Abstract
Trypanosoma brucei is a member of the early-diverged, protistan family Trypanosomatidae and a lethal parasite causing African Sleeping Sickness in humans. Recent studies revealed that T. brucei harbors extremely divergent orthologues of the general transcription factors TBP, TFIIA, TFIIB and TFIIH and showed that these factors are essential for initiating RNA polymerase II-mediated synthesis of spliced leader (SL) RNA, a trans splicing substrate and key molecule in trypanosome mRNA maturation. In yeast and metazoans, TFIIH is composed of a core of seven conserved subunits and the ternary cyclin-activating kinase (CAK) complex. Conversely, only four TFIIH subunits have been identified in T. brucei. Here, we characterize the first protistan TFIIH which was purified in its transcriptionally active form from T. brucei extracts. The complex consisted of all seven core subunits but lacked the CAK sub-complex; instead it contained two trypanosomatid-specific subunits, which were indispensable for parasite viability and SL RNA gene transcription. These findings were corroborated by comparing the molecular structures of trypanosome and human TFIIH. While the ring-shaped core domain was surprisingly congruent between the two structures, trypanosome TFIIH lacked the knob-like CAK moiety and exhibited extra densities on either side of the ring, presumably due to the specific subunits.
INTRODUCTION
In eukaryotes, specific initiation of RNA polymerase (pol) II-mediated (class II) transcription is directed by the general transcription factors (GTFs) TFIIA, TFIIB, the TATA-binding protein (TBP)/TFIID, TFIIE, TFIIF and TFIIH. These factors form the transcription pre-initiation complex (PIC) at core promoters, recruit RNA pol II to the DNA, separate the DNA strands at the transcription initiation site (TIS) and, by phosphorylating the C-terminal domain (CTD) of the largest enzyme subunit RPB1, facilitate the escape of the polymerase from the promoter. Disregarding the ∼13 TBP-associated factors of TFIID, the conserved GTFs comprise 18 polypeptides in Saccharomyces cerevisiae and 19 subunits in mammals (1). In comparison, the archaeal PIC is much less complex: it consists of only three polypeptides, namely TBP, the TFIIB orthologue TFB and TFE which corresponds to the N-terminal portion of the eukaryotic TFIIEα subunit (2,3). Gene sequences of flagellated protists such as the dinoflagellate Giardia lamblia and the trypanosomatids Trypanosoma brucei, Trypanosoma cruzi and Leishmania major (Tritryps) are the most divergent among eukaryotes indicating that the phylogenetic lineages of these organisms have diverged from the main eukaryotic lineage very early in evolution (4,5). Interestingly, most GTFs were not recognized in the completed genomes of both G. lamblia (6) and the Tritryps (7) raising the possibility that these early-diverged eukaryotes harbor a simplified transcription machinery (6,8). However, since the majority of genes in these protistan organisms could not be annotated thus far, it is equally possible that sequence divergence has prevented in silico identification of GTFs.
