synthesized transcripts (60 ng) from pRFL4 mixed with ssRNA fractions from virus-free Δdcl2 (lane pRFL4) and in vitro synthesized transcripts from pRFL4 (60 ng) and SC57 (370 pg) mixed with ssRNA (940 ng) from virus-free Δdcl2 (lane pRFL4 + SC57). The molar ratio of pRFL4 to SC57 in RNA template pRFL4 + SC57 was adjusted to 20:1. Primers used in cDNA synthesis are SC1 for RACE-1 and RACE-2 shown by asterisks in Figure 1A, while primers used in PCR following oligo (dC) tailing were NS22b and abridged anchor GI for RACE-1 and SC4 and abridged anchor GI for RACE-2. Sequences of primers are shown in Table S1. Positions of NS22b and SC4 are shown by asterisks in (A).
To further confirm the absence of subgenomic mRNA, we conducted classic 5′-RACE of viral RNA obtained from CHV1-infected fungal colonies and in vitro transcription. We first roughly quantified full-length CHV1 mRNA by northern blot analysis using serial dilutions of known amounts of in vitro synthesized CHV1 RNA (data not shown). Consequently, we found that 60 ng of in vitro synthesized full-length viral RNA (pRFL4) provided the same band intensity as CHV1 mRNA in infected mycelia regardless of whether host ssRNA (940 ng/lane) was mixed or not (Figure 1C, lanes pRFL4 + and –). Two sets of 5′-RACE (RACE-1 and RACE-2) were carried out for each of four template RNA preparations: ssRNA fractions (1 μg) from CHV1- (lane CHV1) and Δp69b-infected mycelia (lane Δp69b), and in vitro synthesized viral full-length transcripts (60 ng, lane pRFL4) mixed with ssRNA fractions (940 ng) from virus-free mycelia and in vitro synthesized transcripts from pRFL4 (60 ng) and SC57 (370 pg) mixed with ssRNA (940 ng) from virus-free mycelia (lane pRFL4 + SC57) (Figure 1D). SC57 is expected to have the 5′ end at similar map positions to putative ORF B subgenomic mRNA (Figure 1A). While RACE-1 was designed for amplification of the 5′ end of presumable subgenomic RNA, RACE-2 was for amplification of the 5′ terminal ends of viral genomic transcripts. RACE-2 provided DNA fragments of an expected size (250 bp) with four tested RNA preparations (Figure 1D, RACE-2), but not with RNA from virus-free colonies (data not shown). However, RACE-1 allowed only amplification of DNA fragments of 800 bp for Δp69b and of ∼400 bp for in vitro synthesized SC57 + pRFL4. It should be noted that even lower concentration of in vitro synthesized SC57 present at a molar ratio of 1/20 relative to pRFL4 led to amplification of the DNA fragments of 400 bp (Figure 1D RACE-1, lane pRFL4 + SC57). The 400-bp fragments were shown to be derived from SC57 by sequence analysis. Importantly, no DNA fragment was amplified with ssRNA isolated from CHV1-infected mycelia. The combined results clearly indicated that no subgenomic mRNA is produced during CHV1 infection.
Development of a reporter system in C. parasitica for investigation of translation of ORF B
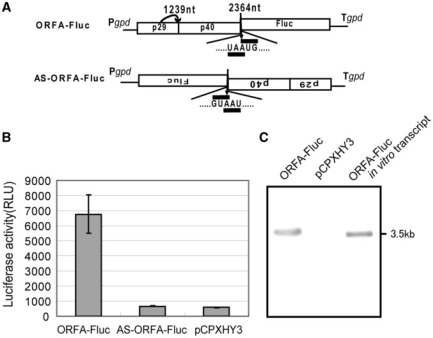
To explore the mechanism underlying translation of CHV1 ORF B, we tested whether Fluc could be expressed from the downstream ORF of CHV1 at a sufficient level for enzymatic detection. The basic construct ORFA-Fluc shown in Figure 2A was prepared and used for transformation of EP155, in which the Fluc gene was linked to the pentamer in frame –1, the same frame as the authentic ORF B, to serve as a reporter. Transcription of ORFA-Fluc was driven by the GPD promoter. Consequently, the chemiluminescence (∼7000 RLU/s) was observed in all hygromycin-resistant transformants with ORFA-Fluc, which showed much higher activities than did transformants with the empty vector, pCPXHY3, or with an antisense construct, AS-ORFA-Fluc (Figure 2B). In order to show that the detected Fluc activities were not from shorter versions of transcripts presumably resulting from internal promoter activities or alternative splicing events, we performed northern blot analysis (Figure 2C). Results showed only one single band at a slightly slower migration position than that of in vitro synthesized transcripts of ORFA-Fluc (∼3.5 kb), indicating no production of shorter forms of Fluc mRNA that may contribute to the Fluc activities in Figure 2B. RT-PCR analyses further confirmed this notion (Figure S1). Together, these results indicate that Fluc is a suitable reporter for investigation of the translation of CHV1 dicistronic mRNA.
Figure 2.
Schematic representation of the basic reporter construct, and luciferase activities of transformants with the constructs. (A) Diagram of the starting construct ORFA-Fluc. ORFA-Fluc, for ORF B translation assay, contains the ORF A domain and the Fluc gene in place of the ORF B sequence, separated by the pentanucleotide. The sequence was inserted into the cloning site of a derivative of pCPXHY1 (pCPXHY3). In this and subsequent figures, Pgpd and Tgpd refer to the promoter and terminator, respectively, of the C. parasitica glyceraldehyde-3-phosphate dehydrogenase gene. AS-ORFA-Fluc is an antisense variant of ORFA-Fluc. (B) Comparison of the Fluc activities of extracts from transformants with the constructs ORFA-Fluc, AS-ORFA-Fluc, and empty vector (pCPXHY3). Mean values and standard deviations were taken from three sets of experiments in each of which at least six independent transformants were used for a single construct. For comparison of translation efficiencies of different constructs, average Fluc activities shown by transformants with the basic construct ORFA-Fluc were defined as 100%. (C) Northern blot analysis of Fluc transcripts. Poly(A) RNA fractions were isolated from transformants with ORFA-Fluc (ORFA-Fluc) and the empty vector (pCPXHY3) using an mRNA isolation kit (Roche). Isolated RNA was electrophoresed and blotted onto nylon membrane as in Figure 1B. The blot was probed with DIG-labeled DNA sequence specific for the p48 sequence (map positions 1493-1653). ORFA-Fluc transcripts synthesized in vitro using plasmid pBS-ORFA-Fluc (Table S2) as template (ORFA-Fluc in vitro transcript) were analyzed in parallel.
