Abstract
Poly(ADP-ribose) polymerase 1 (PARP1) synthesizes poly(ADP-ribose) (PAR) using nicotinamide adenine dinucleotide (NAD) as a substrate. Despite intensive research on the cellular functions of PARP1, the molecular mechanism of PAR formation has not been comprehensively understood. In this study, we elucidate the molecular mechanisms of poly(ADP-ribosyl)ation and identify PAR acceptor sites. Generation of different chimera proteins revealed that the amino-terminal domains of PARP1, PARP2 and PARP3 cooperate tightly with their corresponding catalytic domains. The DNA-dependent interaction between the amino-terminal DNA-binding domain and the catalytic domain of PARP1 increased Vmax and decreased the Km for NAD. Furthermore, we show that glutamic acid residues in the auto-modification domain of PARP1 are not required for PAR formation. Instead, we identify individual lysine residues as acceptor sites for ADP-ribosylation. Together, our findings provide novel mechanistic insights into PAR synthesis with significant relevance for the different biological functions of PARP family members.
INTRODUCTION
Poly(ADP-ribose) polymerases (PARPs) use nicotinamide adenine dinucleotide (NAD) as substrate to synthesize poly(ADP-ribose) (PAR) (1). On the cellular level, PAR formation has been implicated in a wide range of processes, such as maintenance of genomic stability, transcriptional regulation, energy metabolism and cell death (2).
PARP1 was the first protein described to catalyze PAR formation in response to mitogenic stimuli or genotoxic stress (3–7). It contains three functionally distinct domains: an amino-terminal DNA-binding domain (DBD), an auto-modification domain (AD) and a carboxyl-terminal PARP homology domain that includes the catalytic domain (CAT) responsible for PAR formation (8). The DBD extends from the initiator methionine to threonine 373 in human PARP1. It contains two structurally and functionally unique zinc fingers (FI: aa, amino acid, 11–89; FII: aa 115–199) (2,9). Recently, a third and so far unrecognized zinc-binding motif was discovered (FIII: aa 233–373) (10,11). The DBD also contains a bipartite nuclear localization signal (NLS) of the form KRK-X(11)-KKKSKK (aa 207–226) that targets PARP1 to the nucleus (12). The PARP1 zinc fingers FI and FII are thought to recognize altered structures in DNA rather than particular sequences and have also been reported to be involved in protein–protein interactions (13). PARP1 strongly associates with DNA single and double strand breaks generated either directly by DNA damage or indirectly by the enzymatic excision of damaged bases during DNA repair processes (2,9). Several studies indicate that the first zinc finger is required for PARP1 activation by both DNA single and double strand breaks, whereas the second zinc finger may exclusively act as a DNA single strand break sensor (2,9).
The AD of PARP1 is located in the central region of the enzyme, between residues 373 and 525 of human PARP1 (14,15). It was identified as the domain containing acceptor amino acids for the covalent attachment of PAR (16). In addition, several recent studies identified a weak leucine-zipper motif in the amino-terminal region of the AD, which suggests that this motif might be involved in homo- and/or hetero-dimerization (9). The AD of PARP1 also comprises a breast cancer 1 protein (BRCA1) C-terminus (BRCT) domain (from aa 386 to 464 in human PARP1) as well as an unstructured loop that connects the AD with the PARP homology domain.
PARP1 contains an 80–90 amino acid long tryptophane-, glycine-, arginine-rich (WGR) domain carboxyl terminal of the AD. The WGR domain is named after the most conserved central motif of tryptophane (W), glycine (G), arginine (R) residues and may represent a nucleic-acid-binding domain (2). This region of PARP1 has not been extensively characterized and its function is still unknown. The CAT has been suggested to catalyze at least three different enzymatic reactions: the attachment of the first ADP-ribose moiety onto an acceptor amino acid (initiation reaction), the addition of further ADP-ribose units onto already existing ones (elongation reaction) and the generation of branching points (branching reaction) (8). The active site is formed by a phylogenetically well-conserved sequence of ∼50 residues (aa 859–908 of hPARP1). This ‘PARP signature’ contains the NAD acceptor sites and critical residues involved in the initiation, elongation and branching of PAR.
Like PARP1, both PARP2 and PARP3 also contain a WGR as well as a CAT (16). PARP2 and PARP3 lack, however, most motifs present in the amino-terminal half of PARP1. Neither zinc-binding motifs nor leucine-zippers or BRCT domains have been described for PARP2 or PARP3. PARP2 contains an amino-terminal SAP/SAF motif/module [named after scaffold-associated protein/scaffold-associated factor SAF-A/B, Acinus and PIAS; (17)] and a eukaryotic module proposed to be involved in sequence- or structure-specific DNA and RNA binding (18). Furthermore, PARP2 contains an amino-terminal NLS which targets the protein to the nucleus. PARP3 is the least studied and smallest PARP identified so far (19). The protein domain structure of PARP3 is very similar to that of PARP2, featuring a small putative DBD consisting of only 54 residues and apparently containing a targeting motif that is sufficient to localize the enzyme to the centrosome (19,20).
Attempts to obtain structural information on the full-length proteins PARP1, PARP2 and PARP3 by X-ray crystallography or by nuclear magnetic resonance (NMR) have not been successful up to now. The 3D structures of single domains, however, have been solved and allow for a structure-based comparison of different PARP family members (8,21) (PDB: 1A26, 1GS0 and 2PA9). Although the amino acid identity between PARP1 and PARP2 or PARP3 is only moderate (40% and 32% in the CAT, respectively), the overall structure of the CATs of these three proteins is nearly identical. This conservation suggests, in general, similar capabilities to generate PAR. Both PARP1 and PARP2 have been shown to synthesize very complex branched polymers at least in vitro (2). The enzymatic activity of PARP3 and its isoforms has not yet been investigated in detail.
An unresolved issue regarding the mechanism of poly(ADP-ribosyl)ation is how DNA binding in the amino-terminal DBD triggers enzyme activation in the carboxyl-terminal CAT and how the different domains of the different PARPs are coordinated during this process. Furthermore, earlier studies suggested that the auto-modification activity targets between 4 and 28 acceptor residues located in the AD and in the DBD of PARP1 (14,22,23). For histone H1, a major target for trans-poly(ADP-ribosyl)ation by PARP1, glutamic acid residues have been described to function as acceptors for PAR (24). This, together with the reported chemical similarity between the ADP-ribose-PARP1 linkage and carboxyl esters in mono-ADP-riboslyated histones (23), led to the hypothesis that multiple glutamic acid residues present in the AD of PARP1 might function as acceptor sites for auto-poly(ADP-ribosyl)ation (16). However, despite intensive research during the last 40 years, the acceptor amino acids in PARP1 have not been confirmed by mutational studies.
Here, we comprehensively analyze PAR formation by PARP1, PARP2 and PARP3 and find a close cooperativity between the amino-terminal portions of the proteins and their corresponding CATs. We define the DBD (aa 1–373) and the WGR/CAT domain (aa 533–1014) as the minimal domains of PARP1 required for PAR formation. The DNA-dependent interaction between the DBD and the CAT increased Vmax and decreased the Km for NAD. Furthermore, by amino-acid substitutions, we establish that glutamic acid residues within the AD are not required for PAR formation and thus do not function as acceptor amino acids for PAR. Instead, we identify lysine residues within the AD of PARP1 as acceptor sites for ADP-ribosylation.
MATERIALS AND METHODS
Chemicals and antibodies
3H-NAD and protein A sepharose were purchased from GE Healthcare and 32P-NAD was from PerkinElmer. NAD was obtained from Sigma-Aldrich. Anti-PAR antibody LP96-10 was from Alexis Biochemicals or Becton Dickinson, anti-PARP1cat antibody H250 from Santa Cruz Biotechnology and anti-haemagglutinin (HA) antibody 16B12 from Covance.
Plasmids
The baculovirus expression vectors pQE-TriSystem (Qiagen) and BacPak8 (Clontech) were used for the expression of recombinant proteins in Sf21 insect cells as described previously (25,26).
