Abstract
Hybridization of single-stranded DNA immobilized on the surface of gold nanoparticles (GNPs) into double stranded DNA and its subsequent dissociation into ssDNA were investigated. Melting curves and rates of dissociation and hybridization were measured using fluorescence detection based on hybridization-induced fluorescence change. Two distribution functions, namely the state distribution and the rate distribution, were proposed in order to take interfacial heterogeneity into account and to quantitatively analyze the data. Reaction and activation enthalpies and entropies of DNA hybridization and dissociation on GNPs were derived and compared with the same quantities in solution. Our results show that the interaction between GNPs and DNA reduces the energetic barrier and accelerates the dissociation of adhered DNA. At low surface densities of ssDNA adhered to GNP surface, the primary reaction pathway is that ssDNA in solution first adsorbs onto the GNP, and then diffuses along the surface until hybridizing with an immobilized DNA. We also found that the secondary structure of a DNA hairpin inhibits the interaction between GNPs and DNA and enhances the stability of the DNA hairpin adhered to GNPs.
INTRODUCTION
Hybridization of two single-stranded DNA (ssDNA) into a duplex of double-stranded DNA (dsDNA) and its dissociation back into two single strands play an important part in many life processes, and they are essential to many DNA-related technologies. Dissociation and hybridization processes of random-coil oligonucleotide are well-studied in solution (1–6). The rate-limiting step of oligonucleotide hybridization is the formation of a few base pairs from each strand into a transient intermediate called a nucleus. The remaining bases will then quickly form a complete helix (3). Consequently, the hybridization of two oligonucleotides is a second-order reaction, whereas its dissociation is first-order (6–8).
With the development of biotechnology, many researchers have turned their attention to the behavior of DNA at interfaces. The interactions between the substrate and DNA and between the neighboring DNA molecules play an important role in DNA hybridization at the interface. DNA hybridization with immobilized probes can be significantly enhanced by nonspecific adsorption of ssDNA onto the surface and its subsequent two-dimensional diffusion (9,10). Molecular crowding has been shown to affect the thermodynamics and kinetics of hybridization on DNA microchips (11–14). At the solid-liquid interface many factors such as surface strand density (11–13), surface charge (15,16), brush effect (14), point mismatch (17), DNA length (18) and flatness of the substrate (19) result in interfacial heterogeneity and can influence the kinetics and stability of DNA hybridization. These factors make the hybridization and dissociation kinetics of DNA at the interface much more complicated than for DNA free in solution. Typically, the Sips model (20–22), which is deduced from the well-known Langmuir model and assumes a pseudo-Gaussian distribution on the binding energies, is used to interpret the equilibrium isotherms of DNA hybridization at the interface. Alternatively, Schuck's group uses a ‘model-free’ two-dimensional distribution of rate and affinity constants to describe the heterogeneity of the interfacial reaction (23). Nevertheless, new quantitative treatment to clearly characterize the interfacial behaviors of DNA is still desirable.
The DNA and gold nanoparticles (GNPs) system is especially interesting due to its importance in nano-biotechnology. In this article we study the thermodynamics and kinetics of DNA hybridization/dissociation adhered to the surface of GNPs. Although much research has been focused on the hybridization-induced DNA-Au aggregates (24–27), to our knowledge only two groups (28,29) have investigated the thermodynamics of DNA adhered to GNPs, and no systematic studies of the influence of GNPs on the hybridization and dissociation kinetics of DNA has been reported. In our work, we measured the duplex melting curves and reaction rates of DNA hybridization/dissociation using fluorescently labeled DNA molecules. We maintained a low surface density of DNA on the GNP in order to prevent nearby DNAs from interacting. Two ‘model-free’ distributions, namely the state distribution and the rate distribution, were proposed to quantitatively describe the heterogeneity of DNA at the gold interface. Unlike previous DNA–GNP interfacial experiments, our measurements were performed at several temperatures from 27°C to 60°C. The activation entropies and activation enthalpies of DNA duplex dissociation and hybridization on GNPs were measured and compared with the same quantities in solution without GNPs. Our results show that the interaction between the gold surface and DNA remarkably modifies the behavior of immobilized DNA. First, the interaction reduces the energetic barrier and accelerates dissociation. Second for low surface coverage of ssDNA on the GNP, ssDNA from solution first adsorbs at the gold interface and then undergoes two-dimensional diffusion until it encounters and hybridizes with an immobilized ssDNA. Third, dsDNA formed from an immobilized hairpin probe is more stable than dsDNA formed from an immobilized random-coil probe; a result similar to what was found on silicon surfaces in our previous work (30,31).
MATERIALS AND METHODS
Oligonucleotide sequences and chemicals
The DNA sequences and their labeling are shown in Table 1. T3 and T6 have the same sequence. H1, L2, H4, L5 and N7 all have central regions that are complementary to T3 and T6 but different flanking sequences. H1 and H4 have flanking sequences that form a hairpin structure, whereas L2 and L5 instead are random-coil strands with the same number of bases as H1 and H4. N7 contains only the central complementary sequence that forms a random-coil strand with no additional flanking regions. The thiol-modified sequences (H1 and L2) are attached to GNPs to serve as the ‘probe’ strands and have an A10 spacer at 5′ end. ‘Target’ strands T3 is dye TET labeled at 5′ end. 5′ quencher dabcyl labeled T6 is used to hybridize with H4 and L5, which have dye alexa532 labeled in the middle of strand. H4 and L5 were purchased from IDT Inc. (Coralville, IA, USA), and other oligonucleotide strands were purchased from Sangon Company (Shanghai, China).
Table 1.