Insertion of a stop codon into the ORF A frame abolishes expression of the downstream Fluc gene
We anticipated several possible mechanisms for translation of ORF B from full-length mRNA: IRES-mediated translation, frame-shift or reinitiation. To test these possibilities, point mutations were introduced into the basic construct ORFA-Fluc. A total of four nonsense mutations were introduced at different map positions of the p40-coding domain within ORF A to obtain AAA1669-Fluc, GAC2089-Fluc, GCA2299-Fluc, GTG2329-Fluc and AAA2350-Fluc (Figure 3A). The nucleotide triplet at the position (number) shown in each mutant name was mutated to UAA. These constructs were used for transformation of EP155, and then subjected to Fluc assay. As shown in Figure 3B, all nonsense mutants abolished translatability of the Fluc gene, showing 2.7–4.4% of the Fluc activity of ORFA-Fluc, similar levels to a negative control. The only exception was transformants with AAA2350-Fluc, which showed ∼28% of the Fluc activity of transformants with the starting construct ORFA-Fluc, which is wild type in terms of the ORF A sequence.
The observation that insertion of a stop codon far upstream of the UAAUG pentanucleotide (map positions 2362–2366, ref. 19) had detrimental effects on translation of the Fluc ORF (as assessed by Fluc activity) suggests that in order to translate the downstream ORF ribosomes must translate ORF A until the termination codon at 2362–2364. This implies that CHV1 ORF B is translated by a termination-dependent reinitiation mechanism or by frame-shift, but not by internal initiation, assuming that some point mutations would retain translatability in IRES-mediated translation or ribosomal shunt. The results with AAA2350-Fluc suggest that insertion of termination codons near the pentamer is tolerated to some extent for translation of the reporter gene.
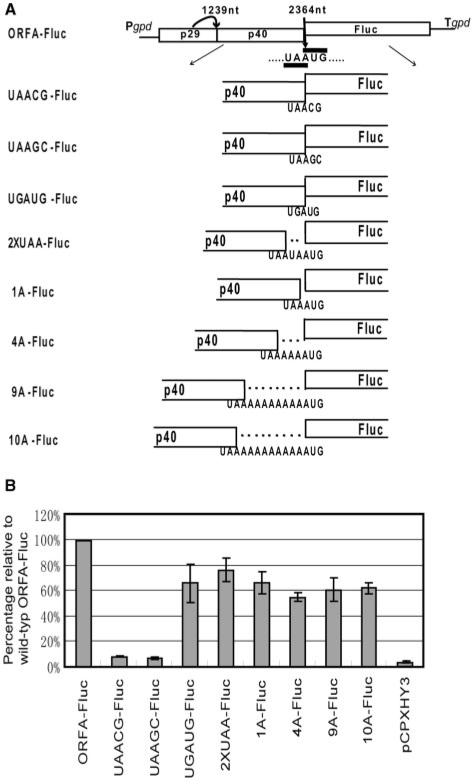
Effects of alterations of the pentamer sequence on Fluc activities expressed from the reporter constructs
If reinitiation is the mechanism for translation of CHV1 ORF B, we anticipated from the previous reports on influenza B virus (9) and retrotransposon SART1 (28) that the UAAUG pentamer is a mediator of translation of ORF B and that the AUG codon in the pentamer serves as the initiator for translation of ORF B. We examined whether the AUG and UAA codons in the pentamer are essential for Fluc activity. To this end, AUG was replaced with ACG or AGC (Figure 4A). Transformants with the two mutants displayed reduced Fluc activities of 8% and 7%, respectively, when compared with the wild-type construct ORFA-Fluc (Figure 4B). Therefore, efficient translation of the Fluc ORF requires an AUG codon in the UAAUG context. Intriguingly, transformants with UGAUG-Fluc, in which the ORF A stop codon UAA had been changed to UGA, or those with the double termination mutant 2XUAA-Fluc, retained 66% and 76.5%, respectively, of the Fluc activity shown by transformants with the wild-type construct (Figure 4B). These results clearly demonstrated that AUG is the initiator for the translation of the Fluc ORF, whereas modification of the UAG triplet is tolerated.
Figure 4.
Effects of modifications of the pentamer on translation of the Fluc ORF in the reporter constructs. (A) Diagram of the pentamer mutant constructs. The pentanucleotide UAAUG was changed in the background of the ORFA-Fluc construct to UAACG, UAAGC, UGAUG and UAAUAAUG to obtain UAACG-Fluc, UAAGC-Fluc, UGAUG-Fluc and 2XUAA-Fluc, respectively. The 1A-Fluc, 4A-Fluc, 9A-Fluc and 10A-Fluc constructs carry insertions of 1 A, 4 As, 9 As and 10 As in the pentanucleotide region, respectively. (B) Luciferase activities detected in mycelial extracts of transformants with the constructs shown in (A). As in Figure 3, fungal transformants with these constructs were examined for Fluc activity, which is shown relative to the activity of transformants with ORFA-Fluc.
We further examined the effect of distance between the UAA termination codon and the AUG reinitiation codon on efficiency of reinitiation. Mutants containing insertions of an A, 4 As, 9 As and 10 As were constructed (Figure 4A). Insertion of a single A resulted in about 30% reduction in Fluc activity relative to the wild-type basic construct ORFA-Fluc, whereas insertions of 4, 9 or 10 As all resulted in similar Fluc activities of 55–62% of the activity with ORFA-Fluc. Interestingly, insertion of 31 nt caused detrimental effects on the luminescence (data not shown). These results indicate that insertion of a limited range of nucleotides, including an additional stop codon, regardless of whether in-frame or out of frame with the downstream ORF, can be tolerated.