The Tritryps are vector-borne, well-characterized human parasites that cause lethal tropical diseases (http://www.who.int/tdr/index.html). In these parasites, protein coding gene expression amongst other cellular processes is unusual: the genes are arranged in long tandem arrays and transcribed polycistronically, and individual mRNAs are resolved from precursors by spliced leader (SL) trans splicing and polyadenylation. Astonishingly, a class II promoter directing transcription of these genes from a concrete initiation site has not been characterized, and it is not understood how RNA pol II is recruited to these gene arrays. The small nuclear SL RNA, the SL donor in trans splicing, is the only small RNA in trypanosomatids that is synthesized by RNA pol II (9). Since SL RNA is consumed in the trans splicing process, trypanosomatids harbor up to 100 tandemly repeated SL RNA gene copies (SLRNAs) per haploid genome which, in contrast to protein coding genes, are transcribed monocistronically from a defined TIS. The SLRNA promoter structure is conserved among trypanosomatids and consists of a bipartite upstream sequence element (10–12) and an initiator (13). While the Tritryp genome annotations revealed, as potential GTFs, only the TBP-related protein 4 (TRF4) and the two TFIIH helicase subunits Xeroderma pigmentosum B (XPB) and XPD (7), recent studies have identified three factors which are indispensable for SLRNA transcription. The first factor was a complex consisting of TRF4, the tripartite small nuclear RNA-activating complex, which has been characterized in humans as a factor specific for small RNA gene promoters, the small subunit of TFIIA (TFIIA-2), and a larger protein whose sequence conservation is too weak for an unambiguous TFIIA-1 assignment (14,15). Since TBP and TFIIH have essential functions in addition to class II transcription initiation, the discovery of TFIIA was the first clear indication that trypanosomatids do possess GTFs. Accordingly, subsequent revisiting of the Tritryp genome sequences identified an extremely divergent TFIIB orthologue whose essential role in SLRNA transcription and its specific interactions with RNA pol II and the SLRNA promoter were confirmed (16,17). Only when T. brucei was recently shown to harbor a transcriptionally relevant TFIIH did it become clear that trypanosomatids also form a PIC similar in complexity to those of yeast and mammals (18).
TFIIH is a multi-subunit GTF and consists of the core and the cyclin-activating kinase (CAK) subcomplexes (19). The core complex comprises XPB and XPD, their respective regulators p52/TFB2 (mammalian/yeast nomenclature) and p44/SSL1 as well as p62/TFB1, p34/TFB4, and the recently discovered small subunit TFB5 (20), while cyclin-dependent kinase 7 (CDK7)/kin28, cyclin H/CCL1, and MAT1/TFB3 constitute the CAK complex. The helicase activity of TFIIH is required for strand separation at the TIS, whereas CDK7 phosphorylates serine 5 of the heptad repeat sequence YSPTSPS in the RPB1 CTD enabling RNA pol II to escape from the promoter. While the heptad motif is completely conserved between yeast and mammals, it does not occur in flagellated protists including T. brucei.
The T. brucei TFIIH core complex has been partially characterized and, in addition to the two known helicases, orthologs of p44 and p52 were unambiguously identified (18,21). Here, we have characterized transcriptionally active T. brucei TFIIH by tandem affinity purification, sedimentation analysis and electron microscopy (EM). While the protein complex harbored a complete core, the trypanosome TFIIH differs from its human counterpart by the presence of two essential trypanosomatid-specific subunits and the lack of a CAK complex component.
MATERIALS AND METHODS
Cell culture
Procyclic cell culture, transfection and selection procedures as well as cloning of cell lines by limiting dilution corresponded precisely to the description published before (18).
DNAs
The PTP tagging construct for XPD, pXPD-PTP-NEO, has been described previously (18). The corresponding plasmid TSP2-PTP-NEO was generated by integrating 531 bp of the 3′ terminal coding sequence into the ApaI and NotI restriction sites of the tagging construct pC-PTP-NEO (22). For transfection, pTSP2-PTP-NEO was linearized with BclI. For RNAi experiments, the coding sequence from position 16 to 515 of either TSP1 or TSP2 was integrated into the standard stem–loop vector according to the published cloning strategy (23). For recombinant expression of TSP2, the complete coding region was cloned into the pGEX-4T-2 vector and fused N-terminally to the GST moiety.
Protein analysis
XPD–PTP and TSP2-PTP were tandem affinity-purified from crude extract as detailed previously (22). For identification, purified proteins were separated on a 10–20% SDS–polyacrylamide gradient gel and stained with Pierce Gelcode Coomassie stain (Pierce, Rockford, IL, USA). Individual protein bands were excised, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry. The identifications were confirmed by a second purification and a MALDI-TOF analysis in which the peptide coverage exceeded 60% in each case (data not shown). Sedimentation of XPD–P and TSP2-P complexes in 4 ml 10–40% linear sucrose gradients by ultracentrifugation was carried out as described before (14). Sypro ruby (Invitrogen, Carlsbad, CA, USA) staining was carried out according to the manufacturer's protocol. For functional in vitro transcription assays, TSP2-P and XPD–P protein was eluted in transcription buffer (150 mM sucrose, 20 mM potassium l-glutamate, 3 mM MgCl2, 20 mM HEPES–KOH, pH 7.7) containing 0.5 mg/ml ProtC peptide and 1 mM CaCl2. Unless stated otherwise, amino acid sequence alignments were carried out with the ClustalW2 program with default parameter values (24).
