Abstract
Transcription factors (TFs) are the key elements responsible for controlling the expression of genes in bacterial genomes and when visualized on a genomic scale form a dense network of transcriptional interactions among themselves and with other protein coding genes. Although the structure of transcriptional regulatory networks (TRNs) is well understood, it is not clear what constrains govern them. Here, we explore this question using the TRNs of model prokaryotes and provide a link between the transcriptional hierarchy of regulons and their genome organization. We show that, to drive the kinetics and concentration gradients, TFs belonging to big and small regulons, depending on the number of genes they regulate, organize themselves differently on the genome with respect to their targets. We then propose a conceptual model that can explain how the hierarchical structure of TRNs might be ultimately governed by the dynamic biophysical requirements for targeting DNA-binding sites by TFs. Our results suggest that the main parameters defining the position of a TF in the network hierarchy are the number and chromosomal distances of the genes they regulate and their protein concentration gradients. These observations give insights into how the hierarchical structure of transcriptional networks can be encoded on the chromosome to drive the kinetics and concentration gradients of TFs depending on the number of genes they regulate and could be a common theme valid for other prokaryotes, proposing the role of transcriptional regulation in shaping the organization of genes on a chromosome.
INTRODUCTION
Products of genes have different functional roles and hence not all genes are used at the same time and for the same purpose. This explains why groups of genes are differentially expressed. For instance, genes encoding for enzymes in krebs's cycle are constitutively expressed in response to most growing conditions while genes responsible for using alternative carbon sources are sporadically required. The decision about which genes should be turned on or off is executed by transcription factors (TFs) that use metabolites/signals as input information from the environmental state and give a transcriptional response as output (1–3). As a result, the notion that different TFs are expressed in different proportions came into existence. For instance, LacI, a repressor of the operon for lactose consumption, is expressed in the order of tens’ of molecules per cell, while global regulators such as CRP (cAMP receptor protein) or IHF (integration host factor) occur in the order of thousands of molecules in the course of the cell cycle (4,5). In bacterial cells, where transcription and translation are coupled to happen in the same compartment these considerations become especially important for regulating gene expression. During transcription, regulatory proteins (TFs) should find and bind to specific DNA sequences on the operator region of their target genes to repress or induce their transcription (6). The protein-DNA interaction is a critical step in gene regulation as TFs find their DNA-binding sites as result of a passive process. Furthermore, TFs do not use energy (e.g. ATP hydrolysis) to get DNA-sequence information (7), which forces these systems to use additional strategies for the optimal performance of different TFs. In the early era of molecular biology, brownian diffusion was thought to be the determining step in DNA-binding site recognition by TFs. However, this assumption was challenged when it was reported that the LacI repressor finds its DNA-targets 90–100 times faster than that predicted by a mere diffusive mechanism (8,9). This observation led to the suggestion of ‘facilitated diffusion’ mechanism. In such a mechanism, TFs alternate between a three dimensional (3D) diffusion in the cell jumping between DNA-strands and one-dimensional (1D) sliding along the DNA to rapidly locate their binding sites (10–12). This hypothesis was corroborated by several works mostly with single molecule studies in which the authors visualized individual TFs interacting with the DNA (4,9,13,14). Several groups have also mathematically modeled the sliding process along the DNA and shown it to be a plausible way of making the search significantly faster than 3D diffusion alone, in particular for TFs in low cellular concentrations (7,15–17). However, it is unclear what factors govern a TF to adopt one or the other strategy discussed above and if there is an interplay between nucleoid structure, genome organization and the biophysical aspects of transcriptional regulation in bacterial systems.
From a genomic perspective, recent works have suggested the importance of chromosomal gene order in bacterial genomes from transcription units and operon organization to divergent and convergent transcriptional control (18–21). Since TFs regulate the expression of genes including themselves and that of other TFs, a functional network of TFs is formed in each organism (22,23). It has been suggested by several groups that biological networks are hierarchical and scale-free in their structure (24,25), however, our understanding on the functional constrains governing this network structure is very limited. Thus, addressing the design behind these architectures in the context of genome organization can provide important insights to a better understanding of genome structure and function. In this study, we first show a link between the out-degree of a TF, their genomic organization with respect to their targets and their concentration at mRNA and protein levels and then propose a conceptual model that explains how the biophysics for the recognition of DNA targets for global and local TFs, concentration gradients for TFs and the gene order of regulons on the chromosome interplay in the larger context of transcriptional networks, using the currently available TRNs of the best characterized gram-negative bacterial model, Escherichia coli (26) and that of the not as well-characterized gram-positive representative, Bacillus subtilis (27).