Taken together, these results clearly eliminate the possibility of a frame-shift mechanism, given that the UAACG and UAAGC mutant constructs impeded translation of the Fluc ORF whereas insertion of a single A did not. If frame-shift were involved in the translation of the Fluc ORF, the opposite results would be expected.
Reinitiation efficiency is < 5%
It was assumed that the sequence upstream and downstream of the UAAUG pentamer would influence the efficiency of reinitiation of ORF B, as observed for the SART1 and calicivirus examples of the same ORF organization (28,38). Therefore, we attempted to use monocistronic (Mp40p48-Fluc) and dicistronic (Dp40p48-Fluc) constructs retaining the ORF A sequence as well as the coding sequence of the papain-like protease p48 to determine the efficiency of naturally occurring reinitiation in infected fungal cells. For this purpose, it was necessary to know the minimum number of amino acid (aa) residues downstream of the p48 cleavage site necessary for complete self-cleavage of p48 when fused with Fluc. Shapira and Nuss (21) showed that aa residues near the p48 cleavage site between G418 and A419 affect the autocatalytic activity. Taking advantage of a baculovirus expression system, we examined self-cleavage activity, as has been performed for p29 (15). Four baculovirus recombinants were prepared that contained no (Bacp48-0aa-Fluc), five (Bacp48-5aa-Fluc), 10 (Bacp48-10aa-Fluc) or 15 (Bacp48-15aa-Fluc) aa residues derived from the downstream sequence between the codons for the p48 cleavage site G and the first methionine residue of Fluc (Figure S2A). These were transfected into insect cells. As shown in Figure S2B, lysates of cells infected with Bacp48-0aa-Fluc contained small amounts of uncleaved product of 108 kDa (indicated by arrowheads in Figure S2B), whereas those infected by the other recombinants showed no 108-kDa product. These results suggested that five downstream aa residues are sufficient for self-cleavage of p48.
The monocistronic (Mp40p48-Fluc) and dicistronic (Dp40p48-Fluc) constructs are identical in sequence, except that Mp40p48Fluc has a two-nucleotide (UA) deletion at the pentamer sequence, resulting in a monocistronic coding structure (Figure 5A). To ensure the autocatalytic activity of p48, Fluc was expressed as a fusion protein containing an additional 10 aa residues derived from the sequence downstream of the p48 cleavage site. The autocatalytic protease activities of p48 in insect cells were confirmed for both constructs; that is, only Fluc product, but not p48/Fluc fusion product, was detected in insect cells infected with the baculovirus constructs (Figure S2C). Using these constructs, the reinitiation ratio was estimated in the fungal host and in insect nonhost cells. Fluc activity in fungal cell lysates of transformants with the Dp40p48-Fluc construct was ∼23-fold lower (4.4%) than that from transformants with Mp40p48-Fluc (Figure 5B). However, the Fluc level exhibited by transformants with Dp40p48-Fluc was much higher than the negative control. In addition, it should be noted that transformants with Dp40p48-Fluc gave a slightly lower Fluc level than those with ORFA-Fluc (data not shown). This observation indicates that the p48-coding region downstream of the pentamer has no important signal for reinitiation.
Figure 5.
Efficiency of CHV1 translation of ORF B in C. parasitica and insect cells as estimated by luciferase assay and western blotting. (A and D) Diagrammatic representation of construct to determine the reinitiation efficiency of translation of ORF B. The entire p40- and p48-coding domains were fused in di- and mono-cistronic organization with the Fluc gene (A). The sequence coding for 10 aa downstream of the p48 cleavage site was inserted between the p48 and Fluc genes. PPH and SV40 pA indicate the polyhedrin promoter and the SV40 polyadenylation signal. For western analysis, the p40-coding domain and a truncated form of p48 (p48t) separated by the pentamer UAAUG (dicistronic) or AUG (monocistronic) were tagged with 3× Flag at both termini (D). These constructs were cloned downstream of the GPD promoter (Dp40p48-Fluc, Mp40p48-Fluc) for transgenic expression in C. parasitica or downstream of the polyhedrin promoter (BacDp40p48-Fluc, BacMp40p48-Fluc, BacDp40p48t-3×Flag, BacMp40p48t-3×Flag) for expression in S. frugiperda cells by the baculovirus system. A baculovirus recombinant harboring the coding domain of p48t tagged with 3× Flag was also prepared (Bacp48t-3×Flag). (B and C) Luciferase activities of lysates of C. parasitica mycelia of transformants and insect cells infected with baculovirus recombinants. Enzymatic activities are shown relative to monocistronic constructs. Averages and standard deviations in (B) were taken as in Figure 3, whereas those in C were taken from three sets of experiments using two independent baculovirus recombinant clones. (E) Western analysis of insect cells infected with baculovirus recombinants. Protein fractions from cells infected with recombinant viruses were serially diluted (as shown at the top of the blot), fractionated in 15% polyacrylamide gel and probed with anti-Flag mouse antibody (Sigma) after transfer onto nitrocellulose membrane (left panel). Lysates derived from cells infected with baculovirus vectors carrying BacMp40p48t-3×Flag and cDNA of MyRV1 S3 deletion mutant ΔC1 (52) were treated in parallel. Proteins in insect cells infected by baculovirus recombinants Bacp48t-3×Flag, BacDp40p48t-3×Flag, BacMp40p48t-3×Flag were also probed on a single membrane sheet with anti-Flag antibody (right panel). Migration positions of p40 tagged with 3× Flag and p48t tagged with 3× Flag are shown by arrows on the right (45 and 23 kDa). M refers to the protein size standards (Bio-Rad): 250, 150, 100, 75, 50, 37, 25 kDa, 20, 15 and 10 kDa.