Cloning, expression and purification of recombinant proteins
Wild-type hPARP1 (NCBI ID: BC037545), hPARP2 (NCBI ID: NM_001042618) and hPARP3 (NCBI ID: BC014260) were cloned and expressed as carboxyl-terminal His-tagged proteins. PARP family chimera were generated by overlapping polymerase chain reaction (PCR) at the position corresponding to amino acid 533 in hPARP1 and expressed as carboxyl-terminal His-tagged proteins. Protein fragments and deletion mutants were generated by PCR and expressed as carboxyl-terminal His-tagged proteins as described before (25,26). Amino-acid substitutions were introduced by site-directed PCR-based mutagenesis and mutant proteins were expressed as described before (25,26). All recombinant proteins were purified by one step affinity chromatography using ProBond resin according to the manufacturer's recommendations (Invitrogen). Expression and purification of all recombinant proteins was analyzed by SDS–PAGE followed by coomassie staining. For the stacking gel a 4.5% acrylamide-bis solution [37.5:1, 40% (w/v), Serva] and for the separating gel a 10–12.5% acrylamide-bis solution was used.
PAR formation assays
3H-NAD time course experiments
One hundred picomoles recombinant purified enzyme and 5 μg of protein fragments in PAR reaction buffer (50 mM Tris–HCl pH 8.0, 4 mM MgCl2, 250 μM DTT, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin) in the presence of 50 pmol annealed double-stranded oligomer (5′-GGAATTCC-3′) were supplemented with 3H-NAD to a final concentration of 400 μM. PAR formation was allowed for 1, 3, 5, 15 and 60 min at 30°C. Reactions were stopped by addition of ice-cold 10% TCA/2% Na4P2O7. Polymers were precipitated for 10 min on ice and then applied onto filter papers. Counts per minute were obtained by liquid scintillation counting. For the determination of Vmax and Km, initial reaction velocities (V0) were obtained by measuring PAR levels generated after 0, 1, 3 and 5 min incubation at different 3H-NAD concentrations and using the GraphPad Prism software for nonlinear regression analysis assuming a one-site binding model. Vmax and Km were calculated from V0 according to Michaelis–Menten.
Anti-PAR western blot
Unless otherwise stated, 10 pmol recombinant purified enzyme and 0.5 μg of protein fragments in PAR reaction buffer in the presence of 5 pmol annealed double-stranded oligomer (5′-GGAATTCC-3′) were supplemented with NAD to a final concentration of 400 μM. PAR formation was allowed for 5 min at 30°C. Reactions were stopped by addition of SDS–PAGE loading buffer and boiling for 5 min at 95°C. Samples were subjected to SDS–PAGE followed by anti-PAR western blot.
32P-NAD auto-modification
Unless otherwise stated, 10 pmol recombinant purified enzyme and 0.5 μg of protein fragments in PAR reaction buffer in the presence of 5 pmol annealed double-stranded oligomer (5′-GGAATTCC-3′) were supplemented with 32P-NAD to a final concentration of 100 nM. Auto-modification was allowed for 10 s at 30°C. Reactions were stopped by addition of SDS–PAGE loading buffer and boiling for 5 min at 95°C. Samples were subjected to SDS–PAGE followed by detection of auto-modification by autoradiography.
PAR detection by silver staining
Following synthesis of PAR as for western blot analysis, PAR chains were purified and separated by modified DNA sequencing gel electrophoresis as described by Fahrer et al. (27).
In vitro co-immunoprecipitation
Ten picomoles recombinant purified enzyme and 0.5 μg of protein fragments were incubated for 5 min at 30°C in Co-IP buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 4 mM MgCl2, 0.2% NP-40, 250 μM DTT, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin) in the absence or presence of 5 pmol annealed double-stranded oligomer (5′-GGAATTCC-3′). The CAT of PARP1 was allowed to bind to the anti-PARP1cat antibody for 1 h at 4°C. Protein A sepharose was added and samples were incubated for another 2 h at 4°C. Samples were washed three times for 5 min in Co-IP buffer containing 300 mM NaCl before being subjected to SDS–PAGE followed by western blot.
RESULTS
Purified full-length human PARP1 and PARP2 are enzymatically active
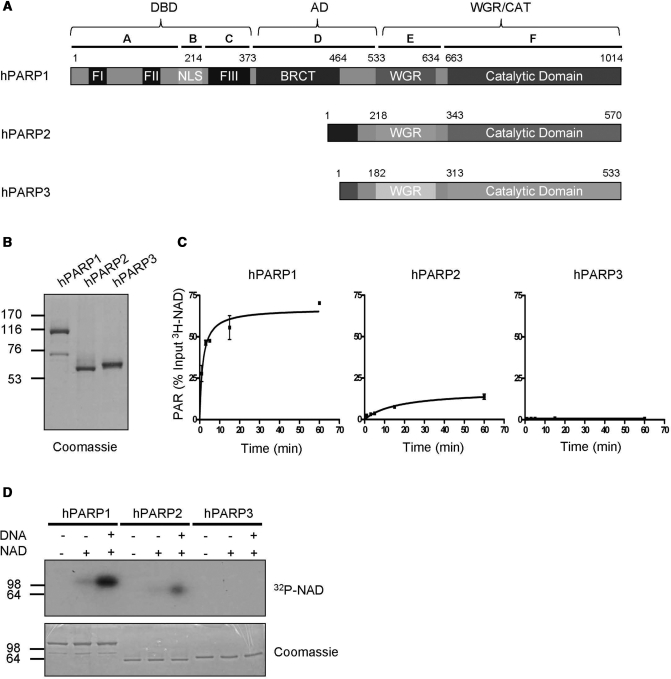
In order to gain detailed insights into the mechanism of PAR formation by different PARP family members, we expressed and purified full-length human PARP1, PARP2 and PARP3 using the baculovirus expression system (Figure 1A and B). PARP3 showed a slower migration velocity than predicted in SDS–PAGE, possibly due to the high content of hydrophobic amino acids in the CAT. To measure PAR formation, the purified proteins were incubated for different time periods with 400 μM tritium-labeled NAD in the presence of double strand break mimicking DNA. Reaction products were precipitated by trichloroacetic acid (TCA) before they were analyzed using a beta counter. PARP1 generated PAR in a time-dependent manner (Figure 1C, left panel). PARP2 also synthesized PAR in a time-dependent manner, however, not as efficiently as PARP1 (Figure 1C, middle panel). The reduced amount of product formed by PARP2 most probably represents a quantitative rather than a qualitative difference, since the length distribution of PAR chains formed by PARP2 was comparable to the length distribution of PAR formed by PARP1 (Supplementary Figure 4B). Human PARP3 did not produce detectable amounts of PAR under the tested conditions (Figure 1C, right panel).
Figure 1.
Purified full-length human PARP1 and PARP2 are enzymatically active. (A) Domain organization of human PARP1, PARP2 and PARP3. Letters A–F indicate domain nomenclature of PARP1 and numbers indicate amino acid positions. (B) Purity of PARP family members after one step affinity chromatography. One microgram of each recombinant, purified protein was used for SDS–PAGE followed by coomassie staining. (C) Time course of PAR formation by different PARP family members. 3H-NAD incorporation into TCA-precipitable polymers was determined by scintillation counts. Substrate concentration: 400 μM 3H-NAD. Reactions were performed in triplicates, error bars represent standard deviations. (D) Auto-modification of different PARP family members detected by autoradiography. Substrate concentration: 100 nM 32P-NAD. Molecular size markers in kilo Daltons are indicated.
Assuming that mono(ADP-ribosyl)ation preceeds PAR formation, we assessed the auto-modification of the three proteins after 10 s incubation with 100 nM radiolabeled NAD (Figure 1D). The short incubation period and the low concentration of NAD were chosen to prevent polymer formation. The discrete bands observed using this approach indeed suggest that under these conditions mostly mono(ADP-ribosyl)ation occurred. In line with the time-course experiments, PARP1 and PARP2 were able to auto-modify themselves in an NAD- and DNA-dependent manner while PARP3 was not (Figure 1D).
