Abstract
RNA triphosphatases (RTPases) are involved in the addition of the distinctive cap structure found at the 5′ ends of eukaryotic mRNAs. Fungi, protozoa and some DNA viruses possess an RTPase that belongs to the triphosphate tunnel metalloenzyme family of enzymes that can also hydrolyze nucleoside triphosphates. Previous crystallization studies revealed that the phosphohydrolase catalytic core is located in a hydrophilic tunnel composed of antiparallel β-strands. However, all past efforts to obtain structural information on the interaction between RTPases and their substrates were unsuccessful. In the present study, we used computational molecular docking to model the binding of a nucleotide substrate into the yeast RTPase active site. In order to confirm the docking model and to gain additional insights into the molecular determinants involved in substrate recognition, we also evaluated both the phosphohydrolysis and the inhibitory potential of an important number of nucleotide analogs. Our study highlights the importance of specific amino acids for the binding of the sugar, base and triphosphate moieties of the nucleotide substrate, and reveals both the structural flexibility and complexity of the active site. These data illustrate the functional features required for the interaction of an RTPase with a ligand and pave the way to the use of nucleotide analogs as potential inhibitors of RTPases of pathogenic importance.
INTRODUCTION
Eukaryotic mRNAs harbor a distinctive m7GpppN cap structure at their 5′ ends (1). The structure is added shortly after the initiation of transcription by a series of three sequential enzymatic reactions (2–4). The first step involves the hydrolysis of the 5′ triphosphate end of the nascent mRNA by an RNA triphosphatase (RTPase) to form a diphosphate extremity. The addition of GMP to the diphosphate end is then mediated by an RNA guanylyltransferase or capping enzyme. Finally, the GpppN cap is methylated by an RNA (guanine-N7) methyltransferase. Since its discovery three decades ago, numerous studies have demonstrated the importance of the cap structure for the stability, transport and translation of mRNAs (2,5).
An important number of enzymes involved in the synthesis of the cap structure have been found in different eukaryotic organisms ranging from fungi, protozoans, viruses, plants and metazoans (6). Numerous structural and functional studies have also contributed to elucidate the basic features of these enzymes (5). Interestingly, significant structural and mechanistic differences are found in the RTPase component of the capping machinery. Metazoan and plant RTPases belong to the cysteine phosphatase family, which also includes numerous protein tyrosine phosphatases (7,8). However, structural and biochemical studies have shown that despite sharing an HCxxxxxR(S/T) motif, a phosphoenzyme intermediate and a core α/β-fold with other cysteine phosphatases, the precise mechanism of phosphoanhydride cleavage by these RTPases differs from the one used by protein phosphatases to hydrolyze phosphomonoesters (7,8). The most important difference is the absence of a carboxylate general acid catalyst in metazoan and plant RTPases (8). Finally, the RTPases of this family are divalent cation independent and are not able to hydrolyze nucleoside triphosphates (NTPs).
Fungi, protozoa and some DNA viruses possess an RTPase that belongs to the triphosphate tunnel family of metal-dependent phosphohydrolases that can also hydrolyze NTPs (9–14). These enzymes harbor two glutamate-containing motifs that are essential for catalysis and that coordinate the essential metal cation (9). The initial crystallization of the Saccharomyces cerevisiae RTPase revealed a novel fold in which the catalytic core is located in a hydrophilic tunnel composed of eight antiparallel β-strands (15). Interestingly, this particular fold appears to be more widely distributed in the various taxa than initially expected—being found in archael and bacterial homologs—thus suggesting a deep evolutionary origin (16). The analysis of the crystal structure of the yeast RTPase revealed the presence of a single sulfate ion, which is coordinated by the side chains of three essential amino acids (Arg393, Lys456 and Arg458). It was suggested that the side-chain interactions with this sulfate ion reflect the contacts made by the protein with the γ-phosphate of the RNA or NTP substrates (15). Numerous mutational studies have also contributed to the identification of a dozen additional residues that are essential for the enzymatic activity through their interactions with the divalent metal ion or through their water-mediated contacts with either the metal ion or the sulfate ion (9,17–19). More recently, analysis of the crystal structure of the RTPase component of mimivirus—a giant virus of amoeba—also revealed a minimized tunnel fold and an active site strikingly similar to the yeast enzyme (20). However, all past efforts to obtain structural information on the interaction between RTPases and their substrates were unsuccessful. This is perhaps not surprising since thermodynamic studies have shown that the binding of the RNA or nucleotide substrates to RTPases results in a destabilization of the enzymes (21,22). Available crystals of the RTPases of the triphosphate tunnel metalloenzyme (TTM) family do not provide any information on the contacts between the enzyme and the triphosphate, sugar or base moieties of the phosphohydrolase substrate.