Reinitiation efficiency was also estimated in insect cells with the same constructs as in fungal cells. The relative activities of insect cells infected with BacMp40p48-Fluc, BacDp40p48-Fluc and a negative control vector were 100%, 2.5% and 0.02%. The p48-Fluc reinitiation efficiency, 2.5%, was similar to that (4.4%) in C. parasitica (Figure 5C). To further confirm this, two additional constructs were made which allowed detection of the reinitiation product of the downstream ORF by western blot analysis (Figure 5D). One construct, BacDp40p48t-3 × Flag, carried a 3× Flag tag at each terminus. Infection of insect cells with the baculovirus recombinant resulted in detection of a major protein band of 45 kDa and a minor band of 23 kDa. These bands were considered to be p40 and a truncated form of p48 tagged by 3× Flag. The absence of the 23-kDa band from cells infected by the baculovirus recombinant containing the monocistronic construct BacMp40p48t-3×Flag strongly suggested that the 23-kDa band was the product translated from the downstream ORF. This is supported by the observation that the 23-kDa protein was detected as the major protein in insect cells infected with the baculovirus recombinant carrying only the p48t-3×Flag portion (Figure 5E, lane Bacp48t-3×Flag). Serial dilution of protein samples from cell lysates revealed the dilution endpoints for the two bands: 16384-fold dilution for the major band and 512-fold dilution for the minor band, allowing calculation of the reinitiation efficiency as 3.1%. This value is very similar to those of 2.5% and 4.4% obtained with the Fluc assay, suggesting that our data are not biased by the assay system.
Modifications of the p40-coding sequence influence the reinitiation efficiency
An indication that the p40-coding domain influences the translation of the Fluc ORF was provided by an observation with an EGFP-Fluc construct (Figure S3A) in which the EGFP and Fluc-coding domains were separated by UAAUG. Although fungal transformants with EGFP-Fluc fluoresced green (Figure S3B), indicating efficient translation of the upstream ORF, they failed to show Fluc activity (Figure S3C). Identical results were obtained from other six independent fungal transformants with EGFP-Fluc. These observations suggest insufficiency of the pentamer for, and involvement of the p40-coding domain in, reinitiation of the downstream Fluc ORF. To test this possibility, three mutants carrying in-frame deletions within the p40-coding sequence, lacking nucleotides at 1388–1564, 2329–2349 and 2350–2361, were constructed for the Fluc assay (Figure S4A). Interestingly, all the constructs caused marked reduction of Fluc activity (Figure S4B). Particularly, the Fluc activities of transformants with Δ2329–2349-Fluc were similar to those of negative controls.
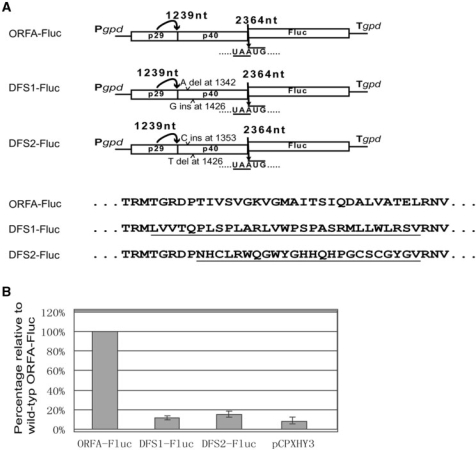
There had been previous indications that the p40 protein might be required for coordinated expression of ORF B from the dicistronic virus mRNA, leading to efficient virus replication (32). These included the observation that a double frame-shift mutation (DFS2) at the two 5′ terminal positions of p40, where nucleotide sequence changes were minimized but 24 aa were modified, resulted in a viable virus mutant undergoing compensatory mutations that converted the virus genome from a dicistronic to a monocistronic structure. It is noteworthy that the mutated positions in DFS2 overlap the deleted region of Δ1388–1564-Fluc. These results raised the interesting question of whether the decrease in translation of Fluc found in Δ1388–1564-Fluc (Figure S4B) was due to the involvement of the deleted region as a protein sequence or a nucleotide sequence. To address this question, the Fluc versions of two double frame-shift mutants, DFS1-Fluc and DFS2-Fluc, were tested (Figure 6A). The mutants each carried single deletions (A at position 1342 for DFS1-Fluc, T at position 1426 for DFS2-Fluc) and single insertions (G downstream of position 1426 for DFS1-Fluc, C downstream of position 1353 for DFS2-Fluc) within the deleted region of Δ1388–1564-Fluc, so that the mutations caused replacement of the aa residues between the two mutated positions with unrelated residues (Figure 6A). Transformants with the DFS1 constructs gave very low Fluc activities, similar to those of the negative control, whereas transformants with DFS2-Fluc showed a slightly higher level of Fluc activity than the negative control (Figure 6B). These results suggested the involvement of the N terminal portion of the p40 protein in translation of the downstream ORF.
Figure 6.
Effects of double-frame-shifting of the p40-coding domain on coupled translation of the Fluc ORF. (A) Schematic drawing of double-frame-shift mutant constructs. The double-frame-shift mutants DFS1-Fluc and DFS2-Fluc each harbor single nucleotide deletions and single nucleotide insertions. The p40 aa sequence coded by DFS1-Fluc and DFS2-Fluc is the same as the authentic p40, except for the aa coded by the sequence between the mutated sites as denoted below the drawing. Modified aa sequences are underlined. These frame-shift mutants would allow translation until the termination codon at the pentamer. (B) Luciferase activities of p40 double-frame-shift mutant constructs. Transformants with the frame-shift constructs were used in the Fluc assay to estimate their relative activities as for Figure 3.
Effects of pentamer mutations on virus viability
It was of great interest to examine the effects of mutations of the pentamer on virus viability. Some of the mutations that affected the efficiency of translation of the Fluc ORF in the reporter system were introduced into the infectious cDNA clone of CHV1-EP713 and then tested for the ability to replicate and induce symptoms. A total of five mutants, CHV1-UGAUG, CHV1-1A (UAAAUG), CHV1-UAACG, CHV1-UAAGC and CHV1-AUG (in each mutant virus cDNA UAAUG was replaced with the nucleotides shown next to CHV1), was used for transfection of EP155 spheroplasts (Table 1). Transfectants with CHV1-UGAUG and CHV1-1A accumulated viral genomic dsRNA at levels similar to that with wild-type CHV1 (data not shown). However, the other mutants were replication defective and no viral dsRNA was detected in transfectants. Even increased amounts of transcripts (15 μg/transfection) derived from the virus cDNA mutants CHV1-UAACG, CHV1-UAAGC and CHV1-AUG were found to be nonviable (Table 1).