MATERIALS AND METHODS
Identification of regulon groups
To identify the different regulon groups based on normalized regulon size and normalized average chromosomal distance between TF and its TGs in a regulon, we used K-means clustering implemented in cluster (28). To find the number of distinct clusters present in the data we first varied the number of clusters (parameter number of clusters in K-means clustering) to identify how many times the optimal solution has been found in 1000 runs using euclidean distance as the similarity metric. We found that when the number of clusters was set to three the optimal solution was found in 350 times out of 1000 runs while when the numbers of clusters was set to 2, 4, 5 the optimal solution was reached in 120, 167 and 84 times, respectively, suggesting that the number of clusters in the set is indeed three. Similar approaches have been used by others in calculating the significance of clusters with other clustering approaches, as principled clustering frequently results in suboptimal solutions in a single run (29).
To determine the composition of the clusters, we ran the K-means clustering algorithm using three as the number of clusters and 1000 as the number of runs. However since different runs of the k-means clustering algorithm may not give the same final clustering solution, we repeated this experiment 10 times and finally took a consensus of the groupings identified in these runs. We repeated the whole approach to identify the distinct clusters in B. subtilis.
Estimating the statistical significance of the regulon groups
To calculate the probability of expecting the chromosomal distances seen in each regulon group by chance, we compared the average chromosomal distance observed in each regulon group against the average chromosomal distances seen in 1000 randomly generated regulon groups obtained by preserving the number of regulatory interactions for each TF in a regulon group. Such a randomization preserves the number of TFs and the interactions in a regulon group but still associates to randomly selected genes in the complete genome thus preserving the topology of the regulon group while shuffling the genomic organization of the targets with respect to their regulating TF.
Statistical significance was assessed based on (i) Z-score, calculated as the number of standard deviations the observed value is away from the randomly expected mean. This is obtained as the ratio between the differences of the observed, x, and random expected, μ, values to the standard deviation, σ i.e. Z = (x – μ)/σ) and (ii) P-values, defined as the fraction of the 1000 random trails which showed a value ≥ what was observed in the real dataset.
RESULTS
Genomic co-localization of TFs and target genes is observed in small regulons
Simple regulons comprise of TFs and the set of genes they regulate and were defined as early as 1964 (30). The functional properties of these sets of genes can be diverse, vary in number and be encoded dispersedly on the chromosome. However, it is unclear if there is any relationship between regulon size and the chromosomal positioning of their genetic components, neither is it known if their relative genome organization has an influence on the cellular concentrations of the TFs.