In the present study, we used computational molecular docking to model the binding of a nucleotide substrate into the yeast RTPase active site. In order to confirm the generated model and to gain additional insights into the molecular determinants involved in substrate recognition and catalysis, we also evaluated the phosphohydrolysis of an important number of nucleotide analogs. Our study highlights the importance of specific residues for the binding of the sugar, base and triphosphate moieties of the nucleotide substrate, and reveals both the structural flexibility and complexity of the active site.
MATERIALS AND METHODS
Molecular docking
Docking calculations were carried out using the Docking Server software and the Dreiding force field was used for energy minimization of guanosine triphosphate (GTP) using built-in Chemaxon tools in Docking Server (23). PM6 semi-empirical charges calculated by MOPAC2007 were added to the ligand atoms. Nonpolar hydrogen atoms were merged and rotatable bonds were defined (24). Docking calculations were carried out using the coordinated of the S. cerevisiae RTPase (Protein Data Bank 1d8h). Essential hydrogen atoms, Kollman united atom type charges and solvation parameters were added with the aid of AutoDock tools (25). Affinity (grid) maps of 20 × 20 × 20 Å grid points and 0.375 Å spacing were generated using the Autogrid program (25). AutoDock parameter set- and distance-dependent dielectric functions were used in the calculation of the van der Waals and the electrostatic terms, respectively. Docking simulations were performed using the Lamarckian genetic algorithm (LGA) and the Solis & Wets local search method (26). Initial position, orientation and torsions of the ligand molecules were set randomly. Each docking experiment was derived from two different runs that were set to terminate after a maximum of 2500 000 energy evaluations. The population size was set to 150. During the search, a translational step of 0.2 Å, and quaternion and torsion steps of 5 Å was applied.
Cet1 expression and purification
The Cet1 protein was expressed and purified as described before (21).
Competition assay
GTPase reactions were performed in reaction mixtures (20 μl) containing 50 mM Tris–HCl, pH 7.0, 5 mM DTT, 2 mM MnCl2, 1 μM of the S. cerevisiae RTPase and 20 μM [γ-32P]GTP. The reactions were incubated for 15 min at 30°C. Initially, the reactions were carried out both in the absence or presence of 100 μM of nucleotide analogs (or tripolyphosphate). The reactions were quenched by the addition of 5 μl of 5 M formic acid. Aliquots of the reactions were spotted on polyethyleneimine–cellulose thin-layer chromatography plates. The plates were developed in a solution of 1 M formic acid and 0.5 M LiCl and the released inorganic phosphate was quantitated by scanning the plates with a PhosphorImager (Molecular Dynamics). The average of at least two single independent experiments is presented.
The IC50 values were evaluated by performing the standard GTPase assay in the presence of increasing concentrations of nucleotide analogs (or tripolyphosphate) ranging from 0 to 200 μM. The Ki values were determined by performing GTP assays with GTP concentrations ranging from 0 to 100 μM in the presence of 0, 5, 10 or 20 μM of analogs (or tripolyphosphate). The average of at least two single independent experiments is presented.
Phosphohydrolase assay
The reaction mixtures (20 μl) containing 50 mM Tris–HCl, pH 7.0, 5 mM DTT, 2 mM MnCl2, 1 μM of the S. cerevisiae RTPase and various concentrations of substrates (GTP, nucleotide analogs or tripolyphosphate) were incubated for 15 min at 30°C. The reactions were quenched by the addition of 400 μl of malachite green reagent (BIOMOL Research Laboratories). The released inorganic phosphate was measured by monitoring the A620. The values were extrapolated to a standard curve for phosphate. Background levels of contaminating phosphate were subtracted in all cases.
RESULTS AND DISCUSSION
Molecular docking
Although important structural information is available from the currently available crystal structures of the members of the TTM family, the substrates (RNA or NTP) are conspicuously absent from all these structures (15,20). We set out to initially use the power of molecular docking to provide information on the interaction between the yeast RTPase and a nucleotide substrate. GTP was selected as the substrate since this purine nucleotide is frequently encountered as the initiating nucleotide in eukaryotic mRNAs (27). Extensive computational docking and structure optimizations were then used to generate a model of the enzyme–GTP complex. More than 2000 000 energy evaluations were performed in order to provide an accurate description of the enzyme–substrate interactions. The model underwent 150 rounds of steepest descent energy minimization and did not contain energetically unfavorable bonds, angles or torsions.