Immunolocalization
Immunolocalizations were essentially carried out as described previously (16). The protein A domains of the PTP tag of XPD–PTP and TSP2-PTP were first probed with a 1:40 000 dilution of a rabbit polyclonal anti-protein A immune serum (Sigma, St Louis, MO, USA) and then with a 1:400 dilution of an Alexa 594-conjugated anti-rabbit secondary antibody (Invitrogen). The cover slips were mounted on glass slides with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) and images with identical exposure settings taken with an Axiovert 200 microscope and prepared with AxioVision software (Zeiss, Oberkochen, Germany).
RNAi analysis
Monitoring cell growth of doxycyline induced and uninduced cells, RNA preparation, semi-quantitative RT–PCR, SL RNA and U2 snRNA analysis by primer extension, and labeling and analysis of nascent RNA using a permeabilized cell system were carried out as described previously (18). RNA signals were quantified by densitometry using ImageJ software (http://rsb.info.nih.gov/ij/).
Anti-TSP2 immune serum
TSP2 was expressed with an N-terminal GST tag in Escherichia coli, and the recombinant protein purified by glutathione affinity chromatography (GE Heathcare, Piscataway, NJ, USA) according to the manufacturer's specifications. The recombinant protein was used for immunizing rats as detailed elsewhere (16).
Transcription analysis
The in vitro transcription system has been described in detail (25,26). Briefly, standard reactions of 40 μl containing 8 μl of extract, 20 mM potassium l-glutamate, 20 mM KCl, 3 mM MgCl2, 20 mM HEPES–KOH, pH 7.7, 0.5 mM of each nucleoside triphosphate (NTP), 20 mM creatine phosphate, 0.48 mg/ml of creatine kinase, 2.5% polyethylene glycol, 0.2 mM EDTA, 0.5 mM EGTA, 4 mM dithiothreitol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 12.5 μg/ml vector DNA, 20 μg/ml GPEET-trm and 7.5 μg/ml SLins19 template were incubated for 1 h at 27°C and stopped by adding 300 μl of 4 M guanidine thiocyanate, 25 mM sodium citrate, pH 7.0 and 0.5% N-lauroylsarcosine. In reactions with antisera, 4 μl of extract was pre-incubated with 1 μl of antiserum for 30 min on ice before reactions were started by adding templates and nucleotides. In assays with extracts from RNAi cells, T7 promoter-free GPEET-trm-V2 and SLins19-V2 templates were used.
Total RNA preparations of transcription reactions were analyzed by primer extension of 32P-end-labeled oligonucleotides Tag_PE and SLtag which hybridize to unrelated oligonucleotide tags of the GPEET-trm and SLins19 RNAs, respectively (18). Primer extension products were resolved on 6% polyacrylamide–50% urea gels and visualized by autoradiography.