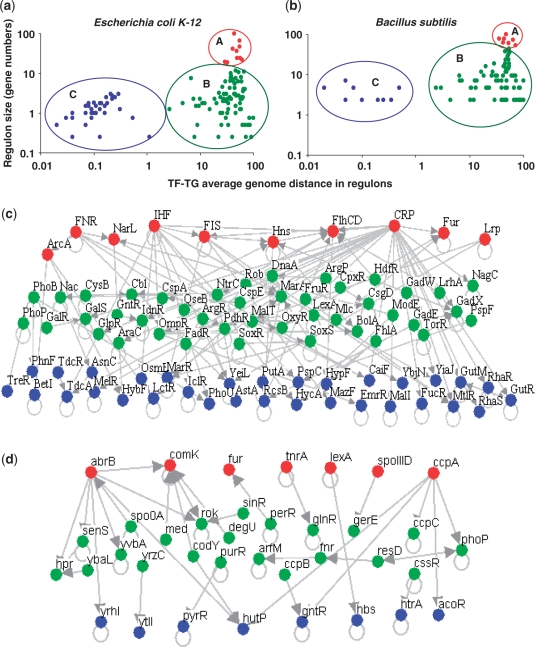
In a previous study we reported a distinct organization of genes coding for TFs and their effector genes (whose products control TFs), depending on whether the effector proteins sense signals from endogenous or exogenous origin in Escherichia coli (21). Here, we analyze if this observed distance, when extended to all members of a regulon, shows any trends depending on the size of the regulon. It should be noted that there is a clear distinction between TF-effector gene pairs and TF-target gene pairs. While the product of the former controls the activity of the TFs the later correspond to the set of genes transcriptionally regulated by the TF (forming part of a regulon). In this work our interest is to understand how the chromosomal distances (measured as number of intervening protein coding genes on a circular genome) between TF and its target genes in different regulons can explain or reflect the network structure. To address these questions, we obtained all regulons wherein TFs regulate at least two genes (excluding auto-regulation) in E. coli and in B. subtilis, taken from regulonDB (26) and DBTBS (31), respectively. We included heterodimeric TFs and excluded auto regulatory interactions. In E. coli K12, our final dataset contained 141 regulons comprising of 1597 regulatory interactions between TFs and their regulated genes; in B. subtilis the dataset contained 54 regulons comprising of 499 genes. First we asked if there is any link between regulon size (number of regulated genes by each TF) and the average chromosomal distance [calculated as the number of intervening protein coding genes on the circular chromosome as described earlier (21)] between the TF and its target genes in each case. As a result of clustering (see ‘Materials and methods’ section), regulons in both organisms can be grouped into three main categories (see Figure 1a and b and Table 1): (A) a few big regulons (10 in E. coli and 7 in B. subtilis) regulating more that 50% of the genes in their transcriptional networks (group A in Figure 1a and b). (B) An intermediate and heterogeneous group of regulons consisting of varying regulon sizes and chromosomal distances (group B); and (C) a group of small regulons having short chromosomal distances (group C). Notably, small regulons (group C) are smaller than the biggest operons of E. coli (15 genes) and B. subtilus (22 genes), possibly suggesting limitations on their sizes to act as functional modules either in the context of co-expression or for horizontal transfer (18,32). The group of 10 TFs in E. coli having the most number of regulated genes, all are classified as global regulators according to one or more previous studies (33) while most of the TFs constituting small regulons were found to sense external fluctuant signals resembling local genetic modules (2). In particular, we found that highly connected TFs were either Nucleoid Associated Proteins (NAPs) like IHF, FIS, HNS or growth condition associated global regulators like CRP (use of carbon sources), Fnr and NarL (anaerobiosis), central intermediary regulators like Lrp, ferric uptake regulator (Fur) or developmental pathway associated factors like FlhDC (responsible for biofilm formation), suggesting that these regulators indeed have key functional roles in controlling the transcriptional responses of the cell depending on the condition of growth. It is interesting to note that several NAPs which are known to act as bacterial analogs of chromatin remodeling factors are enriched in this class (see below). Similarly, a functional analysis of the TFs from group B suggested that several of them are involved in basic cellular activities like regulation of the biosynthesis of amino acids, regulation of cell division and repair, regulation of the uptake of elements, cellular stress and response to antibiotics indicating a limited functional role of these TFs compared to those from group A. Finally, an analysis of TFs from group C suggested that they are involved in the uptake of specific carbon sources, degradation of small molecules and are abundant in two component response regulators (see Supplementary Data for detailed annotations of TFs from different classes). To estimate if the average chromosomal distance seen in each group is significant, we compared this distance against those seen in randomly generated sets as described in ‘Materials and Methods’ section. We found that the observed distances for each of the three groups are significantly smaller than expected by chance, with regulons from group C being the closest (Table 1).
Figure 1.