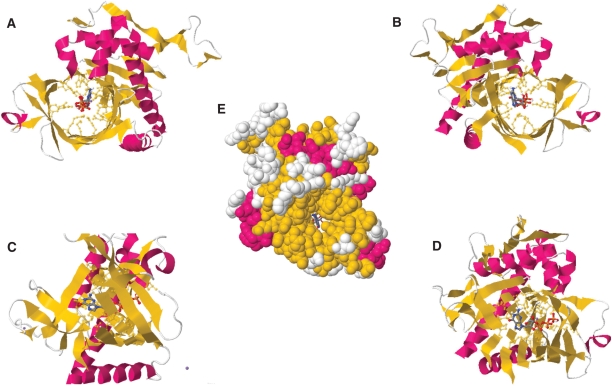
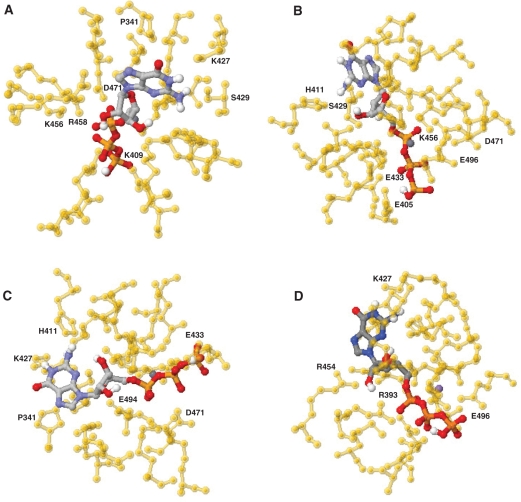
The molecular docking model predicts that the GTP substrate is located in the middle of the tunnel structure of the yeast RTPase (Figure 1). The space-filling analysis suggests that the tunnel can accommodate the ribose, base and phosphate moieties of the GTP molecule, thereby implying that the entire nucleotide—not only the phosphates—is entering the tunnel (Figure 1E). The molecular docking model provides instructive findings on the interaction between specific residues and the nucleotide substrate. For instance, multiple side chains (summarized in Table 1) are contacting the α, β and γ phosphates of GTP. In addition to the Arg393, Lys456 and Arg458 residues that were previously observed in the crystal structure of the S. cerevisiae RTPase (15), six other amino acids appear to be involved in the coordination of the phosphates (Figure 2). Some of these residues such as Arg393, Lys409, Lys456 and Glu494 are solely contacting one phosphate, while others (Glu305, Glu433, Arg458, Asp471 and Glu496) are contacting two adjacent phosphates (Figure 2A–D). Interestingly, all of these phosphate-contacting residues were previously shown to be essential for catalysis through mutational studies, thereby highlighting their importance in catalysis (9,17–19). Based on the crystal structure of the enzyme (15), some of these essential residues were previously proposed to be important for catalysis through their ability to make water-mediated contacts with the phosphate (Glu433) or the essential divalent metal ion (Glu494). Similarly, other residues such as Lys409 were thought to be indirectly involved in catalysis via their interactions with other essential side chains (15,17). Although the role of these amino acids in making important interactions unrelated to the binding of phosphate cannot be excluded, the current model clearly reveals that the side chains of these amino acids are involved in the coordination of the triphosphate moiety of the nucleotide substrate. The previously observed ability of the TTM family of RTPases to hydrolyze tripolyphosphate (28–30), which completely lacks the sugar and base components of the nucleotide, is probably a reflection of the large number of interactions between the active-site residues and the α-, β-, and γ-phosphates.
Figure 1.
Molecular docking model for the binding of GTP to the yeast RNA triphosphatase. (A) Ribbon diagram looking into the tunnel exit of the enzyme with bound GTP. Numerous amino acids are interacting with the nucleotide. (B) A view looking into the triphosphate tunnel entrance. (C) Side view of the enzyme following a 90° rotation to the left depicting both the entrance (left) and exit (right) of the tunnel. (D) A three-quarter exploded view of the tunnel with bound GTP. (E) Space-filling surface view looking into the tunnel entrance of the enzyme with bound GTP.
Table 1.
Key interactions between the active site residues of the yeast RNA triphosphatase and GTP as predicted by the molecular docking model
| Amino acid | Interaction | Distance (Å) |
|---|---|---|
| Lys427 | O6 of guanine | 3.9 |
| Ser429 | NH2 of guanine | 2.6 |
| Asn431 | NH2 of guanine | 3.9 |
| Glu307 | 3′OH of ribose | 4.4 |
| Arg393 | 2′OH of ribose | 2.6 |
| Lys409 | 3′OH of ribose | 3.5 |
| Arg454 | 2′OH of ribose | 3.9 |
| Glu305 | β-PO4 and γ-PO4 | 3.2 and 3.7 |
| Arg393 | α-PO4 | 3.4 |
| Lys409 | α-PO4 | 3.2 |
| Glu433 | β-PO4 and γ-PO4 | 3.1 and 3.6 |
| Lys456 | α-PO4 | 3.6 |
| Arg458 | α-PO4 and β-PO4 | 3.2 and 2.9 |
| Asp471 | α-PO4 and β-PO4 | 4.1 and 4.4 |
| Glu494 | α-PO4 | 4.3 |
| Glu496 | β-PO4 and γ-PO4 | 3.3 and 3.9 |
Figure 2.