Table 1.
Effects of pentamer mutations on CHV1 viability
| cDNA constructs | UAAUG was changed to: | Replication competency | Symptom induction |
|---|---|---|---|
| CHV1-UGA | UGAUG | Yes | Identical to CHV1-WT |
| CHV1-1A | UAAAUG | Yes | Identical to CHV1-WT |
| CHV1-AUG | AUG | No | NA |
| CHV1-ACG | UAACG | No | NA |
| CHV1-AGC | UAAGC | No | NA |
| CHV1-WT | Wild type | Yes | Severe reduction in virulence, sporulation and pigmentation |
NA: not applicable.
Replication competent virus mutants caused symptoms identical to those caused by the wild type CHV1-EP713, characterized by irregular margins and severely reduced pigmentation and sporulation (Table 1). To determine whether the virus mutants were genetically stable during replication, RT-PCR with the primer pair NS21 and NS23 was performed on dsRNA isolated from mycelia infected with each virus mutant at 3 weeks after transfection. RT-PCR fragments (five clones per transfectant) spanning the mutated pentamer region were sequenced (data not shown). Interestingly, both the viable virus mutants (CHV1-UGAUG and CHV1-1A) stably retained their altered sequences (data not shown).
DISCUSSION
Coordinated translational control is essential for virus replication, which relies on the host's translation machinery. CHV1 genomic RNA has two contiguous ORFs, A and B. ORF A encodes two proteins p29 and p40 shown to be nonessential for virus replication, whereas ORF B, which is shifted by –1 with regard to ORF A, encodes a polyprotein of 3165 aa which is processed into mature proteins, including p48 and replicase-related domains (Figure 1A; 12). High-resolution northern and 5′-RACE analyses of ssRNA from CHV1-infected mycelia using in vitro synthesized viral RNAs of different sizes showed no ORF B subgenomic mRNA of the expected size range of 10–11 kb in infected mycelia (Figure 1). This observation argues against the possibility of subgenomic RNA that often allows translation of downstream ORFs on ssRNA genomes and supports the polycistronic nature of the full-length transcript of CHV1.
The finding that introduction of premature nonsense mutations within the p40-coding region abrogates expression of the reporter gene (Figure 3) suggests that translation of ORF A until the natural stop codon is required for translation of the downstream ORF, and eliminates the possibility that the Fluc ORF is translated by IRES or shunt mechanisms. If IRES or ribosomal shunt mediated the downstream translation, it is unlikely that all nonsense mutations at different positions would be able to repress completely Fluc expression, as shown in Figure 3. If a −1 frame-shift was involved, it should happen between map positions 2309 and 2363 (19), given the upstream UAA triplet at positions 2307–2309 on frame –1 with regard to the ORF A frame and the pentamer at positions 2362–2366. However, a single A or 10-A insertion into the pentamer, which would shift the Fluc frame from –1 to 0 (same frame) with regard to ORF A, largely unaffected the translation of the Fluc ORF, which was reduced by 34–38% relative to the wild-type construct ORFA-Fluc (Figure 4). Moreover, p48, but not the p40/p48 fusion product predicted to result from a –1 frame-shift, is detected in CHV1-infected mycelia (39). Together, these data eliminate the possibility of a frame-shift, and clearly leave termination-dependent reinitiation (translation coupling) mediated by the pentamer UAAUG as the mechanism for translation of the Fluc ORF.
Although translational coupling has been established in only a few mRNAs from animal viruses and transposons (Table 2), there are diverse virus and transposon candidates from plants, fungi and animals in which coupled translation is implicated, including mRNAs from two mycoviruses, CHV2 (40) and Helminthosporium victoriae 190S virus (Hv190SV) (41). It is noteworthy that Kojima et al. (28) also demonstrated a possible example of such a cellular mRNA of nonparasitic origin, glutamic acid decarboxylase (GAD), in addition to non-LTR retrotransposons. Extensive bioinformatic and subsequent biochemical analyses may reveal additional examples of cellular mRNAs. Established examples are classified into two groups in terms of the junction sequence: one with overlapping or consecutive termination/start codons typified by UAAUG, AUGA, UAGAUG and AUGucUGA, and the other with a short inter-ORF spacer sequence ranging from 26 to 61 nt. The first group includes mRNAs from several caliciviruses (38,42,43), such as Rabbit hemorrhagic disease virus, Influenza B virus segment 7 (9,27) and Bombyx mori retrotransposon SART1 (28) (Table 2). The second group includes pneumoviruses such as Respiratory syncytial virus (44–46) (Table 2). Despite the difference in the junction sequence, the mechanism of translation of the downstream ORF seems to be fundamentally similar between the two groups (28,47), and is characterized by the following:
dependence of translation initiation for the downstream ORF on translation termination of the preceding ORF (9,28,42);
low efficiency (4.4–20%, Table 2) of translation of the downstream ORF; and
an upstream sequence identified as promoting translation of the downstream ORF (27,42,46,47).
Table 2.
Examples of mRNAs translated by the coupled termination/reinitiation strategy
| Family | Host | Junction sequencea | Efficiency (%) of translation of downstream ORF | Reference | |
|---|---|---|---|---|---|
| CHV1 | Hypoviridae | Fungi | UAAUG | 4.4% | This study |
| Influenza B virus seg. 7 | Orthomyxoviridae | Vertebrate | UAAUG | ∼10% | 9,27 |
| RHDV sgRNA | Caliciviridae | Vertebrate | AUGucUGA | ∼20% | 38 |
| FCV sgRNA | Caliciviridae | Vertebrate | AUGA | 5% | 42 |
| JV genomic RNA | Caliciviridae | Vertebrate | AUGaagAUGacUGA | ND | 43 |
| RSV M2 mRNA | Pneumovirinae | Vertebrate | AUG(n)3AUG(n)9AUG(n)18UGA | ND | 44 |
| MPV M2 mRNA | Pneumovirinae | Vertebrate | AUG(n)60AUGA | ND | 46 |
| AMPV M2 mRNA | Pneumovirinae | Vertebrate | AUG(n)3AUG(n)33UAA | ND | 46 |
| SART1 | Non-LTR retrotransposon | Insect | UAAUG | ND | 28 |
aThe stop codon of the upstream ORF is in bold and italic, while the start codon for the downstream ORF is bold-faced and underlined.