DNA sequences and labeling used in this article
| Symbol | DNA sequence |
|---|---|
| H1 | 5′-HS A10 GTGGGAA TTCTAGCCTGACTTCTTATT TTCCCAC-3′ |
| L2 | 5′-HS A10 AAAAAAA TTCTAGCCTGACTTCTTATT AAAAAAC-3′ |
| T3 | 3′-AAGATCGGACTGAAGAATAA-F1-5′ |
| H4 | 5′-GTGGGAA TTCTAGCCTGACTTCTTATT(F2) TTCCCAC-3′ |
| L5 | 5′-AAAAAAA TTCTAGCCTGACTTCTTATT(F2) AAAAAAC-3′ |
| T6 | 3′-AAGATCGGACTGAAGAATAA-Q-5′ |
| N7 | 5′-TTCTAGCCTGACTTCTTATT-3′ |
The italic letters stand for the bases that form intermolecular duplex, and the underlined letters stand for the bases that form intramolecular base pairs in a hairpin structure. F1 stands for the dye TET, F2 stands for the dye alexa532, and Q stands for the quencher dabcyl.
GNPs coated with 3-mercapto-1-propanol (MCP) reduce interaction with the DNA. It was purchased from Aldrich. GNPs (5 nm and 10 nm diameters) were purchased from Sigma. The buffer solution TE (20×) was purchased from Molecular Probe and diluted with water before use. The composition of TE (20×) is 200 mM Tris-HCl and 20 mM EDTA, with pH 7.5.
Preparation of DNA-functionalized GNPs
The ssDNA attached to GNPs (sspre-DNA-Au) was synthesized as reported previously (32) with minor modifications. Total 400 μl GNPs solution and 8 μl of ssDNA solution were mixed with 10 mM phosphate buffer (pH 7) to a final ssDNA concentration of 80 nM and a final volume of 1000 μl. The mixture was incubated for 24 h at room temperature. NaCl solution was then added to a final concentration of 0.1 M. The solution was incubated for an additional 40 h. Excess thiol-DNA was removed by centrifugation. The red, oily precipitate was washed with 0.1 M NaCl, 10 mM phosphate buffer (pH 7) solution (0.1 M PBS) and then resuspended in 0.1 M PBS. MCP was added to this DNA-Au solution to a final MCP concentration of 3 μM. After 24 h, MCP-blocked DNA-Au was separated by centrifugation and finally redispersed in TE (1×) buffer. The surface density of ssDNA on GNPs was measured according to the fluorescence-based method provided by Demers et al. (32).
dsDNA attached to GNPs (dspre-DNA-Au) was directly synthesized from thiol-dsDNA and GNPs. First, dsDNA was obtained by mixing equal amounts of thiol-probe strands (H1 or L2) and TET-labeled target strands (T3). The MCP-blocked dspre-DNA-Au was then synthesized by mixing the thiol-dsDNA with GNPs according the procedure mentioned above for sspre-DNA-Au. The surface density of dspre-DNA-Au was measured and found to be the same as those of sspre-DNA-Au.
The surface density of the immobilized DNA probe on 5 and 10 nm GNPs was 5.7 ± 0.8 pmol/cm2 (2.7 ± 0.4 DNA strands per GNP) and 7.0 ± 1.1 pmol/cm2 (13.2 ± 2.4 DNA strands per GNP), respectively. Both surface densities were considered low (11,24,29,32,33), and therefore neighboring DNA interactions were negligible. The MCP-blocked sspre-DNA-Au has a high hybridization efficiency (almost 100%) with the complementary target (T3), while the hybridization efficiency of unblocked sspre-DNA-Au was about 30–50%, indicating that blocking with MCP effectively reduces the non-specific interaction between the probe and GNPs (34–36). All of our experiments, unless noted otherwise, were carried out with 3 μM MCP-blocked DNA-Au.
Fluorescence measurement
The procedure of thermodynamic and kinetic measurements was described elsewhere (5). Many dyes can be quenched by GNPs (37) which is convenient for real-time fluorescence measurement. In this work, both the GNP and labeled quencher will quench the fluorescence signal of dye-labeled DNA when dsDNA is formed. The real-time dissociation and hybridization on GNPs and in solution were monitored through the fluorescence intensity recorded on a microscope-mounted spectrometer (Renishaw 1000, Britain) with a temperature controller (THMS 600, Linkam Scientific Instruments Ltd, Britain). The buffer solution used in all measurements was TE (1×) with 0.3 M NaCl.
The duplexes on GNP were made in three ways. The first started from sspre-DNA-Au, which has ssDNA immobilized onto the GNPs and then hybridized to form a duplex with equal molar amounts of complementary ssDNA for 24 h at room temperature. The second named dspre-DNA-Au, dsDNA was formed in solution and then immobilized onto the GNPs. The third named annealed dspre-DNA-Au, dspre-DNA-Au was denatured at a high temperature and then the duplex reformed by annealing at room temperature.
Melting curves were recorded at a linear heating rate of 30°C/h. The concentration of dsDNA was 5 nM. Dissociation rates were measured by the label dilution technique developed by Morrison and Stols (5,6). Briefly, excess non-labeled N7 (50 nM) was mixed with pre-formed dsDNA at a low temperature, and then the time course of dissociation was measured after the mixture was suddenly raised to a temperature near the DNA melting temperature Tm. For the hybridization rate measurement, equal amounts of sspre-DNA-Au and complementary ssDNA at 10 nM was quickly mixed, and the fluorescence signal was collected with time.
RESULTS
Melting curves of duplexes on GNPs and in solution
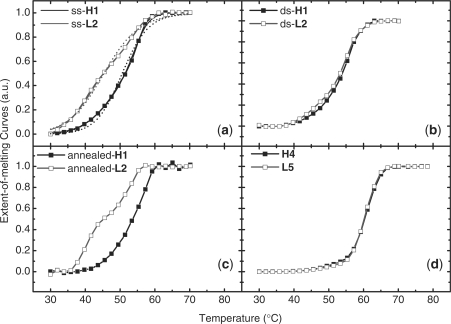
The normalized melting curves of DNA duplexes adhered to 5 nm GNPs and in solution are displayed in Figure 1. The normalization followed a procedure previously reported (38). Our results show that the sspre-DNA-Au duplex containing a hairpin probe has a higher Tm than that of random-coil (Figure 1a), whereas the same duplexes free in solution have Tm values that are almost the same (Figure 1d). The same observation was also made with the 10 nm GNPs over a series of different DNA surface densities (data not shown). The dspre-DNA-Au duplexes, however, have similar Tm values and also a similarly shaped melting curve for the hairpin and random-coil strands (Figure 1b). The difference in melting curves between the hairpin and random-coil probes reappeared after annealing (Figure 1c).