PARP1 synthesized increasing PAR levels in a time- and DNA-dependent manner detected also by western blot and vacuum slot blot using 400 μM NAD (Supplementary Figure 1A and B). PAR formation after 5 min incubation with 400 μM NAD caused a pronounced shift of the coomassie blue-stained proteins in the denaturing gel due to a severely reduced migration velocity of the poly(ADP-ribosyl)ated proteins when compared to unmodified proteins (Supplementary Figure 1C). The observed basal activity of PARP1 in the absence of DNA can be explained either by a contamination with DNA or by the intrinsic DNA-independent activity of the CAT as described by Simonin et al. (28). Analysis of PAR formation by silver staining after modified DNA sequencing gel electrophoresis confirmed PAR formation by PARP1 and PARP2 and no PAR formation by PARP3 (Supplementary Figure 4B).
The carboxyl-terminal domains of PARP1, PARP2 and PARP3 cannot compensate for each other
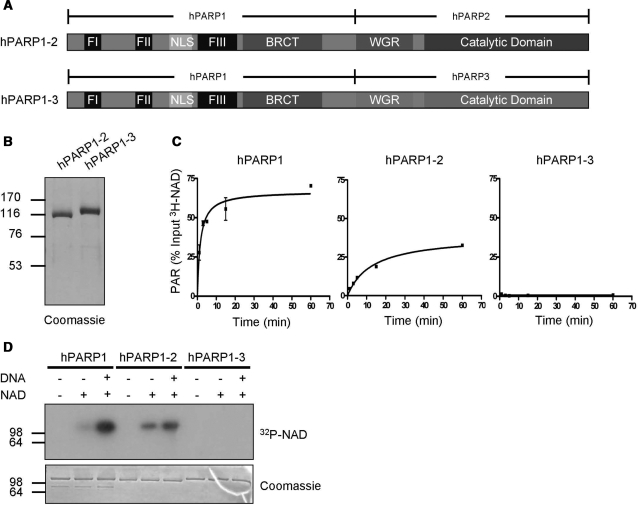
Next, we investigated the crosstalk between the different amino-terminal domains of PARP1, PARP2 and PARP3 with their carboxyl-terminal domains (i.e. WGR/CAT). Therefore, we generated chimera proteins by replacing the WGR/CAT domain of PARP1 with the WGR/CAT domain of PARP2 or PARP3 [named chimera PARP1-2 (aa 1–532 of PARP1 fused to aa 81–570 of PARP2) or chimera PARP1-3 (aa 1–532 of PARP1 fused to aa 48–533 of PARP3)], respectively (Figure 2A and B). We analyzed PAR formation by these proteins and found that replacing the WGR/CAT domain of PARP1 by the one of PARP2 (i.e. chimera PARP1-2) resulted in an active enzyme that showed roughly similar PAR formation in time course experiments as PARP2 (Figure 2C, middle panel). Replacement of the WGR/CAT domain of PARP1 by that of PARP3 (i.e. chimera PARP1-3) resulted in an enzyme that did not produce detectable amounts of PAR under the tested conditions (Figure 2C, right panel and Supplementary Figure 1D). In line with these findings, chimera PARP1-2 was able to auto-modify itself whereas chimera PARP1-3 was not (Figure 2D). Together these results suggest that the WGR/CAT domains of the investigated PARP proteins cannot compensate for each other. The WGR/CAT domains cooperate tightly with their corresponding amino-terminal domains and limit poly(ADP-ribosyl)ation capacity and the ability for auto-modification, despite high levels of structural similarity between the CATs (see Supplementary Figure 2A).
Figure 2.
The carboxyl-terminal domains of PARP1, PARP2 and PARP3 cannot compensate for each other. (A) Domain organization of chimera PARP1-2 and chimera PARP1-3. (B) Purity of chimera PARP1-2 and chimera PARP1-3 after one-step affinity chromatography. One microgram of each recombinant, purified protein was used for SDS–PAGE followed by coomassie staining. (C) Time course of PAR formation by PARP1, chimera PARP1-2 and chimera PARP1-3 as in Figure 1C. (D) Auto-modification of PARP1, chimera PARP1-2 and chimera PARP1-3 detected by autoradiography as in Figure 1D. Molecular size markers in kilo Daltons are indicated.
The carboxyl-terminal domain of PARP1 is not activated by the amino-terminal domains of PARP2 or PARP3
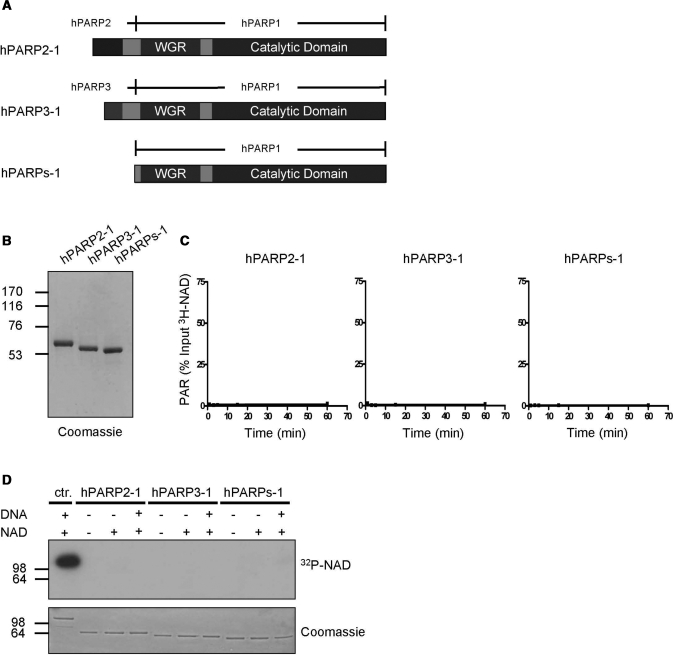
In a second set of chimera proteins we fused the WGR/CAT domain of PARP1 to the amino-terminal domains of PARP2 or PARP3, or deleted the amino-terminal domain of PARP1 [named chimera PARP2-1 (aa 1–80 of PARP2 fused to aa 533–1014 of PARP1), chimera PARP3-1 (aa 1–47 of PARP3 fused to aa 533–1014 of PARP1) or PARPs-1 (aa 533–1014 of PARP1)], respectively (Figure 3A and B). Analysis of these proteins revealed that chimera PARP2-1, chimera PARP3-1 and PARPs-1 did not generate detectable levels of PAR (Figure 3C and Supplementary Figure 1E). Furthermore, no auto-modification of the three proteins was observed under the tested conditions (Figure 3D). These results indicate that the WGR/CAT domain of PARP1 is only stimulated by its corresponding amino-terminal domain, but not by the amino-terminal domains of PARP2 or PARP3.
Figure 3.
The carboxyl-terminal domain of PARP1 is not activated by the amino-terminal domains of PARP2 or PARP3. (A) Domain organization of chimera PARP2-1, chimera PARP3-1 and PARPs-1. (B) Purity of chimera PARP2-1, chimera PARP3-1 and PARPs-1 after one-step affinity chromatography. One microgram of each recombinant, purified protein was used for SDS–PAGE followed by coomassie staining. (C) Time course of PAR formation by chimera PARP2-1, chimera PARP3-1 and PARPs-1 as in Figure 1C. (D) Auto-modification of PARP1 (ctr.), chimera PARP2-1, chimera PARP3-1 and PARPs-1 detected by autoradiography as in Figure 1D. Molecular size markers in kilo Daltons are indicated.