Electron microscopy and single particle image processing
For negative staining, the purified XPD–P complex was diluted 5-fold with transcription buffer and 5 µl of the final mixture were immediately (∼5 s) applied to a carbon-coated grid that had been glow-discharged (Harrick Plasma, Ithaca, NY, USA) for 3 min in air, and the grid negatively stained using 1% uranyl acetate. Grids were examined in a Philips CM120 electron microscope operated at 80 kV. Images were recorded on a 2Kx2K F224HD slow scan CCD camera (TVIPS, Gauting, Germany) at a magnification of 65 000 (0.37 nm/pixel). Images of individual molecules, isolated from neighboring molecules on micrographs, were selected interactively, windowed out (50 × 50 pixels) and imported into the SPIDER program suite (Health Research Inc., Rensselaer, NY, USA) for further processing. For 2D analysis, the reference-free method (27) was used to generate homogeneous class averages, initially processed from 2828 particles. For 3D reconstruction, the initial reference model, taken from human TFIIH (28; filtered to ∼4 nm resolution), was rotated to obtain the most stable view on the carbon; its top-on view was then azimuthally rotated and projected every 4°. Selected images were matched to model projections and refinement cycles were iterated until no structural changes could be detected in 3D reconstruction (29). Eighty percent of the best matching particles, based on the value of the cross-correlation coefficient, were constituted. The resolution of the final reconstruction was determined as ∼2.5 nm estimated from the Fourier shell correlation (FSC) curve using the FSC = 0.5 nm cut-off criterion (data not shown). The presented 3D reconstruction was low pass Gaussian filtered to a resolution of ∼3.5 nm, and then was superimposed onto the human TFIIH 3D for further analysis. UCSF Chimera was used for visualization and comparative analysis of 3D structures as described (30).
RESULTS
Trypanosoma brucei TFIIH consists of a full core and two trypanosomatid-specific subunits
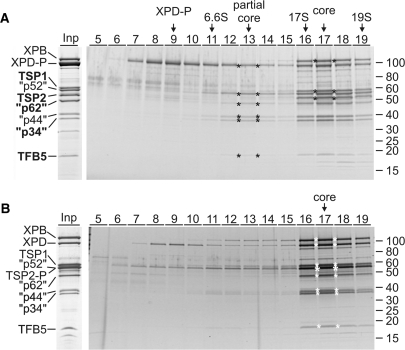
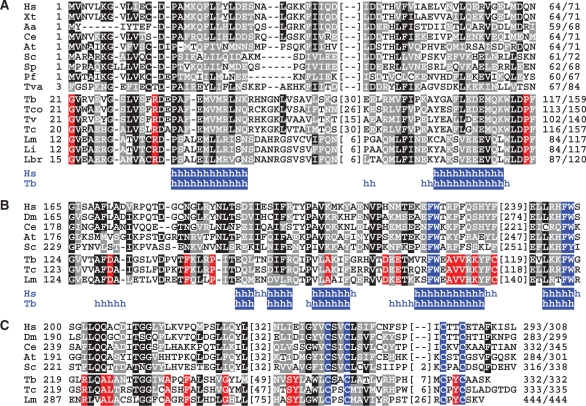
Previously, we have generated a procyclic T. brucei cell line which expressed the essential XPD exclusively as a C-terminal fusion to the composite PTP tag consisting of two protein A domains, a TEV protease cleavage site and the protein C epitope (ProtC) (18). Tandem affinity purification of XPD–PTP revealed a functional TFIIH complex of which XPB and the orthologues of mammalian p44 and p52 were identified (18). Overall, this purification resulted in nine major protein bands (Figure 1A, Input lane). To identify all co-purified proteins, individual bands were excised from the gel and proteins identified by trypsin digest and liquid chromatography-tandem mass spectrometry. Five new proteins were identified which are encoded by genes Tb927.1.1080, Tb11.01.5700, Tb11.01.1200, Tb11.01.7730 and Tb10.61.2600 (Table 1). BLAST searches revealed that the sequences of these proteins are conserved among trypanosomatids but they did not uncover similarities to GTFs (data not shown). However, aligning the trypanosomatid sequences with TFIIH subunit sequences from model organisms identified Tb10.61.2600 as TFB5 (Figure 2A). While the trypanosomatid proteins are larger due to N- and C-terminal extensions and an insertion, the remaining sequence shares clear similarity blocks with TFB5 sequences of other eukaryotes including two conserved α helical domains (Figure 2A). The same alignment strategy showed that Tb11.01.1200 is the orthologue of mammalian p62: the trypanosomatid sequences share conserved residues with their eukaryotic counterparts in most regions (Supplementary Figure S1) exhibiting the highest similarity in an internal region essential for the interaction of p62 with XPD and comprising residues 180-245 of human p62 and conserved FW motifs within predicted helical structures (Figure 2B). While the three remaining subunits are only conserved within trypanosomatids (Supplementary Figures S2–S4), the presence of a C-terminal zinc finger and conserved amino acids around this motif and in an internal motif of unknown function strongly indicate that Tb11.01.7730 is the seventh core subunit and orthologue of human p34 (Figure 2C). The two remaining proteins Tb927.1.1080 and Tb11.01.5700, however, are not orthologues of CAK complex components because they lack highly conserved protein kinase domains and the invariant N-terminal Ring domain of MAT1; they were therefore tentatively and respectively termed trypanosomatid-specific proteins 1 and 2 (TSP1 and 2). Besides an internal CX2CX15CX2C zinc finger in TSP2, which is strictly conserved among trypanosomatids, the TSP sequences did not reveal a recognizable sequence motif. In sum, we concluded that trypanosomatids possess all seven core subunits of the eukaryote-specific GTF TFIIH and that the majority of the isolated TFIIH complexes were not associated with a CAK subcomplex.