Active site of the yeast RNA triphosphatase. The GTP molecule is coordinated by an elaborate network of interactions. The side chains of an important number of amino acids are contacting the phosphates, ribose, and guanine base of the substrate. Different views of the enzyme with bound GTP are shown (A-D).
Analysis of the docking model between the enzyme and GTP suggests that only four amino acids are responsible for the coordination of the hydroxyl groups of the ribose. These are Glu307 and Lys409, which are contacting the 3′OH group, and Arg393 and Arg454, which are coordinating the 2′OH group of the sugar (Figure 2A and D; Table 1). Previous mutational studies have shown that each of these four residues is essential for catalysis (9,17–19). The molecular docking model also highlights key amino acids involved in the binding of the guanine base. These amino acids are either contacting the exocyclic 2-amino (Ser429 and Asn431) or the 6-oxo (Lys427) groups of the pyrimidine ring of guanine (Figure 2A and D). Previous structure–function analyses of the amino acids that are contacting the guanine base in the docking model indicate that these three residues are not essential for catalysis (17,19). Moreover, pi–pi- and cation–pi-stacking interactions are also occurring between His411 (nonessential) and the pyrimidine ring of guanine, while Pro341 is engaged in hydrophobic interactions with the imidazole ring of guanine (Figure 2A and C). The nonessentiality of the amino acids contacting the guanine base is not surprising since the enzyme can efficiently hydrolyze both purine and pyrimidine nucleotides. Both the coordination geometry and the nature of the amino acids contacting the purine/pyrimindine rings are likely modified according to the precise nature of the substrate.
Nucleotide analogs to probe the active site
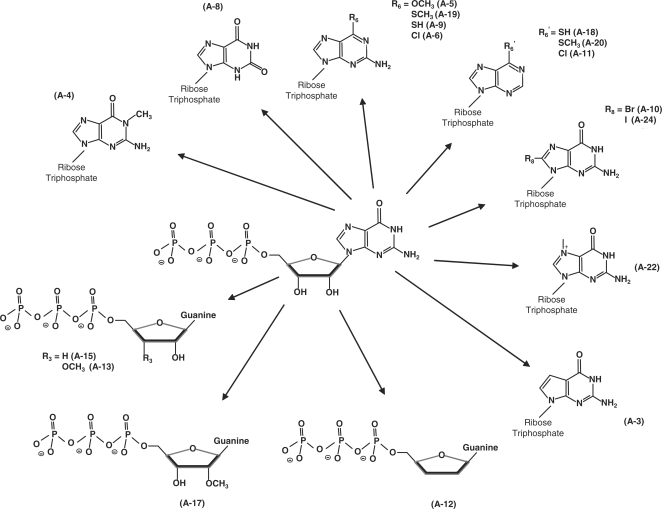
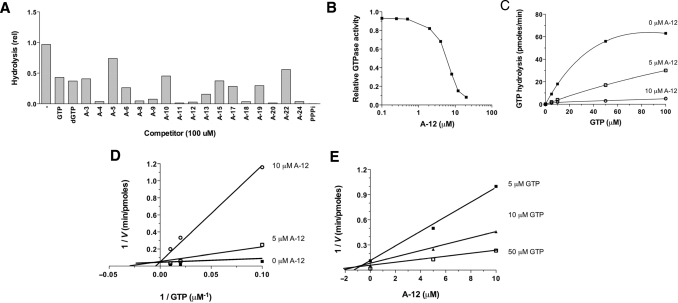
In order to experimentally confirm the docking model and to gain additional insights on the molecular determinants involved in the formation of the enzyme–substrate complex, we have used nucleotide analogs to monitor their effects on the reaction chemistry. The nucleotide analogs displayed various modifications on both the ribose and the guanine base of GTP (Figure 3). We initially monitored the ability of 17 analogs of GTP to inhibit the activity of the yeast RTPase by evaluating both the IC50 and Ki values for each molecule (Table 2). The informative finding is that every analog tested had the ability to inhibit the RTPase reaction, albeit to different extents, thus highlighting the high structural flexibility of the active site (Figure 4A). All the nucleotide analogs used in the current study were competitive inhibitors of the RTPase reaction indicating that they bind to the active site of the enzyme. A typical example using the analog A-12 is shown in Figure 4B–E. In order to gain additional information on the functional flexibility of the catalytic center, we determined the ability of the various nucleotide analogs to be hydrolyzed by the enzyme. The yeast RTPase catalyzed the phosphohydrolysis of all nucleotide analogs tested in the current study with specific activity ranging from 7% to 138% of GTP (Table 2). Interestingly, some of the analogs bound strongly to the enzyme active site (as evidenced from the low IC50 and Ki values) but were not efficiently hydrolyzed. These analogs (A4, A8, A11, A18, A20 and A24) had IC50 values ranging from 2 to 16 μM with low specific phosphohydrolase activities varying from 7% to 18% of the GTP substrate (Table 2).