The DBD of PARP1 is sufficient to stimulate its WGR/CAT domain
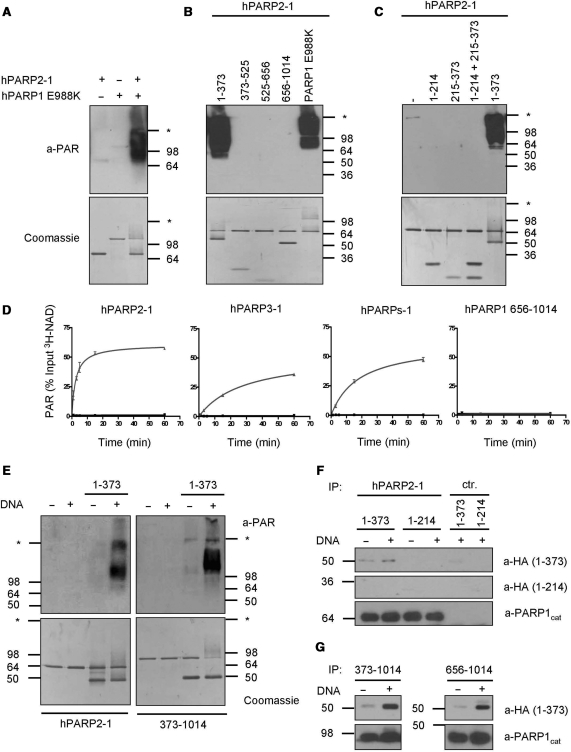
To further investigate the cooperativity between the amino-terminal domain of PARP1 and its WGR/CAT domain, the inactive chimera PARP2-1 was co-incubated with the PARP1 E988K mutant, which lacks the ability to generate PAR. Surprisingly, co-incubation of chimera PARP2-1 with PARP1 E988K strongly induced PAR synthesis, suggesting that PARP1 E988K was able to stimulate the WGR/CAT domain of chimera PARP2-1 (Figure 4A). To map the minimal domain of PARP1, which was able to stimulate the WGR/CAT domain of chimera PARP2-1, we expressed and purified different fragments of PARP1 covering all domains from A to F (see Figure 1A). Analysis of PAR synthesis by western blot upon co-incubation of chimera PARP2-1 with these fragments revealed that the DBD of PARP1 comprising amino acid 1–373 was the only fragment able to stimulate chimera PARP2-1 (Figure 4B). Further dissection of the DBD revealed that only the complete and undisrupted DBD from amino acid 1 to 373 containing FI, FII and FIII was able to stimulate chimera PARP2-1 (Figure 4C). The stimulation of chimera PARP2-1 by the DBD was salt resistant up to 300 mM NaCl (Supplementary Figure 3B, left panel).
Figure 4.
The DBD of PARP1 interacts with and is sufficient to stimulate its WGR/CAT domain. (A) PAR formation by chimera PARP2-1 co-incubated with catalytically inactive PARP1 E988K. PAR was detected by western blot using anti-PAR antibody LP96-10. Substrate concentration: 400 μM NAD. (B) PAR formation by chimera PARP2-1 co-incubated with the indicated fragments of PARP1 or with PARP1 E988K. (C) PAR formation of chimera PARP2-1 co-incubated with the indicated fragments or combination of fragments of PARP1. (D) Time course of PAR formation by chimera PARP2-1, chimera PARP3-1, PARPs-1 and PARP1 656–1014 in the absence or presence of fragment 1–373 as in Figure 1C. Black without fragment 1–373 and grey with fragment 1–373. (E) PAR formation of chimera PARP2-1 or PARP1 373–1014 co-incubated with fragment 1–373 in the absence or presence of DNA. (F) In vitro interaction between chimera PARP2-1 and 1–373. Chimera PARP2-1 was bound to protein A sepharose using an antibody against the CAT of PARP1 (a-PARP1cat) and was then incubated with HA-tagged fragment 1–373 or 1–214 in the absence or presence of DNA. HA-tagged fragments were detected by western blot. PARP1cat antibody coupled to beads without chimera PARP2-1 served as control (ctr.). (G) In vitro interaction between PARP1 373–1014 or 656–1014 with 1–373. Experiments were performed as described in (F). Molecular size markers in kilo Daltons and the border between stacking and separating gel (asterisk) are indicated.
To further assess the specificity of the observed stimulation, the DBD was incubated with different proteins (chimera PARP2-1, chimera PARP3-1, PARPs-1 and PARP1 656–1014) and the time course of PAR formation was analyzed using tritium-labeled NAD. Of note, besides chimera PARP2-1, only chimera PARP3-1 and PARPs-1, but neither the CAT of PARP1 nor full-length PARP2, PARP3, chimera PARP1-2 or chimera PARP1-3, were stimulated by the DBD of PARP1 (Figure 4D and Supplementary Figure 4A and C). Analysis of PAR formation by silver staining after polymer separation using modified DNA sequencing gel electrophoresis confirmed that the observed stimulation in the time course experiments correlated with the synthesis of PAR containing 1 to more than at least 50 ADP-ribose units (Supplementary Figure 4B).
The stimulation of chimera PARP2-1 by the DBD was strongly dependent on DNA (Figure 4E, left panel), which suggests that DNA tightly regulates the interaction necessary for the activation of the CAT. Furthermore, our observation that PARPs-1 but not PARP1 656–1014 together with the DBD was able to generate PAR indicates that the WGR domain of PARP1 is absolutely essential for enzymatic activity.
Since chimera PARP2-1 does not exist physiologically, PAR synthesis by a PARP1 DBD deletion mutant (aa 373–1014) co-incubated with the DBD was analyzed. Interestingly, the DBD was able to stimulate PARP1 373–1014 in a DNA-dependent manner and comparable to PARP2-1 (Figure 4E, right panel and Supplementary Figure 3B, right panel), suggesting that the observed stimulation of PARP2-1 by the DBD represents a physiological regulatory mechanism in the PARP1 full-length context.
The DBD of PARP1 interacts with its CAT domain
The results described above suggest that the DBD of PARP1 interacts with the CAT and/or the WGR domain to stimulate PAR synthesis by the CAT. To test this hypothesis experimentally, in vitro co-immunoprecipitation assays were performed with purified proteins and fragments. The complete DBD (aa 1–373), but not aa 1–214 alone, specifically bound to chimera PARP2-1 in a manner that was stabilized by DNA (Figure 4F). Similarly, the DBD also bound to PARP1 373–1014, and this interaction was enhanced by DNA (Figure 4G, left panel). Interestingly, the CAT domain of PARP1 without the WGR (aa 656–1014) was sufficient for the DNA-dependent interaction with the DBD (Figure 4G, right panel). Since PAR formation was only observed when combining the DBD with PARPs-1 (expressing WGR/CAT) but not with the CAT domain of PARP1 alone (Figure 4D and Supplementary Figure 4A), we conclude that an intact DBD (aa 1–373) interacts with the CAT in a DNA-dependent manner and that the WGR domain is additionally required to allow PAR formation.
The DBD bound to DNA activates the CAT by increasing Vmax and decreasing Km
Next, we determined the enzymatic parameters of chimera PARP2-1 stimulated by the PARP1 DBD in the absence or presence of DNA. We measured the incorporation of tritium-labeled NAD into TCA-precipitable polymers at early reaction time points and obtained initial reaction velocities (V0) for different substrate concentrations by nonlinear regression analysis assuming one substrate-binding site. In the absence of the DBD, chimera PARP2-1 did not generate detectable levels of PAR independent of the addition of DNA (Table 1, second and third column), thus confirming our previous results. In the presence of the DBD, PAR generation was strongly dependent on DNA. Without DNA, the obtained PAR levels were low, but still allowed for curve fitting and calculation of Vmax and Km values (Table 1, fourth column). Addition of DNA increased the maximum reaction velocity Vmax about 4-fold and reduced Km 8-fold (Table 1, compare fifth to fourth column). The reaction efficiency Kcat/Km was thereby increased by more than 30-fold. DNA could thus be considered a V + K-type activator, affecting both turnover rate and substrate affinity. Remarkably, the enzymatic parameters obtained for chimera PARP2-1 together with the PARP1 DBD closely match the values reported for full-length PARP1 (Table 1, compare fifth to first column). Together, these results provide evidence that DNA containing double strand breaks is recognized and bound by the DBD of PARP1, which subsequently binds to the CAT domain to induce structural changes within the catalytic cleft in order to increase the affinity for NAD and stabilize reaction intermediates.
Table 1.