Relationship between size (defined as the number of target genes) and average chromosomal distance for all known regulons in (a) E. coli and (b) B. subtilis. Regulon size is plotted on Y-axis and is normalized with respect to size of the biggest regulon in each genome for the sake of comparison across genomes and the average chromosomal distance between the TF encoding gene and their respective target genes is shown on X-axis. Chromosomal distances were calculated as defined earlier (21) with the maximum distance being half the number of protein coding genes on a circular chromosome. Note that both regulon sizes and average chromosomal distances are normalized with respect to the maximum and both the axes are shown on a logarithmic scale. Flow of regulatory interactions between the TFs heading the regulons, grouped according to their size and chromosomal distance in (c) E. coli and (d) B. subtilis; it can be noted that the regulatory flux among TFs typically follows the order, big to intermediate to small regulons, coloured respectively in red (big), green (intermediate) and blue (small).
Table 1.
Properties of the main groups of regulons identified in the regulatory network of E. coli, based on average chromosomal distance between TF and its target genes
| Regulon group | Number of regulons (% of total regulons) | Regulon size (average no. of genes/regulon) | Total number of regulated genes (% of total regulated genes) | Average distance (in gene numbers) between the TF and the target genes | P-value significance (Z-score) |
|---|---|---|---|---|---|
| A | 10 (7%) | 76–399 (159) | 1595 (99) | 1059.45 | <0.001 (–9.98) |
| B | 73 (52%) | 2–48 (13) | 953 (59) | 889.69 | <0.001 (–10.74) |
| C | 58 (41%) | <12 (4.8) | 281 (17.5) | 2.42 | <0.001 (–20.44) |
Transcriptional regulatory flow in the network of TFs
To find out if there is any coordination between the TFs heading the different groups of regulons identified above, we analyzed the regulatory flow among the TFs constituting the regulatory network (34–36). Figure 1c and d shows the regulatory interactions present between at least two TFs in E. coli and B. subtilis. Note that, all the TFs of group A are at the top of the network hierarchy initiating the regulatory interactions in the network of TFs. The regulatory flow follows an order, from TF members of group A to B to C, and there are no regulatory interactions from members of group C directed to B or A, indicating no feedback at the level of transcriptional regulation from the bottom to the top. However, there are some regulatory interactions between members of the same group and from members of group B towards members of group A. Other approaches for constructing hierarchical networks, such as the bottom-up strategy (36), using TF–TF network did not change our observations that group A shows a preference to occur at the top of the hierarchy while group C appears at the bottom of the hierarchical network (see Supplementary Data). The partitioning of transcriptional network into big, intermediate and small regulons illustrates how the network components could be structured on a chromosome in a scale-free distribution, observed in various biological networks (24,25). It is possible to generalize from our observations, that the TFs at the bottom of this hierarchy often correspond to very specific functional roles like those sensing specific environmental conditions (37) (see Supplementary Data).
Absolute and average mRNA abundance of TFs suggests correlation with regulon size and network hierarchy in E. coli
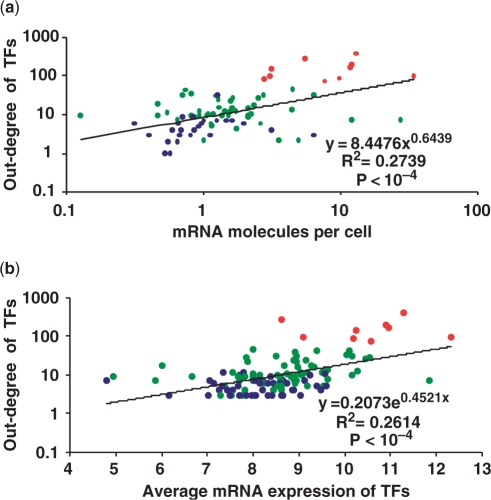
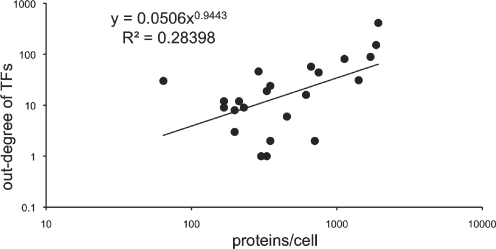
It is believed that global regulators should be present in higher concentrations in the cell compared to local or dedicated TFs (4). In fact, it is known to be valid for nucleoid-associated proteins and other global regulators like CRP, Lrp and Fur in E. coli, whose protein concentrations reach more than 1000 units per cell (5,38). On the other hand, the number of TF proteins of LacI, a dedicated TF for lactose utilization, rises from around 5 to a maximum of 20 upon induction of lactose (39). Indeed, early genomic approaches to study gene expression patterns on a genomic scale which exploited the codon frequency bias of highly expressed cellular machinery like ribosomal, transcription and cheparone associated classes, have shown that sequence specific TFs are generally poorly expressed (40). However, so far, no global analysis has been performed to compare TF protein concentration with their connectivity and network hierarchy. Therefore to address this, we used mRNA profile data from two experiments performed in the M9+glucose medium, in