Figure 1.
Melting curves of duplexes. (a) On 5 nm sspre-DNA-Au, (b) on 5 nm dspre-DNA-Au, (c) on 5 nm annealed dspre-DNA-Au and (d) in solution. Solid dots, duplex composed of hairpin probe; hollow dots, duplex made of random-coil probe. Fitting curves from a two-state model (dash lines) and from the state distribution (solid lines) are shown in (a).
Control experiments showed no changes in the fluorescence signal when sspre-DNA-Au and a non-complementary dye-labeled ssDNA were mixed together, suggesting a negligible adsorption of non-specific ssDNA to the GNP. Consistent with a low surface density of DNA on the GNP, the quenching efficiency of TET-labeled DNA was constant with the hybridization efficiency changed from 100% to 10%. Conversely, Xu and Craig (39) found that the quenching efficiency depends on the hybridization efficiency, which was probably due to their high surface density of DNA.
State distribution of duplexes on GNPs
The dissociation of short dsDNA in solution can be treated with the Helix-Coil two-state model (38). Based on the two-state model, the dissociation enthalpy (Δ H°Sol) and dissociation entropy (Δ S°Sol) can be fitted from the melting curve using (5)
| 1 |
| 2 |
where R is the ideal gas constant, T is the temperature, CdsDNA is the concentration of dsDNA, θ(T) is the fraction of dissociated duplex, and Δ G°Sol is the free energy of duplex dissociation. By assuming that Δ H°Sol and Δ S°Sol are temperature independent, we fitted Equations (1) and (2) to the melting curves of L5–T6 duplexes and found that Δ H°Sol = 138 kcal mol−1 and Δ S°Sol = 375 cal mol−1 K−1.
Duplex dissociation which is adhered to a GNP is more complicated than when it is free in solution. The melting curves of sspre-DNA-Au, dspre-DNA-Au, and annealed dspre-DNA-Au cannot be fitted simply by a two-state model. The complexity of DNA dissociation adhered to GNPs results from the interaction between DNA and heterogeneous GNP interface, whereas solution is homogeneous. Previous work (28,29) shows that, on GNPs, the dissociation enthalpy (Δ H°GNP) and entropy (Δ S°GNP) follow the relation of entropy–enthalpy compensation
| 3 |
To quantitatively analyze the influence of surface heterogeneity on the GNP-duplex stability, we present a simple model with the following assumptions: First, the stability of duplexes adhered to GNPs is not a homogeneous one-state process as in solution, but rather can be described by distribution of sub-states. Second, the dissociation of each sub-state on the GNP satisfies the all-or-none Helix-Coil model. Third, the dissociation enthalpies and dissociation entropies of the ith sub-state satisfy the same entropy–enthalpy compensation equation, i.e. we assume that Δ H°i and Δ S°i satisfy Equation (3), where Δ H°Sol = 138 kcal mol−1 and Δ S°Sol = 375 cal mol−1 K−1.
We denote the proportion of the ith sub-state on GNPs as fi, and require that they be normalized:
| 4 |
Based on the above assumptions, the measured fluorescence signal, I(T), is expressed as
| 5 |
where θi(T) is the fraction of dissociated duplex of the ith sub-state at temperature T and is calculated from Equations (1) and (2) through the values of Δ H°i and Δ S°i.
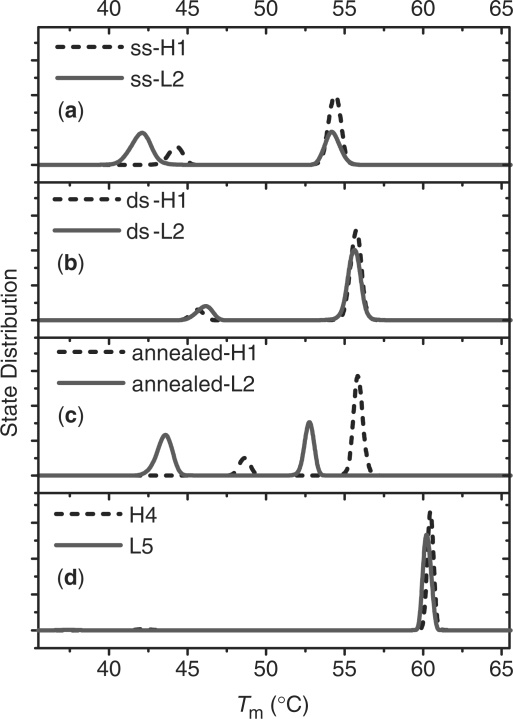
The state distributions of dsDNAs on GNPs as a function of Tm derived from the experimental melting curves are presented in Figure 2. For comparison, the same algorithm is applied to the melting curves in solution, and their state distributions are displayed in Figure 2d. The heterogeneity of DNA duplex stability adhered to GNPs is clearly shown in Figure 2, in which all state distribution curves have two well-separated peaks. In contrast, the homogeneous process of duplex melting in solution has only one narrow peak. The averaged thermodynamic parameters within each component are listed in Table 2.
Figure 2.
State distribution of duplexes. (a) On 5 nm sspre-DNA-Au, (b) on 5 nm dspre-DNA-Au, (c) on 5 nm annealed dspre-DNA-Au and (d) in solution. Dash line, duplex composed of hairpin probe; solid line, duplex made of random-coil probe.
Table 2.