Figure 3.
Nucleotide analogs used in the current study. The GTP analogs harbored various modifications on both the ribose and the guanine base of GTP.
Table 2.
Inhibition of the GTPase activity by nucleotide analogs
| Molecule | IC50 (μM) | Ki (μM) | Phosphohydrolase specific activity (Percentage of GTP)a | Inhibitory potentialb |
|---|---|---|---|---|
| GTP | 87 | 16 | 100 | 1.0 |
| dGTP | 76 | 14 | 99 | 1.1 |
| A-3 | 83 | 15 | 88 | 1.2 |
| A-4 | 8.1 | 1.5 | 18 | 60 |
| A-5 | 150 | 27 | 17 | 3.4 |
| A-6 | 53 | 9.6 | 70 | 2.4 |
| A-8 | 10 | 1.8 | 10 | 86 |
| A-9 | 16 | 2.9 | 41 | 13 |
| A-10 | 93 | 17 | 20 | 4.7 |
| A-11 | 2.0 | 0.4 | 16 | 268 |
| A-12 | 6.1 | 1.1 | 138 | 10 |
| A-13 | 32 | 5.9 | 109 | 2.5 |
| A-15 | 49 | 8.8 | 111 | 1.6 |
| A-17 | 59 | 11 | 116 | 1.3 |
| A-18 | 8.1 | 1.5 | 12 | 89 |
| A-19 | 61 | 11 | 38 | 3.8 |
| A-20 | 2.0 | 0.4 | 7.2 | 597 |
| A-22 | 113 | 21 | 81 | 0.9 |
| A-24 | 8.1 | 1.5 | 7.2 | 149 |
| Tripoly-PO4 | 0.4 | 0.1 | 3.6 | 5965 |
aThe phosphohydrolase specific activities were calculated from the slopes of the titration curves and normalized to the specific activity for the hydrolysis of GTP.
bThe inhibitory potential is defined by the following equation:
(KiGTP/KiMolecule) × (Spec. activityGTP/Spec. activityMolecule)
Figure 4.
Inhibition of the phosphohydrolase activity by nucleotide analogs. (A) The standard GTPase activity was performed in the presence of 20 μM [γ-32P]GTP. The various nucleotide analogs were added at a single concentration of 100 μM and the inhibition of the GTPase activity was evaluated by monitoring the release of radiolabeled inorganic phosphate, which was separated from the GTP substrate by thin-layer chromatography. (B) Dose–response inhibition of the GTPase activity by A12 (2′,3′-dideoxy-GTP). (C and D) Competitive inhibition of the GTPase reaction by A12. The GTPase activity was evaluated in the absence (filled square) or presence of five (open square) or 10 μM (open circle) of A12. (E) Dixon plot of the inhibition performed at each fixed concentration of GTP substrate, five (filled square), 10 (filled triangle), or 50 μM (open square).
Our study indicates that the active center of the yeast RTPase is highly flexible and can accommodate nucleotide substrates displaying a number of unusual chemical modifications. For instance, analogs harboring modifications on the hydroxyl moieties of the ribose (2′OH and 3′OH) were hydrolyzed with a high level of efficiency by the enzyme. The addition of a methyl group to either the 2′ or 3′ hydroxyls (A-13 and A-17) had no significant effect on the phosphohydrolase activity (Table 2). Most strikingly, A-12 which lacks both ribose hydroxyls (2′,3′-dideoxy-GTP) was hydrolyzed very efficiently by the yeast RTPase. This was unexpected since amino acids contacting the ribose hydroxyls (Glu307, Arg393, Lys409, Arg454) were previously shown to be essential for catalysis through mutational studies (9,17–19). However, we observe that two of these residues (Arg393 and Lys409) are also contacting the α-phosphate of the bound GTP, while Glu307 coordinates the essential divalent cation. Moreover, Arg454 forms a salt bridge with Glu492, an interaction that is important for the stabilization of the tunnel architecture (15,17). Therefore, we conclude that the importance of these amino acids is not directly related to their ability to bind to the hydroxyls of the ribose, but rather lies in their other functions namely through the coordination of the α-phosphate and metal ion or in the stabilization of the tunnel structure.