Numerals next to (n) designate spaces composed of the indicated number of nucleotides.
CHV1, Cryphonectria hypovirus 1; RHDV, rabbit hemorrhagic disease virus; FCV, Feline calicivirus; JV, Bovine norovirus-Jena, RSV, Human respiratory syncytial virus; MPV, Murine pneumonia virus; AMPV, Avian metapneumovirus.
ND, not determined.
CHV1, the first example of reinitiation in fungi, is categorized into the first group with a junction sequence, UAAUG, identical to those of Influenza B virus segment 7 and SART1. Translation of the downstream ORF in the reporter constructs shares properties with the examples listed in Table 2. This study determined the reinitiation efficiency to be 2.5–4.4% in homologous and heterologous systems (Figure 5). At least two sequence elements are required for efficient translation coupling of the Fluc ORF: the pentamer and p40-coding sequences upstream of the pentamer. The key element is the pentanucleotide UAAUG at map positions 2362–2366 (19), where ribosomes stop translation of ORF A and resume translation of the reporter ORF. As expected, the efficiency of translation of the Fluc ORF was modulated by altering sequences around the UAAUG pentanucleotide. Substitution of UAACG or UAAGC for UAAUG abolishes translation of the downstream ORF (Figure 4), confirming the AUG triplet in the pentamer as the initiator for translation of the Fluc ORF. Nevertheless, efficiency of translation of the reporter gene was largely unaffected by insertion of 1–10 As into the pentamer, or by changing UAA to UGA or 2XUAA (Figure 4), suggesting flexibility of the junction sequence for translation of the Fluc ORF. It is relevant that, in studies of hypovirus-based fungal expression vector constructs, insertion of foreign genes immediately upstream of the pentamer results in generation of virus progeny with alterations of the sequence near the pentamer (48). For example, several variants of the pentamer are found in the genome sequences of progeny virus, for example—aua-aaa-uuU-AAU-Gua-UAA—(vector constructs Ct2APH and Ct2APHp29Δ25-243; see Figure 6 of 48; pentamer boldfaced, newly created stop codon italicized) versus—aua-aaa—uuu-UAA-UGU-aua—(wild-type, pentamer boldfaced). Thus, the variant has the stop codon UAA (shown by italic letters) two triplets downstream of the authentic position, whereas the start codon was maintained at the original position. This may not be surprising given that a similar configuration is found in a calicivirus, Rabbit hemorrhagic disease virus (RHDV) (AUGucUGA) (Table 2). Together with the results shown in Figure 4, these data may indicate that whether the ORF B start site precedes the stop codon of ORF A is not important for translation of ORF B when their distance is within a few codons.
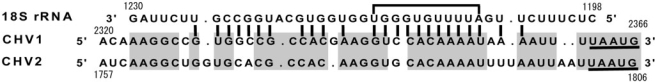
The second sequence element found to promote translation of the downstream ORF is the preceding p40-coding sequence within the preceding ORF A. Discussion of the p40 sequence should be prefaced by consideration of models proposed, on the basis of biochemical analysis, for translational coupling of calicivirus subgenomic mRNAs. Meyers and co-workers (42,49) identified ‘termination upstream ribosomal binding sites (TURBS)’ of ∼70 nt in calicivirus subgenomic RNAs, within which two motifs were shown to play critical roles in reinitiation. Motif 1 is complementary to 18S rRNA (helix 26) and conserved among the TURBS of caliciviruses, whereas motif 2 is not. Recently, Powell et al. (27) also identified a very similar sequence stretch to motif 1 upstream of the pentamer of influenza B virus segment 7 as essential for reinitiation of its downstream ORF. The authors hypothesized that motif 1 allows association with the ribosome via base pairing with 18S rRNA and that motif 2 is involved, by an undefined mechanism, in selection of the correct start site. If this model is applicable to other cases of coupled translation, prediction of similar functional sequences by sequence comparison will be difficult, because the 3′ terminal sequence of the upstream ORF of FCV subgenomic RNA is not conserved in the other mRNAs listed in Table 2. In another model, termination is followed by disassembly of 80S ribosomes by eukaryotic initiation factor 3 (eIF3), which is initially associated with a particular upstream region of 87 nt overlapping the TURBS of calicivirus (FCV) subgenomic mRNA. The 40S subunit is transferred to the standby site slightly upstream from the stop codon to bind eIF3, and eIF3/40S complexes become reinitiation competent after acquiring an eIF2/GTP/Met-tRNA ternary complex (47). How the CHV1 p40-coding sequence is involved in the reinitiation is largely unknown, and systematic mechanistic analysis as performed with the caliciviruses and influenza B virus is needed to define the role of the p40 sequence in CHV1 reinitiation. However, the sequence upstream of the pentamer (map positions 2362–2366, ref. 19) shows sequence similarities to the corresponding region of CHV2, which also has the pentamer and dicistronic genome structure (40). Inspection of Figures S4 and 6 reveals that Fluc activity was drastically reduced in transformants with in-frame deletion and double frame-shift mutants of the p40-coding sequence, in which decoding ribosomes would reach the pentamer. All the constructs caused marked reduction of Fluc activity, and in particular transformants with Δ2329–2349-Fluc showed Fluc activity similar to that of the negative control. Interestingly, this region contains sequence complementary to C. parasitica 18S rRNA (map positions 1205–1224, ref. 50) (Figure 7) and deletion of a p40 sequence spanning this region resulted in elimination of Fluc activities (Figure S4). Thus, this region may be involved in reinitiation in both CHV1 and CHV2 as a cis-acting domain that interacts with 18S rRNA, as is the case for calicivirus subgenomic mRNAs. However, it should be noted that this region is ∼200 nt downstream of helix 26 complementary to TURBS motif1 and helix 26 is not well conserved in the C. parasitica 18S rRNA. Another interesting finding is that the CHV1 p40-coding sequence far from the stop/start site strongly affects reinitiation efficiency (Figures S4 and 6), suggesting that p40 protein, not RNA as a cis-element, is involved in promotion of reinitiation of the downstream Fluc ORF, as discussed by Suzuki and Nuss (32), and that the 5′ p40-coding sequence is required for p40 to enhance coupled translation. The possibility of additional protein factors was also discussed by Pöyry et al. (47).