Melting temperatures and thermodynamic parameters of DNA on GNPs and in solution
| Condition | Duplex | Proportion (%) | ΔH° (kcal mol−1) | ΔS° (cal mol−1 K−1) | Tm (°C) |
|---|---|---|---|---|---|
| sspre- | H1/T3 | 23 | 90 | 245 | 44.2 |
| DNA-Au | 77 | 116 | 315 | 54.4 | |
| L2/T3 | 57 | 86 | 233 | 42.0 | |
| 43 | 116 | 314 | 54.2 | ||
| dspre- | H1/T3 | 11 | 93 | 252 | 45.6 |
| DNA-Au | 89 | 121 | 327 | 55.7 | |
| L2/T3 | 19 | 94 | 255 | 46.1 | |
| 81 | 120 | 326 | 55.6 | ||
| Annealed | H1/T3 | 17 | 100 | 272 | 48.6 |
| dsDNA-Au | 83 | 121 | 329 | 55.9 | |
| L2/T3 | 55 | 89 | 241 | 43.5 | |
| 45 | 111 | 302 | 52.7 | ||
| Solution | H4/T6 | – | 139 | 377 | 60.5 |
| L5/T6 | – | 138 | 375 | 60.3 |
Standard errors for the experimental ΔH°, ΔS° and Tm are 5%, 5% and 0.3°C, respectively.
Distribution of dissociation rates for DNA adhered to GNPs
The dissociation rates of duplexes formed from sspre-DNA-Au, dspre-DNA-Au, or from ssDNA free in solution were measured. The dissociation curves in solution are fitted well by the first-order rate Equation (5)
| 6 |
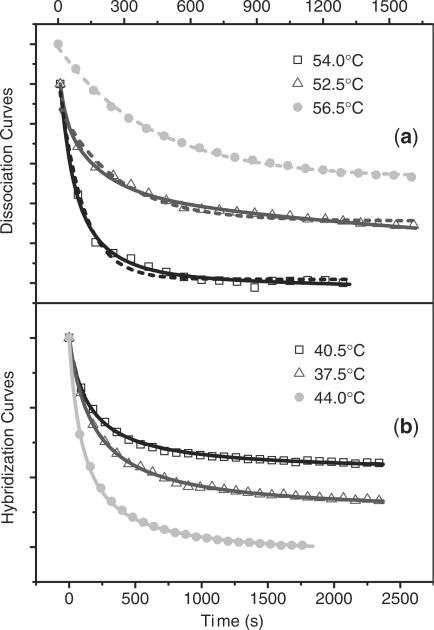
where kd is the dissociation rate, t is the time, I0 is the fluorescence signal at t = 0, It is the fluorescence signal at time t, and I∞ is the signal when fluorescence no longer changes with increasing time. The dissociation curves for duplexes adhered to GNPs cannot be fit by a single exponential (Figure 3a). Similar to the state distribution, we assume that dissociation rate constant of duplexes adhered to GNPs is no longer uniform as in solution, but has a distribution of rates, each of which follows a first-order reaction as in Equation (6).
Figure 3.
Comparison between kinetic data and fittings. (a) Data of dissociation in solution (solid dots) and on 5 nm sspre-DNA-Au (hollow dots). The dashed line is the single exponential fitting, and the solid line is the multiple exponential fitting based on the rate distribution. (b) Data of hybridization in solution (solid dots) and on 5 nm sspre-DNA-Au (hollow dots). The line is the fitting by the second-order reaction.
Based on these assumptions, the dissociation curves of duplexes formed on GNPs can be fitted by considering the contribution of many parallel first-order reactions, each with different reaction rates
| 7 |
where fi (kid) is the proportion of the ith component, whose dissociation rate is kid. fi(kid) satisfies the normalized condition
| 8 |
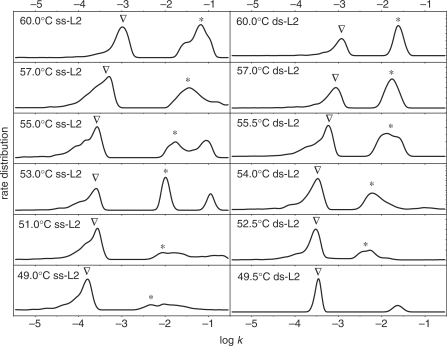
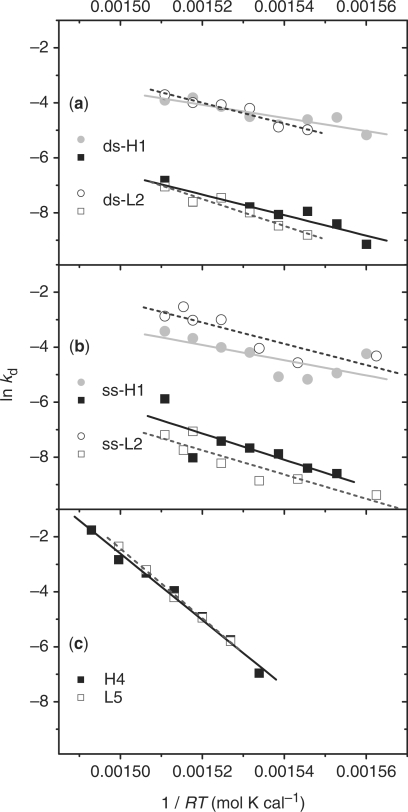
Fitting the experimental data by Equations (7) and (8), the distribution of dissociation rates for duplexes adhered to GNPs can be obtained and are shown in Figure 4. For duplexes bound in dspre-DNA-Au, all of the rate distribution curves of duplex dissociation have two separated peaks in the investigated temperature range. At some temperatures the rate distribution curves for duplexes formed from sspre-DNA-Au have three peaks. The average rate of every component is calculated. By classifying peaks at different temperatures into two major groups, the Arrhenius plots are calculated as shown in Figure 5. From the plots the activation enthalpy (Δ H≠d) and activation entropy (Δ S≠d) are obtained based on Eyring's absolute reaction rate theory form (40–43)
| 9 |
| 10 |
where k is the Boltzmann's constant, h is the Planck's constant, Ead is the activation energy of duplex dissociation, and κ is the transmission coefficient, which was assumed to have a value of 1. All fitting results are listed in Table 3.
Figure 4.