Figure 1.
Characterization of T. brucei TFIIH. (A) The input lane (Inp) shows the final eluate of a XPD–PTP purification separated by SDS–PAGE and stained with Coomassie. On the left, the TFIIH subunit orthologs are specified (quotation marks indicate that the sizes of the trypanosome proteins are different). The five new identifications are indicated by bold lettering. An equivalent eluate was sedimented through a linear 10–40% sucrose gradient which was fractionated from top to bottom. Proteins from each fraction were separated by SDS–PAGE and stained with Sypro ruby. The six proteins of the partial core and the three additional proteins of the TFIIH core complex are marked by asterisks in their peak fractions. Marker proteins with known S-values were analyzed in parallel gradients. Protein marker sizes are specified on the right. (B) A corresponding analysis is shown for TSP2-PTP purified material. Due to the tag, TSP2-P co-migrates with the p52 orthologue (large asterisk).
Table 1.
Trypanosoma brucei TFIIH subunits
| Subunit | GeneDB acc # | Partial core | Mr (kDa) | # aa | th. pI | App. Size (kDa) |
|---|---|---|---|---|---|---|
| XPB/RAD3 | Tb927.3.5100 | No | 105.2 | 938 | 7.5 | 110 |
| XPD/SSL2 | Tb927.8.5980 | Yes | 92.4 | 819 | 7.5 | 98 |
| TSP1 | Tb927.1.1080 | No | 53.6 | 485 | 7.1 | 58 |
| p52/TFB2 | Tb10.70.1900 | Yes | 56.0 | 500 | 6.8 | 56 |
| TSP2 | Tb11.01.5700 | No | 49.1 | 436 | 7.5 | 50 |
| p62/TFB1 | Tb11.01.1200 | Yes | 41.0 | 366 | 6.1 | 48 |
| p44/SSL1 | Tb927.8.6540 | Yes | 38.0 | 351 | 7.0 | 36 |
| p34/TFB4 | Tb11.01.7730 | Yes | 35.5 | 332 | 5.7 | 35 |
| TFB5 | Tb10.61.2600 | Yes | 17.5 | 159 | 5.0 | 18 |
Mammalian/yeast nomenclature.
TSP, trypanosomatid-specific protein; Mr, molecular mass; th. pI, theoretical pI.
Sequences in GeneDB can be accessed through http://www.genedb.org/
Figure 2.