which the absolute number of mRNA molecules were quantified (41–43). We obtained the number of mRNA molecules (per cell) of genes encoding for TFs from this dataset, to see if it correlates with their connectivity and grouping as identified in Figure 1 (see Figure 2a). We found that TFs higher in the network hierarchy had greater number of mRNA molecules per cell associated with them, suggesting that more protein molecules are produced (Figure 2a and Supplementary Data). To investigate further, the relationship between concentration of a TF and its network hierarchy, we compared TF's outdegree against its average gene expression using a large compendium of E. coli microarrays reported recently (44). We found that TF's outdegree and its average mRNA level across experiments follows the hierarchy described above (Figure 2b). Our results suggest that several regulators from group C in Figure 1 are poorly expressed, consistent with previous observations that two-component systems which are enriched in group C and are proximal on the chromosome show poor predicted expression values using codon usage measures (21,40). If we assume that mRNA formation is a determining step in protein synthesis, these data might correspond to the absolute protein concentrations of the respective TFs per bacterial cell implying a correlation between a TF's out-degree and its concentration, extending upon previous studies (22,45,46). These observations clearly indicate that the concentration of a TF is related to the way it is encoded on the chromosome with respect to its target genes, with local TFs regulating few genes present in physical proximity to their target genes and global TFs facilitating the regulation of many genes by increasing their cellular concentration. Indeed it has been postulated using simulations that low copy number TFs need to colocalize with their targets to enable a rapid and reliable gene regulation, confirming the need to place low copy local TFs in physical proximity to their targets in the genome (47). Proteome profiles for TFs were limited to a countable number until recently when two massive proteomic experiments were reported for E. coli (42,48). Excluding the nucleoid-associated proteins which are discussed below, we could obtain protein concentrations for 25 TFs belonging to different levels of E. coli network from these experiments (see Figure 3 and Supplementary Data). Consistent with our observations at mRNA level, TFs with high intracellular levels corresponded with high out-degree when their protein concentration is plotted as a function of the number of target genes (Figure 3). With respect to NAPs, these high-throughput experiments confirm their high abundance reported almost ten years ago using quantitative western blot analyses (49). Indeed, a closer look at the peak expression by the same authors suggested that the production of these NAPs is distributed along the bacterial growth-phases (see Figure 4d). The high cellular levels of these proteins with concentrations varying from 20 000 and 50 000 units made it possible to estimate that on an average each monomer may bind every 500 bp along the genomic DNA (49). In summary, in agreement with the data for mRNAs, we observe that protein abundance corresponds with the out-degree of a TF in the network, with NAPs being particularly abundant and expressed in a growth-phase dependent manner, possibly to re-structure the nucleoid, facilitating the running of particular transcriptional programs depending on growth phase status (see below), (5,49,50). Therefore, one can hypothesize from these results that the scalefree structure of bacterial transcriptional regulatory networks (TRNs) is encoded in the chromosome itself and that genome organization of bacterial chromosomes might indeed be influenced by their TRNs.
Figure 2.
(a) Relationship between mRNA abundance and out-degree of a TF in the regulatory network of E. coli. TFs are colored as per their grouping in Figure 1 with big regulons in red, intermediate ones in green and small regulons in blue. Bigger the regulon, stronger is its tendency to be expressed in higher concentrations. (b) Relationship between out-degree of a TF and its average mRNA level, calculated after processing and normalizing the expression data according to RMA normalization, as reported by the authors (44).
Figure 3.
Number of proteins/cell for TFs as a function of the number of genes transcriptionally regulated by it (excluding NAPs as their protein levels are shown in Figure 4d and ArcA and NarL which are known to be poorly expressed in aerobic condition where the experiment is performed). Data are available as Supplementary Data.