Kinetic parameters of chimera PARP2-1
| hPARP1a | hPARP2-1 −1–373 −DNA | hPARP2-1 −1–373 +DNA | hPARP2-1 +1–373 +DNA | hPARP2-1 +1–373 +DNA | |
|---|---|---|---|---|---|
| Vmax (μmol × min−1 × mg−1) | 0.2–2.4 | NC | NC | 0.114 ± 0.010 | 0.488 ± 0.026 |
| Km (μM) | 59–278 | NC | NC | 1111 ± 127.7 | 140.8 ± 19.72 |
| Kcat (s−1) | 0.41 | NC | NC | 0.121 ± 0.011 | 0.521 ± 0.028 |
| Kcat/Km (s−1 × mM−1) | 1.475–6.949 | NC | NC | 0.109 ± 0.022 | 3.696 ± 0.715 |
NC: not calculable (product levels below detection limit).
aValues as reported in the literature.
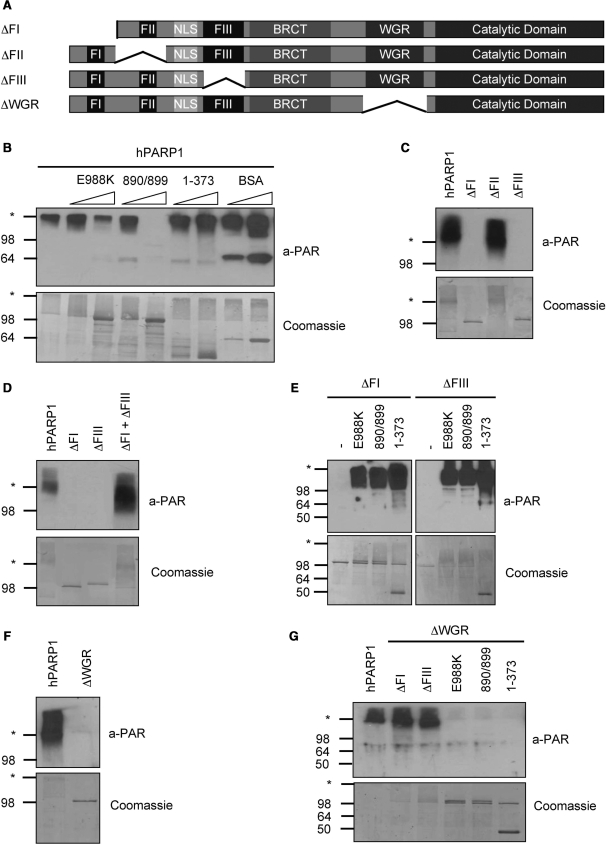
PARP1 forms a catalytic dimer which requires at least one functional FI and FIII domain for activity
The CAT of PARP1 was previously described to dimerize (29). To investigate whether our purified proteins were also able to form dimers, the enzymatic activity of full-length PARP1 was assessed by western blot analysis after co-incubation with different molar ratios of two catalytically inactive PARP1 mutants (E988K or M890V/D899N, respectively) (Supplementary Figure 3A). A molar ratio of 1:5 (wt:mutant) severely reduced PAR formation (Figure 5B), suggesting that the different proteins are indeed able to form dimers and to regulate each other.
Figure 5.
PARP1 forms a catalytic dimer which requires at least one functional FI and FIII domain for activity. (A) Domain organization of the PARP1 deletion mutants used for this figure. (B) PAR formation by PARP1 when co-incubated with the indicated inactive proteins or fragments at a molar ratio of 1:1 or 1:5. According to the manufacturer, the anti-PAR antibody LP96-10 cross reacts with bovine serum albumin (BSA) (band at around 64 kDa). (C) PAR formation by DBD deletion mutants ΔFI, ΔFII and ΔFIII. (D) PAR formation by a combination of the two DBD deletion mutants ΔFI and ΔFIII. (E) PAR formation by the DBD deletion mutants ΔFI and ΔFIII when they were co-incubated with catalytically inactive PARP1 mutants or with fragment 1–373. (F) PAR formation by PARP1 lacking the WGR domain. (G) PAR formation by PARP1 ΔWGR in combination with DBD deletion mutants, catalytically inactive PARP1 mutants or with fragment 1–373. Molecular size markers in kilo Daltons and the border between stacking and separating gel (asterisk) are indicated.
We showed earlier that only the full-length DBD was able to stimulate chimera PARP2-1 (Figure 4). This finding suggests that disruption of the DBD of PARP1 by deleting one of the DBD sub-domains would render the protein inactive due to lost activation of the CAT. Deletion of the regions containing zinc finger FI (ΔFI, aa 1–111) or zinc-binding domain FIII (ΔFIII, aa 279–333) indeed rendered PARP1 inactive, while a mutant lacking FII (ΔFII, aa 117–201) was still able to synthesize PAR as examined by western blot analysis (Figure 5A and C). Thus, the presence of FI and FIII is required for enzymatic activity, while neither FII nor the spacing between FI and FIII seem to be critical for PAR formation. Surprisingly, co-incubation of the two inactive mutants ΔFI and ΔFIII fully restored activity, suggesting that the two proteins can interact and that the lack of the critical domains containing FI and FIII could be inter-molecularly complemented to form a functional active dimer (Figure 5D). In line with this finding, the mutants ΔFI and ΔFIII could also be complemented by co-incubation with the catalytically inactive PARP1 mutants E988K and M890V/D899N or with the DBD alone (Figure 5E).
The WGR domain is vital for enzymatic activity
We showed that the WGR domain is required for enzymatic activity of PARP1 (Figure 4). A PARP1 deletion mutant lacking the WGR domain (ΔWGR, aa 525–656) was indeed not able to generate PAR (Figure 5F). Similarly, a PARP2 deletion mutant lacking the WGR domain was also inactive (data not shown), suggesting that the so far uncharacterized WGR domain of PARP family members is absolutely required for the enzymatic activity. Importantly, the PARP1 ΔWGR mutant could functionally complement the two PARP1 mutants ΔFI and ΔFIII by providing its DBD (Figure 5G). Co-incubation of the PARP1 ΔWGR mutant with the catalytically inactive PARP1 mutants E988K and M890V/D899N, both possessing a functional WGR, however, did not restore enzymatic activity (Figure 5G). Thus, the PARP1 DBD and the CAT can be regarded as independent and flexible protein units in a catalytic dimer, whereas the WGR domain is functionally tightly associated with and required for the activation of the CAT.
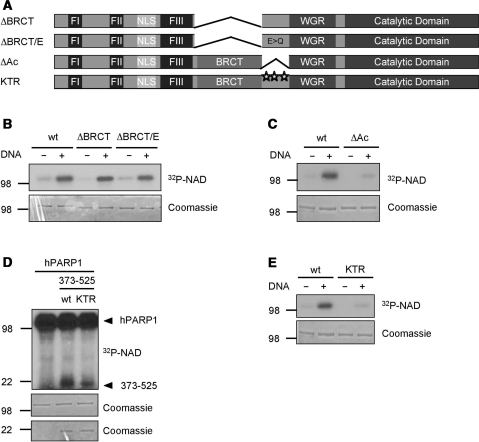
Glutamic acid residues in the AD of PARP1 are not modified
It is widely believed that multiple glutamic acid residues within the AD of PARP1 serve as acceptor sites for the covalent attachment of PAR (16). To our knowledge, however, this assumption has so far not been confirmed by amino acid substitutions. Therefore, we decided to test whether glutamic acid residues within the AD of PARP1 are required for the catalytic activity of the enzyme and function as acceptors for PAR. First, we deleted the BRCT domain as part of the AD (Figure 6A). A PARP1 ΔBRCT mutant was as active as its wild-type counterpart with regard to auto-modification (Figure 6B, first four lanes) and PAR formation (Supplementary Figure 5A, left panel). Next, in the context of the PARP1 ΔBRCT mutant, we additionally mutated all eight glutamic acid residues in the remaining auto-modification loop between amino acids 484 and 557 to glutamine (ΔBRCT/E) (Figure 6A). Surprisingly, these substitutions also did not reduce auto-modification (Figure 6B, last two lanes) or PAR formation (Supplementary Figure 5A, right panel). These results strongly indicate that glutamic acid residues within the AD of PARP1 are not required for enzymatic activity and are unlikely to serve as acceptors for PAR.