Some of the analogs used in the current study contain chemical modifications on the guanine base of the GTP molecule (Figure 3). Although the S. cerevisiae RTPase is active on both purine and pyrimidine nucleotides (ATP, CTP and UTP were used as substrates with specific activities of 103%, 103% and 109% of the GTP substrate, respectively), analysis of the GTP analogs helped to illuminate the flexibility and complexity of the RTPase active site. For instance, the addition of a chemical group to the C8 position of guanine had a negative effect on the phosphohydrolysis activity. Analogs harboring such modification (A-10 and A-24) were used inefficiently by the enzyme. Although it can be argued that the addition of a bromo- or iodo- group at this position can potentially alter the electronic properties of the guanine ring, analysis of the docking model indicates that steric hindrance is the most likely explanation for the limited hydrolysis of these substrates. Analysis of the enzyme–GTP model indicates that the space is occupied by Lys474 and Glu476 of the β10 strand that comprises part of the walls of the tunnel. The presence of halogen substituents with large atomic radius at the C8 position of the guanine base reveals that the conformational importance of amino acids is not directly involved in catalysis. The importance of these amino acids in forming an optimal nucleotide binding pocket could not be inferred from previous mutational analyses. Similarly, the addition of a methyl group at the N1 position of guanine (A-4, N1-methyl-GTP) decreases the catalytic activity of the enzyme by 5-fold (Table 2). In our docking model, Lys427 occupies the space that is filled by the additional methyl group, thereby hindering the optimal positioning of the substrate and the concomitant hydrolysis of the analog (Figure 2A, C and D).
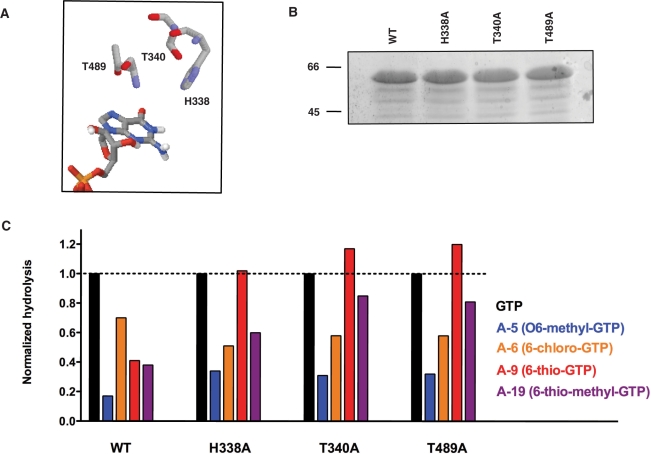
The 6-oxo group of the guanine ring also appears important for substrate recognition/alignment and hydrolysis. The phosphohydrolysis activity of all the analogs, which harbored chemical modifications at this position (A-5, A-6, A-9, A-19), was lower as compared to the hydrolysis of GTP (Table 2). We initially suspected that the inability of these analogs to form hydrogen bonds through this position was the likely explanation to explain their low catalytic usage. However, Lys427 is the only proximal amino acid that can potentially coordinate the 6-oxo group of guanine through hydrogen bonding, and previous studies have shown that the substitution of this residue by alanine has no effect on the catalytic activity (17). Steric hindrance might again be a factor since analogs with larger substituents (A-5, O6-methyl-GTP; A-9, 6-thio-GTP and A-19, 6-thio-methyl-GTP) were less hydrolyzed relative to A-6 (6-chloro-GTP), which harbored a smaller substituent at the same position on the guanine ring. Closer examination of the enzyme–GTP docking model indicates that three amino acids are in the vicinity of the 6-oxo group of guanine (Figure 5A). These are His338 and Thr340 of the β2 strand and Thr489 of the β11 strand, which are just too far removed to hydrogen bond with the 6-oxo group but might interfere with the larger substituents displayed by the analogs. The importance of these amino acids was, therefore, investigated by generating three distinct enzymatic mutants. His338, Thr340 and Thr489 were individually substituted by alanine and the mutant polypeptides were expressed and purified in parallel with the wild-type enzyme (Figure 5B). The GTP substrate was hydrolyzed efficiently by the mutant polypeptides as evidenced from the specific activities of 98, 69 and 79% of wild-type activity for the H338A, T340A and T489A mutant, respectively (data not shown). The informative finding is that the replacement of these lateral side chains by alanine resulted in an increase in the phosphohydrolysis of analogs harboring larger substituents at the C6 position (A-5, A-9 and A-19). For instance, the relative hydrolysis of 6-thio-methyl-GTP (A-19) increased from 0.38 for the wild-type enzyme to 0.60, 0.85 and 0.81 for the H338A, T340A and T489A mutants, respectively (Figure 5C). A similar increase in phosphohydrolysis was also observed for both the A-5 (O6-methyl-GTP) and A-19 (6-thio-methyl-GTP) analogs when the His338, Thr340 and Thr489 lateral side chains were individually replaced by alanine (Figure 5C). However, these substitutions had no positive effect on the phosphohydrolysis of A-6 (6-chloro-GTP), which harbored a smaller substituent at the same position on the guanine ring. Overall, the mutagenesis data are consistent with steric hindrance preventing the phosphohydrolysis of analogs harboring larger substituents at the C6 position.