Figure 7.
Complementarity between hypovirus sequences and the C. parasitica 18S rRNA. Above the CHV1 sequence is a portion of the C. parasitica 18S rRNA sequence (nucleotides 1198–1230, ref. 50) given in a 3′–5′ orientation. Possible base pairing between the 18S rRNA and the CHV1 sequence is indicated by solid lines. A stretch showing perfect complementarity is bracketed. Sequences upstream of the start/stop site that are conserved between CHV1-EP713 (nucleotides 2320–2366, ref. 19) and CHV2-NB58 (nucleotides 1757–1806, ref. 40) are highlighted in gray.
Lastly, this study also provides interesting insights into the relation between virus viability and translation of ORF B. CHV1 variants containing pentamer mutations (CHV1-UAACG, CHV1-UAAGC, CHV1-1A, CHV1-UGAUG and CHV1-AUG) were tested for their replication competency. In these mutant viruses, p40 and p48 are expected to be unaltered in amino acid sequence. Additionally, ACG or AGC are likely to act as initiation codons in place of AUG in methionine-tRNA-dependent translation, since Luttermann and Meyers (43), Pöyry et al. (47) and Powell et al. (27) reported that non-AUG codons could serve as the restart codon. Therefore, whether these mutant viruses are replication competent is likely to depend on the translation efficiency of ORF B. Interestingly, CHV1-1A and CHV1-UGAUG, but not CHV1-UAACG, CHV1-UAAGC or CHV1-AUG, were able to replicate and cause symptoms identical to those caused by wild-type CHV1 (Table 1). Reporter constructs with the same pentamer mutations as the replication-competent virus constructs showed only 34% reduction in reinitiation of ORF B, whereas those carrying the same mutations as the replication-defective constructs showed 92–93% reduction (Figure 4). This observation suggests that there is a threshold level of translation of ORF B that determines CHV1 viability. It is noteworthy that alteration in the translation efficiency of the downstream ORF of another polycistronic viral mRNA has been reported to affect the viability of the virus. Saccharomyces cerevisiae virus L-A (ScV-L-A), a totivirus, utilizes a –1 frame-shift to express the downstream ORF encoding RdRp at an efficiency of 1.9%. Frame-shifting is functionally analogous to coupled translation, although it allows production of a fusion protein encoded by the downstream ORF. Both increased or decreased frame-shift efficiencies resulted in defects in virus (satellite virus, killer factor) replication (51). The nonviability of CHV1-AUG could be explained in the same way as for ScV-L-A. This construct would lead to translation of ORF B at a much higher rate than in the wild-type construct, resulting in expression of p48 fused with p40 while maintaining complete self-cleavage ability of p48 (Figure S2). As an alternative explanation, p40/p48 fusion products may be defective in the hit-and-run function at the initiation step of CHV1 infection (39).
In summary, we developed a reporter system to assess CHV1 ORF B translation in which ORF B was replaced with the Fluc gene. Using the system, we showed that the downstream Fluc ORF was translated by reinitiation at ∼4.4% of ORF A in C. parasitica. Two sequence elements on the p40 sequence, the 5′-terminal and 3′-terminal regions, were identified as promoting Fluc ORF reinitiation. These conclusions drawn with the reporter system are probably applicable for CHV1 ORF B translation. However, it should be noted that the reporter constructs lack the authentic 5′ and 3′ terminal sequences of the CHV1 genomic RNA that may affect ORF B reinitiation. Further, environments of CHV1-infected cells are likely different from those of transformed cells used for the assay in this study. It will be of interest to compare expression levels of CHV1 mature proteins encoded by ORF A and ORF B in infected mycelia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Goho International Fund (to G.L.H.); Yomogi Inc.; and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industries.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Drs. Donald L. Nuss and Bradley I Hillman for their generous gift of a number of plasmid clones and fungal strains used in this study. We also thank Mr K. Maruyama for his excellent technical help.