Figure 6.
Lysine residues within the AD of PARP1 are target sites for poly(ADP-ribosyl)ation. (A) Domain organization of the PARP1 mutants used for this figure. (B) Auto-modification of the indicated PARP1 mutants lacking either the BRCT domain (ΔBRCT) or the BRCT domain and carrying substitutions for all glutamic acid residues in the remaining stretch of the AD (ΔBRCT/E). (C) Auto-modification of a PARP1 deletion mutant lacking aa 466–525 (ΔAc), a region that was previously shown to be acetylated. (D) Trans-poly(ADP-ribosyl)ation of the AD from amino acid 373–525 by PARP1. KTR, K498/521/524R. (E) Auto-modification of a PARP1 K498/521/524R mutant. Molecular size markers in kilo Daltons are indicated.
Lysine residues are acceptor sites in PARP1
In contrast to the deletion of the BRCT domain, deletion of the remaining amino acids in the AD of PARP1 (ΔAc, aa 466–525) (Figure 6A), a region previously reported to be acetylated (26), resulted in severely impaired auto-modification (Figure 6C) and reduced PAR formation (Supplementary Figure 5B), suggesting that acceptor sites are localized in this region of PARP1. As PAR levels generated by PARP1 ΔAc were decreased but did not drop completely, additional PAR acceptor sites are likely to exist in other domains of PARP1. Trans-poly(ADP-ribosyl)ation of different fragments of PARP1 by wild-type PARP1 indeed confirmed that not only the AD but also a fragment containing amino acid 1–214 is modified (Supplementary Figure 5C).
ADP-ribose has earlier been described to be a potent histone glycation and glycoxidation agent in vitro, leading to the formation of ketoamine glycation conjugates (30). When we analyzed the chemical linkage stability of auto-modified PARP1, we found that it was stable up to pH 10 (but not at pH 13.5) and that incubation with 1 M hydroxylamine at pH 7 for 30 min at 30°C or 60°C did not release the modification (Supplementary Figure 5D). This observation suggests that the protein-ADP-ribose linkage indeed might occur on lysines. To exclude that the investigated auto-modification of PARP1 was due to traces of ADP-ribose within the provided NAD, the inactive PARP1 mutant M890V/D899N was incubated with radioactive NAD. Only upon long exposure a faint labeling of PARP1 M890V/D899N was observed while PARP1 E988K was able, as described earlier, to modify itself (Supplementary Figure 5E), confirming that the observed auto-modification of PARP1 was due to its enzymatic activity.
Next, we analyzed the trans-poly(ADP-ribosyl)ation of a fragment comprising the AD (aa 373–525) of PARP1 by full-length PARP1. Although auto-modification of PARP1 was much more efficient than trans-poly(ADP-ribosyl)ation, specific labeling of the AD fragment was observed (Figure 6D, second lane). In order to identify individual lysine residues within the AD which serve as acceptor sites for PAR, we analyzed trans-poly(ADP-ribosyl)ation of the AD fragment containing three lysine to arginine substitutions (K498, K521 and K524, called KTR). These sites were previously reported to be targets for acetylation (26). Modification of the 373–525 KTR fragment by full-length PARP1 was reduced as compared to 373–525 wt (Figure 6D, third lane). Since trans-poly(ADP-ribosyl)ation of a protein fragment might lead, due to structural constrains, to unspecific modification of amino acids and might thus not be comparable to modification of the full-length protein, we generated a full-length PARP1 mutant which contains the three lysine to arginine substitutions at position 498, 521 and 524. Importantly, this mutant showed strongly reduced auto-modification, very much comparable to the levels observed for the PARP1 ΔAc mutant (Figure 6E). Overall, these experiments provide evidence that not glutamic acid residues but instead at least three lysine residues within the auto-modification loop (aa 466–525) and additional residues within the first 214 amino acids of PARP1 are target sites for enzymatic auto-ADP-ribosylation.
DISCUSSION
In this study we analyzed the poly(ADP-ribosyl)ation capacity of PARP1 and the closely related proteins PARP2 and PARP3 under standardized reaction conditions and investigated the molecular mechanism of PAR formation. Human PARP1 and PARP2 were able to auto-modify themselves and generate PAR, although to different levels. Neither polymer formation nor auto-modification was observed for PARP3 under the tested conditions.
PARP1 deletion mutants and fusion proteins had been successfully employed before to study different aspects of poly(ADP-ribosyl)ation (31,32). Here, we have generated PARP family chimera to analyze the molecular mechanism of PAR formation. The PARP chimera revealed that the WGR/CAT domains of PARP1, PARP2 and PARP3 tightly cooperate with their corresponding amino-terminal domains. Closer examination of PARP1 revealed that FI, FIII and the WGR/CAT domain of PARP1 are required and sufficient for PAR formation. FII and the BRCT domain, however, were not essential for the enzymatic activity. The DBD interacted directly with the CAT domain of PARP1. DBD bound to DNA increased Vmax and reduced the Km of the CAT for NAD. We also provide evidence that PARP1 forms a catalytic dimer in which lack of either FI or FIII could be functionally complemented by a protein containing these domains. Finally, we identified three lysine residues within the AD and additionally the first 214 amino acids of the DBD as target sites for enzymatic covalent auto-poly(ADP-ribosyl)ation by PARP1.
We employed three different methods to assess poly(ADP-ribosyl)ation. First, 3H-NAD at a concentration of 400 μM was used to measure TCA-precipitable polymer formation in time course experiments. Second, 32P-NAD at a concentration of only 100 nM was used to measure auto-modification after short incubation periods (10 s). This approach resulted in distinct bands corresponding to the modified protein and most likely representing mono(ADP-ribosyl)ation or short oligomers of ADP-ribose attached to the labeled protein. Third, unlabeled NAD at a concentration of 400 μM was used to measure PAR formation detected by western blot. The anti-PAR antibody typically detected high molecular weight polymers, most of which remained as a smear at the top of the separating gel or even in the stacking gel. This approach was not very suitable to make quantitative statements but could be readily applied to analyze whether a protein was active or not.
Human PARP3 was previously described by Augustin et al. to be an active enzyme, as detected by autoradiography after 15 min incubation with 10 μM 32P-NAD (19). Augustin and co-workers did not, however, compare the activity of PARP3 to that of PARP1 or any other PARP family member under these conditions. We analyzed PARP3 in comparison to PARP1 and PARP2 under standardized reaction conditions and could not observe any activity for this protein. However, when we applied the conditions provided by Augustin et al. to measure PARP3 activity by autoradiography, we could also observe PARP3 auto-modification (data not shown), suggesting that the protein possesses some degree of activity under certain well-defined conditions. Further investigations are needed to analyze the extent of PAR formation by PARP3 as well as its physiological relevance.
The DBD of PARP1 interacted in a coordinated and DNA-dependent manner with the CAT domain of PARP1, but not with that of PARP2 or PARP3. Thus, despite the high level of structural similarity between the CATs of PARP1, PARP2 and PARP3, these domains cannot compensate for each other and may possess unanticipated intrinsic regulatory functions. Since the PARP1 DBD is not or only partially present in other PARP family members, the newly identified intra-molecular interaction might provide a promising surface for the development of PARP1 specific inhibitors.
In our study, several enzymatic dead mutants with deletions in the DBD could be functionally complemented by another inactive PARP1 mutant containing the missing domain, thus implicating that PARP1 is forming a dimer for poly(ADP-ribosyl)ation. The existence of catalytically active protein dimers in which each monomer is lacking a domain required for enzymatic activity was surprising and suggests that PARP1 is a highly flexible molecule with rather loose domain architecture.
Consistent with earlier reports (33,34) our results showed that zinc finger FI is absolutely required for the DNA-dependent activation of the protein, whereas zinc finger FII is dispensable. Zinc finger FII may, however, determine the binding specificity for DNA single strand breaks as suggested previously by Gradwohl et al. (35). Our data revealed that the recently discovered zinc-binding motif FIII is essential for the interaction of the DBD with the CAT and thus also for the activation of the enzyme. Furthermore, the so far uncharacterized WGR domain is an indispensable prerequisite for PAR formation, although this domain is not necessary for the interaction between the DBD and the CAT.