Rate distribution curves of duplex (L2/T3) on 5 nm sspre-DNA-Au (ss-L2) and 5 nm dspre-DNA-Au (ds-L2) at different temperatures. Peaks are classified into two groups with triangle or asterisk.
Figure 5.
Arrhenius plots of duplex dissociation rates. (a) on 5 nm dspre-DNA-Au, (b) on 5 nm sspre-DNA-Au, and (c) in solution. Solid dots, duplex composed of hairpin probe; hollow dots, duplex made of random-coil probe.
Table 3.
Activation enthalpies and activation entropies of duplex dissociation on GNPs and in solution
| Condition | Duplex | Component with high dissociation rates |
Component with low dissociation rates |
||
|---|---|---|---|---|---|
| Δ H≠d (kcal mol−1) | Δ S≠d (cal mol−1 K−1) | Δ H≠d (kcal mol−1) | Δ S≠d (cal mol−1 K−1) | ||
| sspre-DNA-Au | 5 nm H1/T3 | 38 ± 3 | 48 ± 9 | 48 ± 16 | 69 ± 48 |
| 5 nm L2/T3 | 39 ± 10 | 50 ± 29 | 44 ± 9 | 57 ± 27 | |
| dspre-DNA-Au | 5 nm H1/T3 | 36 ± 7 | 39 ± 21 | 41 ± 8 | 49 ± 25 |
| 5 nm L2/T3 | 38 ± 6 | 45 ± 17 | 49 ± 6 | 72 ± 18 | |
| 10 nm H1/T3 | 39 ± 12 | 50 ± 37 | 48 ± 16 | 71 ± 49 | |
| Solution | H4/T6 | 121 ± 6 | 293 ± 17 | – | – |
| L5/T6 | 127 ± 3 | 312 ± 10 | – | – | |
Rate of duplex formation on GNPs
The DNA hybridization rates for ssDNA adhered to 5 and 10 nm sspre-DNA-Au and ssDNA free in solution were measured. All the hybridization curves were well fit by a second-order rate equation (Figure 3b) (5)
| 11 |
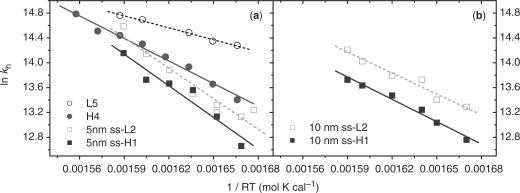
where kh is the hybridization rate and c°ssDNA is the concentration of one ssDNA at initial time t = 0. If the hybridization rate data is analyzed using a distribution of states, they exhibit a single sharp peak (data not shown). The Arrhenius plot of hybridization rates is linear (Figure 6), from which the hybridization activation enthalpies and activation entropies are obtained and listed in Table 4.
Figure 6.
Arrhenius plots of hybridization rates. (a) L5/T6 (hollow circle) and H4/T6 (solid circle) pairs in solution, L2/T3 (hollow square) and H1/T3 (solid square) pairs on 5 nm sspre-DNA-Au. (b) L2/T3 (hollow square) and H1/T3 (solid square) pairs on 10 nm sspre-DNA-Au.
Table 4.
Activation enthalpies and activation entropies of duplex hybridization on GNPs and in solution
| Condition | Duplex | Δ H≠hyb (kcal mol−1) | Δ S≠hyb (cal mol−1 K−1) |
|---|---|---|---|
| 5 nm sspre-DNA-Au | H1/T3 | 16.3 ± 2.3 | 21.1 ± 7.4 |
| L2/T3 | 15.7 ± 2.0 | 19.7 ± 6.6 | |
| 10 nm dspre-DNA-Au | H1/T3 | 11.4 ± 0.9 | 4.6 ± 2.8 |
| L2/T3 | 10.7 ± 1.2 | 3.2 ± 3.8 | |
| Solution | H4/T6 | 11.5 ± 0.5 | 6.2 ± 1.7 |
| L5/T6 | 5.8 ± 0.3 | −11.0 ± 1.0 |
DISCUSSION
Influence of GNPs on duplex melting stability
Consistent with Xu and Craig's work (28), our experiments show that the melting temperature of duplexes formed on GNPs is lower than the melting temperature of the same duplex formed in solution. As shown in Table 2, dissociation enthalpy of duplexes adhered to GNPs (Δ H°GNP) is always smaller than the dissociation enthalpy of duplexes in solution (Δ H°Sol). This agrees with previous studies by Stevens et al. (44) on micro-particles.
Melting curves of the DNA duplexes with hairpin and with random-coil strands are almost the same in solution. For duplexes formed on GNP surfaces, however, the Tm values for duplexes containing random coil sequence are lower than those containing hairpins. In our previous study on silicon surfaces (30,31), we also found that duplexes composed of hairpins are more stable than those of random-coils. These phenomena can be explained by structure-dependent non-specific interactions between the DNA probe and the gold surface. As shown in previous reports (35,45), DNA bases can non-specifically interact with gold nanoparticles. For the hairpin probe H1, seven base pairs are formed and the DNA strand adopts a compact configuration that prevents these bases from interacting with the nanoparticle surface. In contrast, for the random-coil probe L2, exposed nucleic bases along the flexible single strand can easily interact with the nanoparticle. Although formation of a MCP monolayer inhibits the non-specific adsorption (34–36) of ssDNA to the GNP, interaction between the bases and nanoparticle still exist because some strands are adsorbed onto the nanoparticle and form partial double helixes. The presence of these partial duplexes will reduce the Tm of duplexes formed from random coil DNA as found in our study. Fiche and Livache's (46) also found a broad melting range and low Tm of DNA adhered to a surface, which they also suggest should result from the dispersion caused by partial hybridization of segments of the probe DNA.