Sequence alignments of TFIIH core subunits. (A) Alignment of TFB5 sequences from Homo sapiens (Hs, accession number Q6ZYL4), Xenopus tropicalis (Xt, NP_001017298), Aedes aegypti (Aa, NP_00166033), Caenorhabditis elegans (Ce, NP_497212), Arabidopsis thaliana (At, NP_172702), Saccharomyces cerevisiae (Sc, YDR079C-A), Schizosaccharomyces pombe (Sp, SPBC32F12.15), Plasmodium falciparum (Pf, PF14_0398), Trichomonas vaginalis (Tva, XP_001276879) and from the trypanosomatids T. brucei (Tb, Tb10.61.2600), Trypanosoma congolense (Tco, congo855a04.p1k_14), Trypanosoma vivax (Tv, tviv639h03.p1k_13), T. cruzi (Tc, Tc00.1047053511283.50), L. major (Lm, LmjF16.1145), Leishmania infantum (Li, LinJ16_V31190) and Leishmania braziliensis (Lbr, LbrM16_V2.1180). Positions with >50% of identical and conserved residues were shaded in black and gray, respectively. Red shading indicates positions in which trypanosomatid sequences are invariant and no conservation is present in the other sequences. Numbers of amino acids without significant similarity are specified in parentheses. Below, positions in the human and T. brucei sequences with a high probability of α-helical (h) structure are indicated. (B) Corresponding alignment of an internal domain of p62 orthologs. Shading as above except that the FW motifs are shaded in blue. Hs (P32780), Drosophila melanogaster (Dm, NP_610957), Ce (NP_499880), At (NP_175971), Sc (YDR311W), Tb (Tb11.01.1200), Tc (Tc00.1047053508851.79), Lm (LmjF36.3110). (C) Alignment of the C-terminal sequences of p34 orthologs. Shading as above except that the cysteine residues of the putative zinc finger are shaded in blue. Hs (Q13889), Dm (NP_608574), Ce (NP_499249), At (NP_564050), Sc (YPR056W), Tb (Tb11.01.7730), Tc (Tc00.1047053508707.149), Lm (LmjF32.2885).
To determine whether the nine co-purified proteins form a single complex, we subjected the final eluate of the XPD–PTP purification to sucrose gradient sedimentation, fractionated the gradient from top to bottom, and detected co-purified proteins by SDS–PAGE and Sypro ruby staining (Figure 1A). While unbound, tagged XPD–P (XPD–PTP is reduced to XPD–P after TEV protease cleavage) was most abundant in fraction 9, a partial core complex, containing all subunits except XPB, sedimented around fraction 13. The stable association of these proteins in the absence of other proteins confirmed their identification as TFIIH core subunits. Interestingly, XPB completed the core complex only in the presence of TSP1 and TSP2 with a common sedimentation peak in fraction 17 suggesting that the TSPs are bona fide TFIIH subunits in T. brucei. To validate this notion, we tagged TSP2 C-terminally with the PTP tag and tandem affinity-purified this subunit. As expected, the same nine proteins co-purified in apparently stoichiometric amounts, and, when subjected to sucrose gradient sedimentation, they co-sedimented in fraction 17 as before (Figure 1B). This confirmed that both TSPs are quantitatively and stably associated with the trypanosome TFIIH core.
XPD and TSP2 exhibit similar nuclear localization patterns
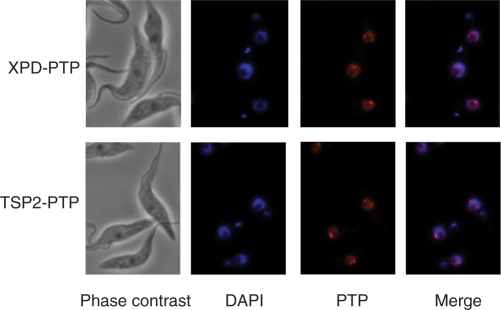
Detection of PTP-tagged XPD and TSP2 by indirect immunofluorescence with a highly specific polyclonal immune serum recognizing the protein A domains of the tag showed that both proteins were exclusively localized to the nucleus (Figure 3). Nuclei were diffusely stained and one or two bright spots, which most likely correspond to the two alleles of the SLRNA tandem array locus, were apparent. These results corresponded to the previous localizations of the p52 orthologue of trypanosome TFIIH (18) and of a differently tagged XPD (21), and they support the finding that TSP2 is in an integral part of TFIIH.