The interaction between the DBD bound to DNA and the CAT domain increased the maximum reaction velocity Vmax by a factor of four and reduced the Km for NAD roughly from 1 mM to 140 μM (Table 1). The reaction efficiency Kcat/Km was thereby increased by a factor of more than 30. The total cellular NAD concentration was previously estimated to be around 350 μM (36). Zhang et al. argued that NAD cofactors should readily pass through nuclear pores, which would suggest that cytoplasmic NAD levels reflect nuclear NAD concentrations (37). The same group estimated the free nuclear NAD concentration to be around 70 μM (38). Despite this uncertainty in the estimation of free nuclear NAD concentrations, we believe that increasing the affinity of PARP1 for NAD by binding to DNA double strand breaks might be an important regulatory step to allow PAR formation at physiological NAD concentrations. Release of PARP1 from DNA would consequently reduce the affinity of PARP1 for NAD and terminate PAR formation. Importantly, the nuclear concentration of NAD can be modulated by NMN adenylyl transferase 1 (NMNAT-1), which catalyzes the final step of NAD biosynthesis. A recent study revealed that NMNAT-1 is able to interact with and stimulate PARP1 (39). It is thus tempting to speculate that PARP1 activation by its binding to DNA strand breaks is supported by the localized action of NMNAT-1.
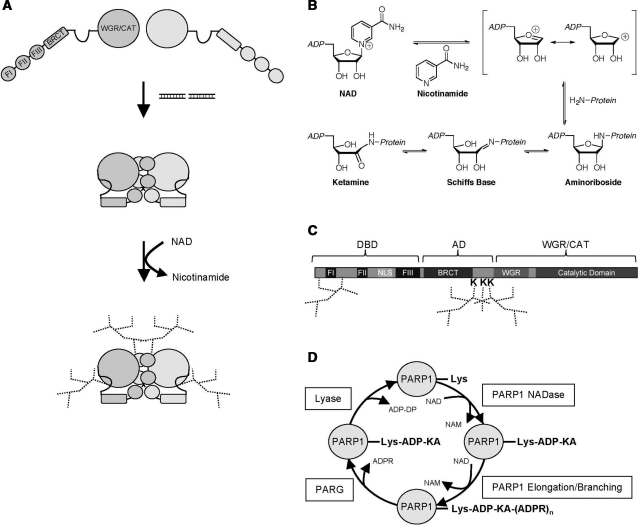
Our results suggest that activation of PARP1 occurs in defined sequential steps (Figure 7A). First, the DBD binds to certain damages within the DNA. This enhances the interaction between the DBD and the CAT domain. As a consequence, minor structural rearrangements within the catalytic cleft occur, resulting in an increased affinity for NAD. Increasing substrate affinity and additionally substrate turnover rates then allows for high reaction efficiency and very rapid auto-modification at distinct lysine residues followed by PAR chain elongation. An analogous model can be envisioned for the protein chimera PARP2-1, which is activated by the PARP1 DBD in the presence of double strand breaks mimicking DNA (Supplementary Figure 6A).
Figure 7.
Model for PARP1 activation and ADP-ribosylation of lysine residues. (A) Model for the sequential activation and regulation of PARP1. The DNA-dependent interaction between the DBD and the WGR/CAT induces a state of high substrate affinity and high turnover rate in PARP1. Subsequently, acceptor amino acids in the auto-modification loop as well as in the DBD are poly(ADP-ribosyl)ated. (B) Proposed reaction mechanism for NADase-dependent auto-ADP-ribosylation of lysine residues by PARP1 via Schiff base formation. (C) Scheme depicting the identified PAR acceptor lysine residues in PARP1. (D) A revised view of ADP-ribose metabolism. PARP1 catalyzes lysine mono(ADP-ribosyl)ation via its NADase activity and subsequently PAR chain elongation. PARG cleaves glycosidic ribose–ribose bonds to generate PARP1-Lys-ADP-KA, which is then substrate for an ADP-ribosyl protein lyase. See discussion for details. NAM, nicotinamide; Lys, lysine; ADP-KA, ADP-ketamine; ADPR, ADP-ribose; ADP-DP, ADP-3″-deoxypentose-2″-ulose.
The assumption that PARP1 is modified at glutamic acid residues was based mainly on the chemical stability of the ADP-ribose-PARP1 linkage, which was very heterogeneous but in part of a similar type as carboxyl esters in mono-ADP-riboslyated histones isolated from cells (23). The presented mutation analysis studies revealed that neither deletion of all glutamic acid residues in the BRCT domain (aa 385–476, containing nine glutamic acid residues) nor additional mutation of the remaining glutamic acid residues to glutamine in the AD (aa 477–557, containing eight glutamic acid residues) affected auto-modification or PAR formation and thus provide strong evidence that these amino acids in the AD are not the acceptor sites for poly(ADP-ribosyl)ation. Interestingly, mutation of the three lysines K498, K521 and K524 in the AD of PARP1 to arginines strongly reduced the auto-modification of the enzyme, suggesting that these residues in fact are acceptors for PAR. A longer exposure of the gel revealed a weak labeling of PARP1 KTR (data not shown) and it may well be that additional lysine residues serve as acceptor sites in this domain. Furthermore, acceptor sites can also be expected in the DBD of PARP1. Whether these sites are also lysine residues or whether outside the AD other amino acids serve as PAR acceptors is currently not known.
Modification of proteins by ADP-ribose can be characterized according to their chemical properties. ADP-ribosylated lysine residues were described to be stable in the presence of 1 M hydroxylamine at pH 7, while chemically modified glutamic and aspartic acid residues would rapidly release the ADP-ribose moiety (40,41). Our chemical analysis of modified PARP1 revealed that the observed linkage most likely corresponds to the glycation linkage described above. Thus we propose that ADP-ribosylation of PARP1 is catalyzed by its NADase activity, which subsequently allows modification of lysine residues positioned close to the catalytic active site to Lys-ADP-ribose ketamine (Figure 7B, C and D). This moiety could then serve as acceptor for the elongation reaction, which is catalyzed by glutamic acid residue E988 in human PARP1. We are currently investigating whether other ADP-ribose acceptor proteins are modified by PARP1 in the same manner.
To date two enzymes, poly(ADP-ribose) glycohaydrolase (PARG) and ADP-ribosyl protein Lyase, have been described to be involved in PAR catabolism (42,43). While PARG possesses both exo- and endoglycosidic activities, the Lyase was described to cleave the bond between proteins and mono(ADP-ribose). ADP-ribosylation of lysines creates a chemical bond, which is not a substrate for PARG, which cleaves the ribose–ribose bonds. Breaking a lysine-ADP-ribose linkage would instead require the activity of a Lyase (Figure 7D). Alternatively, the last ADP-ribose moieties might remain on PARP1 to serve as elongation sites for the next round of PAR formation or to mark the chromatin to memorize the location of previous DNA damage repair.
Lysine residues K498, K521 and K524 were previously identified as targets for acetylation by p300 and P300/CBP-associated factor (PCAF) in a stimulus-dependent manner (26). Remarkably, simple addition of PCAF reduced poly(ADP-ribosyl)ation by PARP1 (unpublished observation), suggesting that the interaction domain of PARP1 with PCAF is overlapping with the ADP-ribose acceptor sites. Furthermore, we recently showed that acetylation of PARP-2 strongly reduced the enzymatic activity (44). Already more than 20 years ago, a possible interrelation between poly(ADP-ribosyl)ation reactions and post-translational protein acetylation had been discussed (45,46). Our finding that acetylation of lysine residues interferes with ADP-ribosylation supports this idea and points at an interesting crosstalk between acetylation of and ADP-ribosylation by PARP family members. This crosstalk hypothesis is further strengthened by the finding that the enzymatic activity of PARP1 is not required for the function as transcriptional co-activator of NF-κB, a role which requires acetylation of PARP1 (47).