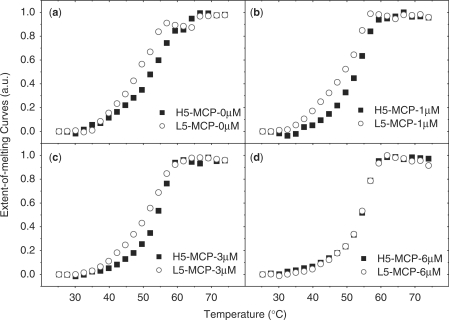
Similar to a hairpin structure, the helix of dsDNA prevents the interaction between the bases and the nanoparticle surface. Duplexes of DNA formed in solution and then attached to the GNP (dspre-DNA-Au) inhibit non-specific adsorption to the GNP, thus diminishing the difference between duplexes composed of hairpins and random-coils. When dspre-DNA-Au is denatured by heating and then cooled to reform the duplexes, the annealed dspre-DNA-Au formed duplexes similar to the sspre-DNA-Au because after denaturing the released bases can interact with the nanoparticle surface and lead to partial hybridization as in the case of sspre-DNA-Au. To further validate our hypothesis, we increased the concentration of blocking reagent to reduce absorption. When the concentration of MCP is increased to 6 μM, the difference between hairpin and random-coil duplex formation from sspre-DNA-Au disappeared, and the influence of secondary structure on GNP was the same as that in solution (Figure 7). Our result is consistent with Gao et al.'s work (47) on planar surfaces. This extraordinary behavior of the DNA hairpin probe on a substrate has unique applications shown in our previous work (30,31).
Figure 7.
Melting curves of duplexes on 5 nm sspre-DNA-Au. (a) 0 μM MCP, (b) 1.0 μM MCP, (c) 3.0 μM MCP and (d) 6.0 μM MCP. Solid dots, duplex composed of hairpin probe; hollow dots, duplex made of random-coil probe.
Duplex state distribution on GNPs
The heterogeneity of interfacial reactions is well known, but nevertheless, it has not been quantitatively well characterized. In this article, we propose temperature-dependent state and rate distributions to quantitatively analyze the interfacial heterogeneity. These distributions are similar to the one published by Juraj Svitel et al. (23), where a two-dimensional distribution of rate and affinity constants are applied to surface binding kinetics in an optical biosensor.
The state distribution curves of DNA duplexes adhered to GNPs have two peaks. We propose that each peak represents a sub-group, with different interactions between the heterogeneous GNP substrate and DNA strand. The sub-group with higher Tm should correspond to a full duplex and that with lower Tm is probably due to the formation of a partial duplex. Clearly, the proportion of partial duplexes in the dspre-DNA-Au case is smaller than for the sspre-DNA-Au, because the helix formation prevents the nonspecific adsorption of DNA bases onto the GNP surface. Similarly, DNA hairpin structures also inhibit adsorption. In the annealed dspre-DNA-Au, however, adsorption is restored after denaturing the adhered duplex and the proportion between full duplex and partial duplex becomes similar to that in sspre-DNA-Au. State distribution curves of duplex formed on GNPs with different blocking reagent concentration were calculated from melting curves (Supplementary Data). By increasing the MCP concentration, the proportion of partial duplexes decreases because of the reduced interaction between GNP surface and DNA probe. When the GNP surface is completely shielded by 6 µM MCP, the contact interaction between the DNA and Au is eliminated and random-coil and hairpin probes become identical. Also, the melting temperature of full duplexes decreases with decreasing MCP concentration. Those phenomena suggest that (i) even the probe, which has the ability to form a full duplex, has reversible interactions with the GNP surface; (ii) hairpin structures can suppress the DNA–substrate interaction, therefore the melting temperature variation of the hairpin is less sensitive to MCP concentration than that of random-coil probe.
We examined the influence of neighboring DNA–DNA interactions by applying state distribution analysis to duplex formation by sspre-DNA-Au with different probe densities (Supplementary Data). The random-coil has widespread multiple peaks at high probe density, and only two peaks at low probe density. The enhancement of heterogeneity at high probe density for random-coil should result from neighboring DNA–DNA interaction. The hairpin, however, has two peaks at all probe densities, suggesting that secondary structure also inhibits the neighboring interaction.
Duplex dissociation on GNPs
Our rate distribution indicates that DNA dissociation on GNPs should be a multiple-pathway process with multiple steps in each reaction pathway. Two separated peaks represent two distinguishable dissociation pathways. Interestingly, the activation enthalpies (Δ H≠GNP) of both dissociation pathways are much lower than the dissociation enthalpy (Δ H°GNP), while Δ H≠Sol and Δ H°Sol are similar in the solution. The high value of Δ H≠Sol in solution suggests that, except for the helix and coil states, there is no significant intermediate state during DNA dissociation. Low values of Δ H≠GNP than Δ H°GNP suggest, however, that the duplex dissociation on GNPs is a multi-step process that reduces the apparent activation energy. It is reasonable to suggest that GNPs play the key role. During dissociation of adhered duplex, the denatured bases of both probe and target DNA are non-specifically adsorbed on the substrate. Then, the adsorbed target ssDNA is released from the GNPs surface after the duplex becomes fully dissociated. According to this picture, the adsorption energy between the substrate and dissociated ssDNA compensates the energy required for duplex dissociation and effectively reducesΔ H≠GNP. The adsorption of ssDNA on the GNP surface restricts the movement of the dissociated strand and reduces the accessible configurations, which should lead to the decrease of the activation entropy (Δ S≠GNP) of duplex dissociation. This deduction is consistent with our results (Table 3).