Figure 5.
Steric hindrance caused by analogs harboring large substituents at the 6-oxo position of the guanine ring of GTP. (A) The His338, Thr340 and Thr489 residues of the enzyme are in the vicinity of the 6-oxo group of the GTP substrate. (B) Aliquots (2 µg) of the purified wild-type (WT, lane 1), H338 A (lane 2), Thr340A (lane 3) and Thr489A (lane 4) mutant proteins were analyzed by electrophoresis through a 12.5% polyacrylamide gel containing 0.1% SDS and visualized with Coomassie Blue Dye. The positions and sizes (in kDa) of the size markers are indicated on the left. (C) Normalized phosphohydrolase activities of the WT, H338A, T340A and T489A mutants. The phosphohydrolase-specific activities were calculated from the slopes of the titration curves and normalized to the specific activity for the hydrolysis of GTP by the WT enzyme. GTP, O6-methyl-GTP (A-5), 6-chloro-GTP (A-6), 6-thio-GTP (A-9) and 6-thio-methyl-GTP (A-19) were used as substrates.
One of the most striking features of the GTP–enzyme complex formation is the importance of the 2-amino group of guanine. In that regard, A-11, A-18 and A-20 are particularly interesting since they only differ with A-6, A-9 and A-19 by the lack of the 2-amino moiety (Figure 3). The absence of the amino group drastically reduces the hydrolysis of the analogs (compare A-6 with A-11, A-9 with A-18 and A-19 with A-20) highlighting its importance for efficient catalysis (Table 2). The importance of this functional group is also underscored by the relative inability of the enzyme to hydrolyze A-8, which only differs with GTP by harboring a 6-oxo group at this position instead of the amino group (Table 2). The most likely explanation for the importance of the 2-amino group is that it can act as hydrogen-bond donor with specific amino acids. Analysis of our docking model with bound GTP reveals that two amino acids (Ser429 and Asn431) are potentially contacting the 2-amino group of guanine through hydrogen bonding. We hypothesize that Ser429 is likely the key amino acid responsible for coordination of the 2-amino group. Previous conservative substitution analyses have revealed the importance of the hydroxyl moiety of the lateral side chain of Ser429 for the activity of the protein (18). Moreover, earlier in vivo studies have indicated that the substitution of this residue by alanine elicits a cold-sensitive phenotype, thereby highlighting the importance of this residue in the proper folding of the enzyme (18). As for Asn431, previous mutational analyses have shown that it is not critical for hydrolysis (17), and its distance from the 2-amino group suggests that it makes a rather weak hydrogen bond with guanine.
The RTPase component of the capping machinery is an attractive target for the future development of inhibitors targeting pathogenic fungi, viruses and protozoans since these RTPases are structurally and mechanistically different from their human counterpart. Some of the analogs identified in the current study appear as good starting blocks for the design of more specific inhibitors. These analogs bound very tightly to the yeast RTPase active site but were not efficiently hydrolyzed. In that respect, both A-11 and A-20, which are structurally related with a substitution of the 6-oxo group and the lack of the 2-amino group of guanine, possess high inhibitory potential (Table 2). Although tripolyphosphate, which was previously shown to inhibit other RTPases of the TTM family (31), displays even higher inhibitory potential in vitro, it is unlikely that such a molecule could eventually display any specificity toward pathogenic RTPase in vivo. Because our study revealed the intrinsic flexibility of the RTPase active site, we believe that additional chemical modifications of A-11 and A-20 could lead to the development of molecules displaying a very robust and specific inhibition of RTPases of pathogenic importance.
CONCLUSIONS
The current study highlights both the complexity and flexibility of the TTM family active site. It is clear that the RTPase reaction requires a very precise alignment between the active-site residues, the substrate and the metal ion cofactor. Two observations support this conclusion. First, the hydrolysis of RNA triphosphate ends is activated by magnesium, but not by manganese or cobalt, whereas the NTPase activity is supported by manganese and cobalt, but not magnesium. Second, unnatural substrates such as nucleotide analogs or tripolyphosphate can still be hydrolyzed even if the conformation of the active site is less than optimal, lacking several important contacts with the substrate. In summary, our structural docking model—coupled with the use of an important number of nucleotide analogs—highlights the importance of specific residues for the binding of the nucleotide substrate, and reveals both the structural flexibility and complexity of the active site. Our study illustrates the important structural and functional features for the interaction of an RTPase with a ligand and opens the way to the use of nucleotide analogs as potential inhibitors of the RTPases of pathogens.