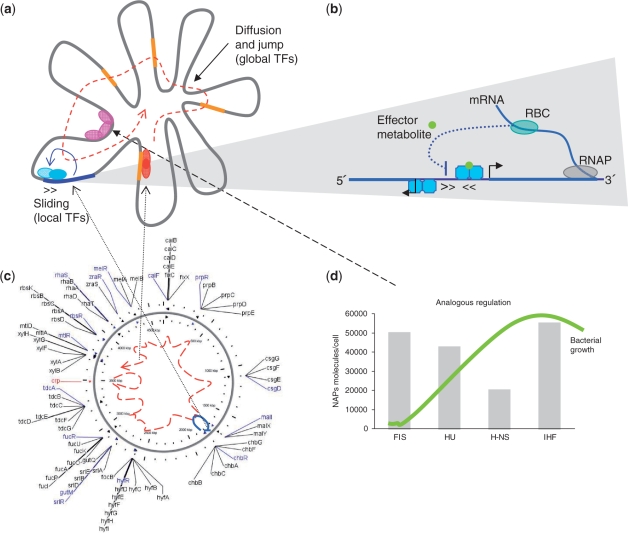
Figure 4.
Integrated model of TRN in bacteria (a) combined model representing various factors involved (b) activity and mechanistic basis for the functioning of local TFs (c) an example of global and local TFs co-regulating genes involved in the uptake of carbon sources in E. coli (d) protein abundance of different nucleoid-associated proteins along the growth-phases, acting as analog regulators.
A conceptual model for the structuring of regulatory networks in bacteria
In the integrated model we propose here (Figure 4), the biophysical aspects of TFs for reaching their DNA-binding sites might be the main driving force for structuring the regulatory networks in bacteria as we know presently. This conceptual model is supported by the following observations and evidences:
TFs governing small regulons are located close to their regulated genes on the chromosome and this spatial arrangement together with the fact that transcription and translational mechanisms occur simultaneously, should favor that the newly synthesized protein can contact quickly its target DNA through the sliding and hopping mechanism as was shown in the case of LacI (4,9) (Figure 4c). These local regulators are normally expressed in lower cellular concentrations as they would be required sporadically to regulate few operons whose products have dedicated functions. For instance, regulation of alternative carbon sources in E. coli is mainly governed by the global regulator CRP and a group of local TFs controlling small regulons which are located proximally on the chromosome with respect to their target genes (Figure 4b). The role of the products encoded in these small regulons is to transport and carry out the first catabolic steps of alternative sugars until their catabolism converges in the glycolysis pathway. Additionally, note that most of these TFs in bacteria are autoregulated (22). Thus, this sliding mechanism could be a generalized strategy for a quicker and tighter control of TFs over their own expression (51).
In contrast, global regulators which are distantly located with respect to the large number of genes they regulate employ a different strategy. Targeting DNA seems to be accurately managed by raising the concentration of the respective TFs and the actual mechanism used for binding DNA would be 3D diffusion and jumping between the DNA strands (Figure 4a and CRP path in Figure 4b). The large cellular concentrations of these proteins might be maintained, in part, given that most global regulators are autoregulated in both positive and negative manner (22). Such a mechanism would also make sure that the concentrations of these proteins are maintained at high intracellular levels.
A third major player for gene regulation in bacteria is the way the DNA molecule is packed into nucleoids (5,49,52). Recent studies provide evidence that the DNA molecule is organized into loops of different lengths (10–100 kbp) which make it possible for some DNA regions to be spatially proximal which would otherwise be distant on a linear molecule of DNA (53–56). Although the exact co-ordinates of these DNA loops is yet to be unveiled even in well-studied systems like E. coli, it is known that nucleoid associated proteins (NAPs) are specifically engaged in structuring DNA depending on the growth condition. These proteins bridge or bend the DNA molecule facilitating DNA loops and nucleoid's structuring (5,52). In particular, NAPs are shown to express in growth-phase dependent manner with FIS at the beginning of stationary phase, HNS in the mid-exponential and IHF in the arrested phase (see Figure 4d) (49). These observations suggest that NAPs might structure the DNA molecule in a different way depending on the growth phase and this action should facilitate or predispose off only a section of the DNA template for the activity of global and local regulators and the running of specific transcriptional programs. Accordingly, it has been suggested that NAPs act as analog regulators whereas the rest of the TFs responding to specific conditions (e.g. by binding signal effectors) act as digital regulators (50,57) (Figure 4c).