Duplex formation on GNPs
As in solution, duplex formation on GNPs follows a second-order reaction. Previous work indicates that hybridization at interfaces can proceed either directly through three-dimensional diffusion of the target DNA through solution to the probe DNA bound to the GNP, or through nonspecific reversible adsorption of the target DNA to regions not covered with immobilized probe DNA, followed by two-dimensional diffusion of the target to the probe (9,10). Our results show that the hybridization rate of a hairpin strand on a GNP is lower than that of a random-coil strand, despite having almost the same hybridization activation enthalpies (Δ H≠hyb). This phenomenon not only confirms that the secondary structure of the hairpin persists at the interface and inhibits the hybridization, but it also implies that the hybridization is a reaction-controlling step. Kelso's group reaches the same conclusion based on the DNA hybridization on a system of micro-particles (48). Our previous results showed that, in solution, destruction of the hairpin structure is involved in the rate-limiting step at low temperatures (5) leading to the increase of both activation enthalpy (Δ H≠hyb) and activation entropy (Δ S≠hyb) of hybridization. On GNPs, however, the values of Δ H≠hyb and Δ S≠hyb are the same between hairpin and random coil hybridization within experimental error, indicating that the destruction of the hairpin structure is not involved in the rate-limiting step of DNA hybridization on GNPs. Combining all of our results, we propose that the primary reaction pathway of adhered duplex formation on GNP is non-specific adsorption of target ssDNA, two-dimensional diffusion, and hybridization. The last step, hybridization reaction between immobilized probe DNA and adsorbed target DNA, is the rate limiting step of duplex formation on GNP. As a result, Δ H≠hyb and Δ S≠hyb are not influenced by secondary structure of the DNA probes, but rather they depend on the diameter of the GNPs as observed (Table 4).
CONCLUSIONS
In our experiment, DNA-modified GNPs were chosen to investigate how the substrate–DNA interaction influences both thermodynamics and kinetics of DNA hybridization. The state distribution and the rate distribution were introduced to quantitatively describe the heterogeneity of DNA resulting from interaction between DNA and heterogeneous GNP surface. Different duplex melting properties between hairpin and random-coil sequence DNAs come from the structure-influenced interaction between the bases and GNPs. Steric hindrance induced by the compact configuration in a DNA hairpin probe prevents its bases from interacting with the surface and leads to a higher stability of hairpin-formed duplexes on the GNPs. Nonspecific adsorption of a DNA strand onto a GNP dominates both the duplex dissociation and formation processes. The nonspecific interaction reduces duplex dissociation activation enthalpy and accelerates the dissociation process. In hybridization the complementary target DNA in the solution does not directly react with the immobilized probe, but rather it is first adsorbed onto the GNP surface, followed by two-dimensional diffusion until it finally hybridizes with an immobilized probe DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Basic Research Support Foundation of China (NKBRSF) (2006CB910304) and National Natural Science Foundation of China (NSFC) (20673002, 20733001). Funding for open access charge: National Key Basic Research Support Foundation of China (NKBRSF) (2006CB910304).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Mr John F. Beausang and Mr Shashank Bharill for many valuable comments and suggestions.
REFERENCES
- 1.Craig ME, Crothers DM, Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 2.Eigen M, Porschke D. Co-operative non-enzymic base recognition. 1. Thermodynamics of helix-coil transition of oligoriboadenylic acids at acidic pH. J. Mol. Biol. 1970;53:123–141. doi: 10.1016/0022-2836(70)90049-5. [DOI] [PubMed] [Google Scholar]
- 3.Porschke D, Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of helix-coil transition of oligoribouridylic oligoriboadenylic acid system and of oligoriboadenylic acid aloe at acidic pH. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 4.Pörschke D, Uhlenbeck OC, Martin FH. Thermodynamics and kinetics of helix-coil transition of oligomers containing gc base pairs. Biopolymers. 1973;12:1313–1335. [Google Scholar]
- 5.Chen CL, Wang WJ, Wang Z, Wei F, Zhao XS. Influence of secondary structure on kinetics and reaction mechanism of DNA hybridization. Nucleic Acids Res. 2007;35:2875–2884. doi: 10.1093/nar/gkm177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison LE, Stols LM. Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry. 1993;32:3095–3104. doi: 10.1021/bi00063a022. [DOI] [PubMed] [Google Scholar]
- 7.Wetmur JG, Davidson N. Kinetics of renaturation of DNA. J. Mol. Biol. 1968;31:349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- 8.Blake RD, Klotz LC, Fresco JR. Polynucleotides.9. Temperature dependence of kinetics of complex formation in equimolar mixtures of polyriboadenylate and polyribouridylate. J. Am. Chem. Soc. 1968;90:3556–3562. doi: 10.1021/ja01015a047. [DOI] [PubMed] [Google Scholar]
- 9.Chan V, Graves DJ, McKenzie SE. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys. J. 1995;69:2243–2255. doi: 10.1016/S0006-3495(95)80095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axelrod D, Wang MD. Reduction-of-dimensionality kinetics at reaction-limited cell-surface receptors. Biophys. J. 1994;66:588–600. doi: 10.1016/s0006-3495(94)80834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shchepinov MS, CaseGreen SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watterson JH, Piunno PAE, Wust CC, Krull UJ. Effects of oligonucleotide immobilization density on selectivity of quantitative transduction of hybridization of immobilized DNA. Langmuir. 2000;16:4984–4992. [Google Scholar]
- 14.Halperin A, Buhot A, Zhulina EB. Brush effects on DNA chips: thermodynamics, kinetics, and design guidelines. Biophys. J. 2005;89:796–811. doi: 10.1529/biophysj.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vainrub A, Pettitt BM. Sensitive quantitative nucleic acid detection using oligonucleotide microarrays. J. Am. Chem. Soc. 2003;125:7798–7799. doi: 10.1021/ja035020q. [DOI] [PubMed] [Google Scholar]