FUNDING
Canadian Institutes for Health Research (CIHR) grant. CIHR and Université de Sherbrooke grant to RNA group. M.B. is a Chercheur Boursier Junior 2 from the Fonds de recherche en santé du Québec and a member of the Infectious diseases group of the Centre de Recherche Clinique Étienne-Lebel. Funding for open access charge: Canadian Institutes for Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 4.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 5.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic. Acids Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 6.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 7.Tagaki T, Moore CR, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 8.Changela A, Ho CK, Martins A, Shuman S, Mondragon A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho CK, Pei Y, Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J. Biol. Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 10.Ho CK, Shuman S. A yeast-like mRNA capping apparatus in Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 2001;98:3050–3055. doi: 10.1073/pnas.061636198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hausmann S, Altura MA, Witmer M, Singer SM, Elmendorf HG, Shuman S. Yeast-like mRNA capping apparatus in Giardia lamblia. J. Biol. Chem. 2005;280:12077–12086. doi: 10.1074/jbc.M412063200. [DOI] [PubMed] [Google Scholar]
- 12.Hausmann S, Vivares CP, Shuman S. Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi. J. Biol. Chem. 2002;277:96–103. doi: 10.1074/jbc.M109649200. [DOI] [PubMed] [Google Scholar]
- 13.Myette J, Niles EG. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleotide triphosphate phosphohydrolase activities. Demonstrate that both activities are carried out at the same active site. J. Biol. Chem. 1996;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- 14.Martins A, Shuman S. Mutational analysis of baculovirus capping enzyme Lef4 delineates an autonomous triphosphatase domain and structural determinants of divalent cation specificity. J. Biol. Chem. 2001;276:45522–45529. doi: 10.1074/jbc.M107615200. [DOI] [PubMed] [Google Scholar]
- 15.Lima CD, Wang LK, Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 16.Gong C, Smith P, Shuman S. Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins. RNA. 2006;12:1468–1474. doi: 10.1261/rna.119806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisaillon M, Shuman S. Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 2001;276:17261–17266. doi: 10.1074/jbc.M100980200. [DOI] [PubMed] [Google Scholar]
- 18.Bisaillon M, Shuman S. Functional groups required for the stability of yeast RNA triphosphatase in vitro and in vivo. J. Biol. Chem. 2001;276:30514–30520. doi: 10.1074/jbc.M104936200. [DOI] [PubMed] [Google Scholar]
- 19.Pei Y, Ho CK, Schwer B, Shuman S. Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J. Biol. Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- 20.Benarroch D, Smith P, Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. doi: 10.1016/j.str.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Bisaillon M, Bougie I. Investigating the role of metal ions in the catalytic mechanism of the yeast RNA triphosphatase. J. Biol. Chem. 2003;278:33963–33971. doi: 10.1074/jbc.M303007200. [DOI] [PubMed] [Google Scholar]
- 22.Bougie I, Bisaillon M. Inhibition of a metal-dependent viral RNA triphosphatase by decavanadate. Biochem. J. 2006;398:557–567. doi: 10.1042/BJ20060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayo SL, Olafson BD, Goddard WA. DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 1990;94:8897–8909. [Google Scholar]
- 24.Stewart JPP. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 26.Solis FJ, Wets RJB. Minimization by Random Search Techniques. Math. Operat. Res. 1981;6:19–30. [Google Scholar]
- 27.Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei Y, Schwer B, Hausmann S, Shuman S. Characterization of Schizosaccharomyces pombe RNA triphosphatase. Nucleic Acids Res. 2001;29:387–396. doi: 10.1093/nar/29.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keppetipola N, Jain R, Shuman S. Novel triphosphate phosphohydrolase activity of Clostridium thermocellum TTM, a member of the triphosphate tunnel metalloenzyme superfamily. J. Biol. Chem. 2007;282:11941–11949. doi: 10.1074/jbc.M611328200. [DOI] [PubMed] [Google Scholar]
- 30.Jain R, Shuman S. Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme. J. Biol. Chem. 2008;283:31047–31057. doi: 10.1074/jbc.M805392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong C, Shuman S. Chlorella virus RNA triphosphatase. Mutational analysis and mechanism of inhibition by tripolyphosphate. J. Biol. Chem. 2002;277:15317–15324. doi: 10.1074/jbc.M200532200. [DOI] [PubMed] [Google Scholar]