DISCUSSION AND CONCLUSIONS
In this study we take advantage of the wealth of information about the transcriptional networks in the best characterized bacterial models and integrate a series of observations to understand the principles behind the structure of transcriptional networks. We show that the regulatory flux is driven from big to small regulons in both E. coli and B. subtilis. Using data from independently reported studies in E. coli we demonstrate that higher a TF is in the transcriptional hierarchy more are its detected number of mRNA and protein molecules per cell, reflecting its need to be expressed in higher concentrations to regulate target genes located dispersedly on the chromosome. In contrast to big regulons, local or dedicated TFs (lower in the network hierarchy) were found to be expressed in much lower concentrations explaining the reasons for their proximity on the chromosome to their target genes. These observations give insights into how the scale-free structure of transcriptional networks can be encoded on the chromosome to drive the kinetics and concentration gradients of TFs, depending on the number of genes they regulate and could facilitate the horizontal transfer of local environment-specific transcriptional modules. Although our distance calculations do not take into account the three dimensional topology of the chromosome under a given cellular condition, it is easy to note that the chromosomal proximity of TFs to their targets in the case of small regulons can not be explained due to chance alone. While in the case of global TFs one can argue that as they regulate several genes, their average linear chromosomal distance could be an over-estimation of intracellular proximity considering the dynamic nature of the nucleoid. However, global TFs with their fuzzy binding sites in contrast to local TFs could complement their affinity to targets by increasing their concentrations to a sufficient degree when needed (46,47). Thus, our results suggest that TRNs play an important role in genome organization by shaping the organization of genes in genomes. These observations illustrate how bacteria as simple biological systems fit predicted theoretical principles in order to optimize their cellular performance in a compacted genome.
SUPPLEMENTARY DATA
Supplementary Data available at http://www.mrc-lmb.cam.ac.uk/genomes/sarath/ScalefreeTRN/.
FUNDING
Medical Research Council and Cambridge Commonwealth Trust (to S.C.J.); A.M.A. acknowledges support and discussions which resulted from a workshop on ‘Dynamics of Regulatory Networks’ at Centro Internacional de Ciencias AC held in Cuernavaca, México in spring 2007. Funding for open access charge: Department of Genetic Engineering (CINVESTAV Irapuato)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andrew Emili, Gabriel Moreno-Hagelsieb and Denis Thieffry for providing helpful suggestions and comments on previous versions of this manuscript.
REFERENCES
- 1.Jacob F. La Logique du Vivant, Une Histoire de L'Hérédité. Paris: Gallimard; 1970. [Google Scholar]
- 2.Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 4.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 7.Hu L, Grosberg AY, Bruinsma R. Are DNA transcription factor proteins Maxwellian demons? Biophys. J. 2008;95:1151–1156. doi: 10.1529/biophysj.108.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 9.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 10.Richter PH, Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor – operator association and membrane bound enzymes. Biophys. Chem. 1974;2:255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- 11.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 12.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor – operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 13.Shimamoto N. One-dimensional diffusion of proteins along DNA. Its biological and chemical significance revealed by single-molecule measurements. J. Biol. Chem. 1999;274:15293–15296. doi: 10.1074/jbc.274.22.15293. [DOI] [PubMed] [Google Scholar]
- 14.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu. Rev. Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 15.Murugan R. Generalized theory of site-specific DNA-protein interactions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:011901. doi: 10.1103/PhysRevE.76.011901. [DOI] [PubMed] [Google Scholar]
- 16.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl Acad. Sci. USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherstvy AG, Kolomeisky AB, Kornyshev AA. Protein–DNA interactions: reaching and recognizing the targets. J. Phys. Chem. B. 2008;112:4741–4750. doi: 10.1021/jp076432e. [DOI] [PubMed] [Google Scholar]