Figure 3.
XPD–PTP and TSP2-PTP exhibit similar nuclear localization patterns. Cells exclusively expressing XPD–PTP or TSP2-PTP were fixed and stained with DAPI, and the tagged proteins detected with a polyclonal rabbit anti-protein A antibody and an Alexa 594-conjugated anti-rabbit secondary antibody.
Silencing of TSP2 expression is lethal and affects SL RNA synthesis
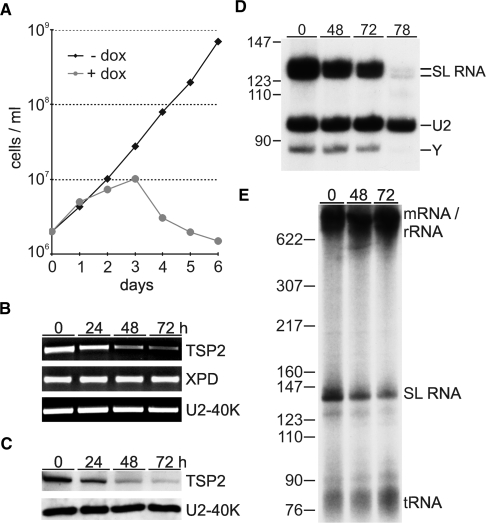
Since TFIIH is essential for trypanosome growth and SLRNA transcription (18,21), it was possible that TSP1 and 2 were important for these functions. We first focused on TSP2 and generated procyclic cell lines for doxycycline-inducible expression of TSP2 dsRNA (31). RNAi-mediated knock-down of TSP2 started to affect cell growth between 24 and 48 h postinduction and led to rapid cell death after 72 h (Figure 4A). The induction of TSP2 dsRNA synthesis resulted in a specific loss of TSP2 mRNA and did not affect the abundance of XPD mRNA or the mRNA of the spliceosomal U2-40K protein within the analysis period of 72 h (Figure 4B). This result was confirmed at the protein level (Figure 4C) with a polyclonal antibody of high specificity which was raised against recombinant, GST-tagged TSP2 (Supplementary Figure S5). Together, these results suggested that the growth defect was a direct consequence of silencing TSP2 expression.
Figure 4.
The role of TSP2 in parasite viability and SLRNA transcription in vivo. (A) Growth curves of a representative clonal procyclic cell line, transfected with a construct for inducible TSP2 dsRNA expression, in the presence or absence of the inducing reagent doxycycline. (B) Semi-quantitative RT–PCR analysis of TSP2, XPD and U2-40K mRNA in total RNA preparations of cells which were doxycycline-induced for 0, 24, 48 and 72 h. (C) Immunoblot of whole cell lysates derived from the same cells was probed with TSP2 and U2-40K-specific polyclonal antisera. (D) Primer extension analysis of SL RNA and, as a control, of U2 snRNA in total RNA preparations of induced cells. As indicated on the right, extension products of cap methylated and unmethylated SL RNA differ by 4 nt. Y specifies the SL RNA intron-specific extension product of the branched Y structure generated in the first trans splicing step. (E) Radio-labeled nascent RNA of induced cells was separated on a 6% polyacrylamide-50% urea gel and visualized by autoradiography. Pre-m/rRNA, SL RNA and tRNA are indicated on the right and pBR322-MspI marker sizes on the left.
To investigate whether TSP2 silencing affected SL RNA synthesis, we first monitored the abundance of SL RNA in steady state RNA by primer extension. In comparison to U2 snRNA, which in T. brucei is synthesized by RNA pol III, both mature SL RNA and the SL RNA intron splicing product decreased 48 and 72 h after RNAi induction and were nearly undetectable 78 h postinduction (Figure 4D). This decrease in steady-state SL RNA was most likely caused by a synthesis defect because labeling of nascent SL RNA was reduced by 69.3% 72 h post induction (Figure 4E). Interestingly, this result is in close accordance with knockdowns of XPD (18), TFIIB (16) and TRF4 (32) suggesting that a mere 2- to 3-fold inhibition of SL RNA synthesis suffices to effectively kill trypanosomes. Importantly, these SL RNA synthesis defects are specific; they do not represent a general death phenotype and they cannot be attributed to the recently reported stress-induced SL RNA silencing phenotype (33) because silencing the expression of class I transcription factors inhibited cell growth with similar kinetics without affecting SL RNA abundance (34,35).