During apoptosis, PARP-1 is cleaved by different caspases to generate 89-kDa and 24-kDa fragments, a well-characterized hallmark of apoptosis. The data shown provide a functional explanation for the observed inactivation of PARP1 upon caspase cleavage, as this cleavage is separating FI and FII (aa 1–214) from FIII (aa 214–373), thus no longer allowing the DBD to interact as an intact polypeptide with the CAT domain for subsequent activation.
Our chemical and mutational analyses provide evidence that lysine residues are acceptor sites for auto-modification by PARP1 in vitro (Figure 6B). As PARP1 is the main acceptor protein for poly(ADP-ribosyl)ation in vivo (48,49), our findings are most likely also relevant in vivo. Confirming acceptor sites in vivo, however, is very difficult for different reasons. First, mutations within the DBD to eliminate the acceptor sites in this region and to allow only the analysis of the three lysine residues in the auto-modification domain would affect the activation of PARP1 by DNA. Second, PARP1 is known to be modified by PARP2 and possibly by other PARP family members. Whether theses proteins are modifying PARP1 also at the auto-modification sites or at other residues is currently not known. In any case, however, this crosstalk would interfere with in vivo analysis of PARP1 auto-modification.
In conclusion, we propose that PARP1 forms a catalytic dimer that allows the interaction of the DNA-binding domain with the CAT to modify distinct lysine residues as ADP-ribose acceptor sites in the AD as well as additional acceptor sites in the DNA-binding domain. These insights will allow further investigations to elucidate the biological functions of PARP1 and its enzymatic activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [PA00A-115384 to P.O.H., 31-109315 and 31-122421 to M.A. and S.M.], VonTobel Stiftung and Kanton of Zurich to M.A., S.M., M.F. and M.O.H. Funding for open access charge: Swiss National Science Foundation (31-122421).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Jay Siegel (University of Zurich, Switzerland), Dr Mike Jacobson (University of Arizona, USA) and Dr Mathias Ziegler (University of Bergen, Norway) for their critical and constructive comments on the manuscript. The help of Anina Nagy regarding cloning and protein purifications is deeply appreciated. We thank all members of the Institute of Veterinary Biochemistry and Molecular Biology (University of Zurich, Switzerland) for their advice and critical reading of the manuscript.
REFERENCES
- 1.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki H, Uchida K, Shima H, Sato T, Okamoto T, Kimura T, Miwa M. Molecular cloning of cDNA for human poly(ADP-ribose) polymerase and expression of its gene during HL-60 cell differentiation. Biochem. Biophys. Res. Commun. 1987;146:403–409. doi: 10.1016/0006-291x(87)90543-2. [DOI] [PubMed] [Google Scholar]
- 4.Cherney BW, McBride OW, Chen DF, Alkhatib H, Bhatia K, Hensley P, Smulson ME. cDNA sequence, protein structure, and chromosomal location of the human gene for poly(ADP-ribose) polymerase. Proc. Natl Acad. Sci. USA. 1987;84:8370–8374. doi: 10.1073/pnas.84.23.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kameshita I, Matsuda M, Nishikimi M, Ushiro H, Shizuta Y. Reconstitution and poly(ADP-ribosyl)ation of proteolytically fragmented poly(ADP-ribose) synthetase. J. Biol. Chem. 1986;261:3863–3868. [PubMed] [Google Scholar]
- 6.Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat. Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 7.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 8.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc. Natl Acad. Sci. USA. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342 (Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 11.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber V, Molinete M, Boeuf H, de Murcia G, Menissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992;11:3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pion E, Ullmann GM, Ame JC, Gerard D, de Murcia G, Bombarda E. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005;44:14670–14681. doi: 10.1021/bi050755o. [DOI] [PubMed] [Google Scholar]
- 14.Desmarais Y, Menard L, Lagueux J, Poirier GG. Enzymological properties of poly(ADP-ribose)polymerase: characterization of automodification sites and NADase activity. Biochim. Biophys. Acta. 1991;1078:179–186. doi: 10.1016/0167-4838(91)99007-f. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. SAP – a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 18.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol. Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 20.Urbanek P, Paces J, Kralova J, Dvorak M, Paces V. Cloning and expression of PARP-3 (Adprt3) and U3-55k, two genes closely linked on mouse chromosome 9. Folia Biol. (Praha) 2002;48:182–191. [PubMed] [Google Scholar]
- 21.Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza-Alvarez H, Alvarez-Gonzalez R. Biochemical characterization of mono(ADP-ribosyl)ated poly(ADP-ribose) polymerase. Biochemistry. 1999;38:3948–3953. doi: 10.1021/bi982148p. [DOI] [PubMed] [Google Scholar]
- 23.Kawaichi M, Ueda K, Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J. Biol. Chem. 1981;256:9483–9489. [PubMed] [Google Scholar]
- 24.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 25.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 26.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 27.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonin F, Hofferer L, Panzeter PL, Muller S, de Murcia G, Althaus FR. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. J. Biol. Chem. 1993;268:13454–13461. [PubMed] [Google Scholar]
- 29.Mendoza-Alvarez H, Alvarez-Gonzalez R. The 40 kDa carboxy-terminal domain of poly(ADP-ribose) polymerase-1 forms catalytically competent homo- and heterodimers in the absence of DNA. J. Mol. Biol. 2004;336:105–114. doi: 10.1016/j.jmb.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 30.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J. Biol. Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- 31.Smulson M, Istock N, Ding R, Cherney B. Deletion mutants of poly(ADP-ribose) polymerase support a model of cyclic association and dissociation of enzyme from DNA ends during DNA repair. Biochemistry. 1994;33:6186–6191. doi: 10.1021/bi00186a018. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal D, Hong T, Cherney B, Zhang S, Shima T, Danielsen M, Smulson M. Expression and characterization of a fusion protein between the catalytic domain of poly(ADP-ribose) polymerase and the DNA binding domain of the glucocorticoid receptor. Biochem. Biophys. Res. Commun. 1994;202:880–887. doi: 10.1006/bbrc.1994.2012. [DOI] [PubMed] [Google Scholar]
- 33.Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- 34.Menissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J. Mol. Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- 35.Gradwohl G, Menissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 38.Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc. Natl Acad. Sci. USA. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc. Natl Acad. Sci. USA. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods Enzymol. 1997;280:275–287. doi: 10.1016/s0076-6879(97)80119-x. [DOI] [PubMed] [Google Scholar]
- 41.Cervantes-Laurean D, Minter DE, Jacobson EL, Jacobson MK. Protein glycation by ADP-ribose: studies of model conjugates. Biochemistry. 1993;32:1528–1534. doi: 10.1021/bi00057a017. [DOI] [PubMed] [Google Scholar]
- 42.Ueda K, Oka J, Naruniya S, Miyakawa N, Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 1972;46:516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- 43.Oka J, Ueda K, Hayaishi O, Komura H, Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 1984;259:986–995. [PubMed] [Google Scholar]
- 44.Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Wong M, Smulson M. A relationship between nuclear poly(adenosine diphosphate ribosylation) and acetylation posttranslational modifications. 2. Histone studies. Biochemistry. 1984;23:3726–3730. doi: 10.1021/bi00311a024. [DOI] [PubMed] [Google Scholar]
- 46.Malik N, Smulson M. A relationship between nuclear poly(adenosine diphosphate ribosylation) and acetylation posttranslational modifications. 1. Nucleosome studies. Biochemistry. 1984;23:3721–3725. doi: 10.1021/bi00311a023. [DOI] [PubMed] [Google Scholar]
- 47.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J. Biol. Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 48.Yoshihara K, Hashida T, Yoshihara H, Tanaka Y, Ohgushi H. Enzyme-bound early product of purified poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 1977;78:1281–1288. doi: 10.1016/0006-291x(77)91431-0. [DOI] [PubMed] [Google Scholar]
- 49.Adamietz P. Poly(ADP-ribose) synthase is the major endogenous nonhistone acceptor for poly(ADP-ribose) in alkylated rat hepatoma cells. Eur. J. Biochem. 1987;169:365–372. doi: 10.1111/j.1432-1033.1987.tb13621.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.