- 18.Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 19.Warren PB, ten Wolde PR. Statistical analysis of the spatial distribution of operons in the transcriptional regulation network of Escherichia coli. J. Mol. Biol. 2004;342:1379–1390. doi: 10.1016/j.jmb.2004.07.074. [DOI] [PubMed] [Google Scholar]
- 20.Menchaca-Mendez R, Janga SC, Collado-Vides J. The network of transcriptional interactions imposes linear constrains in the genome. Omics. 2005;9:139–145. doi: 10.1089/omi.2005.9.139. [DOI] [PubMed] [Google Scholar]
- 21.Janga SC, Salgado H, Collado-Vides J, Martinez-Antonio A. Internal versus external effector and transcription factor gene pairs differ in their relative chromosomal position in Escherichia coli. J. Mol. Biol. 2007;368:263–272. doi: 10.1016/j.jmb.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. J. Mol. Biol. 2008;381:238–247. doi: 10.1016/j.jmb.2008.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janga SC, Collado-Vides J. Structure and evolution of gene regulatory networks in microbial genomes. Res. Microbiol. 2007;158:787–794. doi: 10.1016/j.resmic.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 25.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 26.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Slonim N, Elemento O, Tavazoie S. Ab initio genotype-phenotype association reveals intrinsic modularity in genetic networks. Mol. Syst. Biol. 2006;2:20060005. doi: 10.1038/msb4100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maas WK, Maas R, Wiame JM, Glansdorff N. Studies on the mechanism of repression of arginine biosynthesis in escherichia coli. I. Dominance of repressibility in zygotes. J. Mol. Biol. 1964;78:359–364. doi: 10.1016/s0022-2836(64)80199-6. [DOI] [PubMed] [Google Scholar]
- 31.Ishii T, Yoshida K, Terai G, Fujita Y, Nakai K. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res. 2001;29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Ma HW, Buer J, Zeng AP. Hierarchical structure and modules in the Escherichia coli transcriptional regulatory network revealed by a new top-down approach. BMC Bioinformatics. 2004;5:199. doi: 10.1186/1471-2105-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobrin R, Beg QK, Barabasi AL, Oltvai ZN. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics. 2004;5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl Acad. Sci. USA. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagomarsino MC, Jona P, Bassetti B, Isambert H. Hierarchy and feedback in the evolution of the Escherichia coli transcription network. Proc. Natl Acad. Sci. USA. 2007;104:5516–5520. doi: 10.1073/pnas.0609023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Hao Z, Bieniek E, Calvo JM. Modulation of Lrp action in Escherichia coli by leucine: effects on non-specific binding of Lrp to DNA. J. Mol. Biol. 2001;314:1067–1075. doi: 10.1006/jmbi.2000.5209. [DOI] [PubMed] [Google Scholar]
- 39.Droge P, Muller-Hill B. High local protein concentrations at promoters: strategies in prokaryotic and eukaryotic cells. Bioessays. 2001;23:179–183. doi: 10.1002/1521-1878(200102)23:2<179::AID-BIES1025>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 42.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 43.Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- 44.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seshasayee AS, Fraser GM, Babu MM, Luscombe NM. Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res. 2009;19:79–91. doi: 10.1101/gr.079715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J. Mol. Biol. 2008;379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolesov G, Wunderlich Z, Laikova ON, Gelfand MS, Mirny LA. How gene order is influenced by the biophysics of transcription regulation. Proc. Natl Acad. Sci. USA. 2007;104:13948–13953. doi: 10.1073/pnas.0700672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marr C, Geertz M, Hutt MT, Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. London. UK: Chapman & Hall/CRC; 2007. [Google Scholar]
- 52.Zimmerman SB. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 2006;156:255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 53.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kepes F. Periodic transcriptional organization of the E.coli genome. J. Mol. Biol. 2004;340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 55.Riva A, Carpentier AS, Barloy-Hubler F, Cheron A, Henaut A. Analyzing stochastic transcription to elucidate the nucleoid's organization. BMC Genomics. 2008;9:125. doi: 10.1186/1471-2164-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 57.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.