REFERENCES
- 1.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fütterer J, Hohn T. Translation in plants—rules and exceptions. Plant Mol. Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. An analysis of vertebrate mRNA sequences: intimations of the translational control. J. Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fütterer J, Kiss-Laszlo Z, Hohn T. Non-linear ribosome migration on cauliflower mosaic virus 35S mRNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 6.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 8.Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl Acad. Sci. USA, 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath CM, Williams MA, Lamb RA. Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuss DL, Hillman BI, Rigling D, Suzuki N. Family Hypoviridae. In: Fauquet CM, et al., editors. Virus Taxonomy: Eighth Report of the International Committee for the Taxonomy of Viruses. San Diego: Academic Press; 2005. pp. 597–601. [Google Scholar]
- 11.Anagnostakis SL. Biological control of chestnut blight. Science. 1982;215:466–471. doi: 10.1126/science.215.4532.466. [DOI] [PubMed] [Google Scholar]
- 12.Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 2005;3:632–642. doi: 10.1038/nrmicro1206. [DOI] [PubMed] [Google Scholar]
- 13.Faruk MI, Eusebio-Cope A, Suzuki N. A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J. Virol. 2008;82:740–754. doi: 10.1128/JVI.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruk MI, Izumimoto M, Suzuki N. Characterization of mutants of the chestnut blight fungus, Cryphonectria parasitica that show unusual hypovirus symptoms. J. Gen. Plant Pathol. 74:425–433. [Google Scholar]
- 15.Sun L, Nuss DL, Suzuki N. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 2006;87:3703–3714. doi: 10.1099/vir.0.82213-0. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Suzuki N. Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1-EP713. RNA. 2008;14:2557–2571. doi: 10.1261/rna.1125408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng F, Allen TD, Hillman BI, Nuss DL. Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut blight fungus Cryphonectria parasitica. Eukaryot Cell. 2007;6:1286–1298. doi: 10.1128/EC.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin EV, Choi GH, Nuss DL, Shapira R, Carrington JC. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl Acad. Sci. USA. 1991;88:10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapira R, Choi GH, Nuss DL. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 1991;10:731–739. doi: 10.1002/j.1460-2075.1991.tb08004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi GH, Shapira R, Nuss DL. Co-translational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl Acad. Sci. USA. 1991;88:1167–1171. doi: 10.1073/pnas.88.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapira R, Nuss DL. Gene expression by a hypovirulence-associated virus of the chestnut blight fungus involves two papain-like protease activities. Essential residues and cleavage site requirements for p48 autoproteolysis. J. Biol. Chem. 1991;266:19419–19425. [PubMed] [Google Scholar]
- 22.Dawe AL, Nuss DL. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Ann. Rev. Genet. 2001;35:1–29. doi: 10.1146/annurev.genet.35.102401.085929. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N, Maruyama K, Moriyama M, Nuss DL. Hypovirus papain-like protease p29 is an enhancer of viral dsRNA accumulation and vertical transmission. J. Virol. 2003;77:11697–11707. doi: 10.1128/JVI.77.21.11697-11707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillman BI, Suzuki N. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 2004;63:423–472. doi: 10.1016/S0065-3527(04)63007-7. [DOI] [PubMed] [Google Scholar]
- 25.Turina M, Rostagno L. Virus-induced hypovirulence in Cryphonectria parasitica: still an unresolved conundrum. J. Plant Pathol. 2007;89:165–178. [Google Scholar]
- 26.Shapira R, Choi GH, Hillman BI, Nuss DL. The contribution of defective RNAs to the complexity of viral-encoded double-stranded RNA populations present in hypovirulent strains of the chestnut blight fungus Cryphonectria parasitica. EMBO J. 1991;10:741–746. doi: 10.1002/j.1460-2075.1991.tb08005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell ML, Napthine S, Jackson RJ, Brierley I, Brown TD. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA. 2008;14:2394–2406. doi: 10.1261/rna.1231008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima KK, Matsumoto T, Fujiwara H. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell Biol. 2005;25:7675–7686. doi: 10.1128/MCB.25.17.7675-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 30.Choi GH, Nuss DL. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science. 1992;257:800–803. doi: 10.1126/science.1496400. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki N, Chen B, Nuss DL. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 1999;73:9478–9484. doi: 10.1128/jvi.73.11.9478-9484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki N, Nuss DL. The contribution of p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 2002;76:7747–7759. doi: 10.1128/JVI.76.15.7747-7759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuura Y, Possee RD, Overton HA, Bishop DHL. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J. Gen. Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki N, Sugawara M, Kusano T, Mori H, Matsuura Y. Immunodetection of rice dwarf phytoreoviral proteins in both insect and plant hosts. Virology. 1994;202:41–48. doi: 10.1006/viro.1994.1320. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. pp. A8.40–A8.45. [Google Scholar]
- 36.Churchill ACL, Ciufetti LM, Hansen DR, Van Etten HD, Van Alfen NK. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 1990;17:25–31. [Google Scholar]
- 37.Chen B, Choi GH, Nuss DL. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science. 1994;264:1762–1764. doi: 10.1126/science.8209256. [DOI] [PubMed] [Google Scholar]
- 38.Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 2003;278:34051–34060. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- 39.Deng F, Nuss DL. Hypovirus papain-like protease p48 is required for initiation but not for maintenance of virus RNA propagation in the chestnut blight fungus Cryphonectria parasitica. J. Virol. 2008;82:6369–6378. doi: 10.1128/JVI.02638-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hillman BI, Halpern BT, Brown MP. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology. 1994;201:241–250. doi: 10.1006/viro.1994.1289. [DOI] [PubMed] [Google Scholar]
- 41.Soldevila AI, Ghabrial SA. Expression of the Totivirus Helminthosporium victoriae 190S virus RNA-dependent RNA polymerase from its downstream open reading frame in dicistronic constructs. J. Virol. 2000;74:997–1003. doi: 10.1128/jvi.74.2.997-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luttermann C, Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- 43.McCormick CJ, Salim O, Lambden PR, Clarke IN. Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 2008;82:8917–8921. doi: 10.1128/JVI.02362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmadian G, Randhawa JS, Easton AJ. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gould PS, Easton AJ. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 2005;280:21972–21980. doi: 10.1074/jbc.M502276200. [DOI] [PubMed] [Google Scholar]
- 46.Gould PS, Easton AJ. Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae. J. Virol. 2007;81:8488–8496. doi: 10.1128/JVI.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki N, Geletka LM, Nuss DL. Essential and dispensable virus-encoded elements revealed by efforts to develop hypoviruses as gene expression vector. J. Virol. 2000;74:7568–7577. doi: 10.1128/jvi.74.16.7568-7577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen B, Chen C-H, Bowman BH, Nuss DL. Phenotypic changes associated with wild-type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology. 86:301–310. [Google Scholar]
- 51.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;6:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Supyani S, Hillman BI, Suzuki N. Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyltransferase-encoding segment. J. Gen. Virol. 2007;88:342–350. doi: 10.1099/vir.0.82318-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Nuss DL. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl Acad. Sci. USA. 2008;105:16749–16754. doi: 10.1073/pnas.0807225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hillman BI, Supyani S, Maruyama K, Suzuki N. A Reovirus of the fungus Cryphonectria parasitica that is infectious as particle and related to the Coltivirus genes of animal pathogens. J. Virol. 2004;78:892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.