- 16.Vainrub A, Pettitt BM. Surface electrostatic effects in oligonucleotide microarrays: Control and optimization of binding thermodynamics. Biopolymers. 2003;68:265–270. doi: 10.1002/bip.10271. [DOI] [PubMed] [Google Scholar]
- 17.Tawa K, Knoll W. Mismatching base-pair dependence of the kinetics of DNA-DNA hybridization studied by surface plasmon fluorescence spectroscopy. Nucleic Acids Res. 2004;32:2372–2377. doi: 10.1093/nar/gkh572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stillman BA, Tonkinson JL. Expression microarray hybridization kinetics depend on length of the immobilized DNA but are independent of immobilization substrate. Anal. Biochem. 2001;295:149–157. doi: 10.1006/abio.2001.5212. [DOI] [PubMed] [Google Scholar]
- 19.Glazer MI, Fidanza JA, McGall GH, Trulson MO, Forman JE, Frank CW. Kinetics of oligonucleotide hybridization to DNA probe Arrays on high-capacity porous silica substrates. Biophys. J. 2007;93:1661–1676. doi: 10.1529/biophysj.106.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sips R. On the structure of a catalyst surface. J. Chem. Phys. 1948;16:490–495. [Google Scholar]
- 21.Peterson AW, Wolf LK, Georgiadis RM. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 22.Vijayendran RA, Leckband DE. A quantitative assessment of heterogeneity for surface-immobilized proteins. Anal. Chem. 2001;73:471–480. doi: 10.1021/ac000523p. [DOI] [PubMed] [Google Scholar]
- 23.Svitel J, Balbo A, Mariuzza RA, Gonzales NR, Schuck P. Combined affinity and rate constant distributions of ligand populations from experimental surface binding kinetics and equilibria. Biophys. J. 2003;84:4062–4077. doi: 10.1016/S0006-3495(03)75132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akamatsu K, Kimura M, Shibata Y, Nakano S, Miyoshi D, Nawafune H, Sugimoto N. A DNA duplex with extremely enhanced thermal stability based on controlled immobilization on gold nanoparticles. Nano Lett. 2006;6:491–495. doi: 10.1021/nl0524748. [DOI] [PubMed] [Google Scholar]
- 25.Jin RC, Wu GS, Li Z, Mirkin CA, Schatz GC. What controls the melting properties of DNA-linked gold nanoparticle assemblies? J. Am. Chem. Soc. 2003;125:1643–1654. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 26.Harris NC, Kiang CH. Defects can increase the melting temperature of DNA – nanoparticle assemblies. J. Phy. Chem. B. 2006;110:16393–16396. doi: 10.1021/jp062287d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurst SJ, Hill HD, Mirkin CA. “Three-dimensional hybridization” with polyvalent DNA-gold nanoparticle conjugates. J. Am. Chem. Soc. 2008;130:12192–12200. doi: 10.1021/ja804266j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Craig SL. Thermodynamics of DNA hybridization on gold nanoparticles. J. Am. Chem. Soc. 2005;127:13227–13231. doi: 10.1021/ja052352h. [DOI] [PubMed] [Google Scholar]
- 29.Lytton-Jean AKR, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 30.Wei F, Chen CL, Zhai L, Zhang N, Zhao XS. Recognition of single nucleotide polyrnorphisms using scanning potential hairpin denaturation. J. Am. Chem. Soc. 2005;127:5306–5307. doi: 10.1021/ja043661v. [DOI] [PubMed] [Google Scholar]
- 31.Wei F, Qu P, Zhai L, Chen CL, Wang HF, Zhao XS. Electric potential induced dissociation of hybridized DNA with hairpin motif immobilized on silicon surface. Langmuir. 2006;22:6280–6285. doi: 10.1021/la060602h. [DOI] [PubMed] [Google Scholar]
- 32.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R, Viswanadham G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 33.Wong ELS, Chow E, Gooding JJ. DNA recognition interfaces: the influence of interfacial design on the efficiency and kinetics of hybridization. Langmuir. 2005;21:6957–6965. doi: 10.1021/la050725m. [DOI] [PubMed] [Google Scholar]
- 34.Park S, Brown KA, Hamad-Schifferli K. Changes in oligonucleotide conformation on nanoparticle surfaces by modification with mercaptohexanol. Nano Lett. 2004;4:1925–1929. [Google Scholar]
- 35.Herne TM, Tarlov MJ. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 36.Levicky R, Herne TM, Tarlov MJ, Satija SK. Using self-assembly to control the structure of DNA monolayers on gold: A neutron reflectivity study. J. Am. Chem. Soc. 1998;120:9787–9792. [Google Scholar]
- 37.Maxwell DJ, Taylor JR, Nie SM. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J. Am. Chem. Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 38.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Craig SL. Influence of the extent of hybridization on the hydrodynamic radius of DNA-functionalized gold nanoparticles. Langmuir. 2007;23:2015–2020. doi: 10.1021/la0622830. [DOI] [PubMed] [Google Scholar]
- 40.Eyring H. The activated complex in chemical reactions. J. Chem. Phys. 1935;3:107. [Google Scholar]
- 41.Glasstone S, Laidler KJ, Eyring H. The Theory of Rate Processes: The Kinetics of Chemical Reactions, Viscosity, Diffusion and Electrochemical Phenomena. New York: McGraw-Hill; 1941. [Google Scholar]
- 42.Foltastogniew E, Russu IM. Sequence dependence of base-pair opening in a DNA dodecamer containing the Caca/Gtgt sequence motif. Biochemistry. 1994;33:11016–11024. doi: 10.1021/bi00202a022. [DOI] [PubMed] [Google Scholar]
- 43.Dewey TG, Turner DH. Laser temperature jump study of solvent effects on poly(adenylic acid) stacking. Biochemistry. 1980;19:1681–1685. doi: 10.1021/bi00549a025. [DOI] [PubMed] [Google Scholar]
- 44.Stevens PW, Henry MR, Kelso DM. DNA hybridization on microparticles: determining capture-probe density and equilibrium dissociation constants. Nucleic Acids Res. 1999;27:1719–1727. doi: 10.1093/nar/27.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storhoff JJ, Elghanian R, Mirkin CA, Letsinger RL. Sequence-dependent stability of DNA-modified gold nanoparticles. Langmuir. 2002;18:6666–6670. [Google Scholar]
- 46.Fiche JB, Buhot A, Calemczuk R, Livache T. Temperature effects on DNA chip experiments from surface plasmon resonance imaging: Isotherms and melting curves. Biophys. J. 2007;92:935–946. doi: 10.1529/biophysj.106.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Y, Wolf LK, Georgiadis RM. Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henry MR, Stevens PW, Sun J, Kelso DM. Real-time measurements of DNA hybridization on microparticles with fluorescence resonance energy transfer. Anal. Biochem. 1999;276:204–214. doi: 10.1006/abio.1999.4344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.