Together, these data strongly indicated that TSP2 is encoded by an essential gene and required for SLRNA transcription. TSP1 appears to be of similar relevance because a corresponding TSP1 knockdown analysis affected trypanosome growth and SL RNA abundance to similar extents (Supplementary Figure S6).
TSP2 is essential for SLRNA transcription in vitro
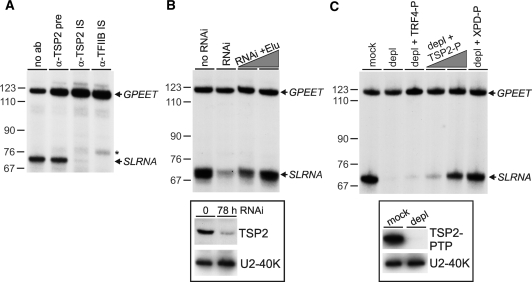
To confirm the essential role of TSP2 in SLRNA transcription, we co-transcribed in a homologous cell-free extract the SLRNA promoter template SLins19 and the control template GPEET-trm, which contains the GPEET procyclin promoter and recruits RNA pol I (25). In a first set of reactions, we pre-incubated extract with anti-TSP2 immune serum and found that the antibody, presumably by interacting with TSP2, abolished class II transcription from the SLRNA promoter whereas it did not affect RNA pol I-mediated transcription from the GPEET promoter (Figure 5A). This effect was as strong as that of an immune serum directed against TFIIB and specific because the corresponding pre-immune serum did not inhibit transcription. In a second set of experiments, extract from cells in which TSP2 expression was silenced was found to be impaired in SLRNA but not in GPEET transcription (Figure 5B). This defect was directly caused by the TSP2 knockdown because adding back PTP-purified TSP2 fully reconstituted SLRNA transcription. Third, we generated a procyclic cell line which exclusively expressed TSP2-PTP and depleted TSP2 from the corresponding extract by IgG affinity chromatography (Figure 5C). The depleted extract did not support SLRNA transcription, whereas GPEET transcription was unaffected. Again, this was a specific effect because SLRNA transcription could be reconstituted by TSP2-PTP and XPD–PTP purified TFIIH but not by the PTP-purified and active TRF4/SNAPc/TFIIA complex (14). Together, these in vitro transcription results showed unambiguously that the purified and characterized TFIIH complex was functionally active and that TSP2 is a TFIIH subunit which is essential for SLRNA transcription.
Figure 5.
TSP2 is essential for SLRNA transcription in vitro. (A) Templates GPEET-trm and SLins19 were co-transcribed in extract which was preincubated without serum (no ab), with anti-TSP2 preimmune serum (α-TSP2 pre), or with immune sera against TSP2 (α-TSP2 IS) and TFIIB (α-TFIIB IS). GPEET-trm and SLins19 transcripts were detected by primer extension assays of total RNA prepared from transcription reactions. GPEET and SLRNA products are specified on the right and pBR322-MspI marker sizes on the left. The asterisk indicates extension products of aberrantly initiated SLRNA transcripts which were specifically detected upon TFIIB inhibition. (B) Corresponding transcription reactions were carried out with extracts prepared from cells in which TSP2 expression was normal (no RNAi) or silenced for 78 h (RNAi). The latter extract was reconstituted with PTP-purified TSP2 to approximately 25% and 100% of its original level. In the insert below, an immunoblot shows the TSP2 level in both extracts. As a loading control, the nuclear protein U2-40K was detected on the same blot. (C) Extract was prepared from cells which exclusively expressed TSP2-PTP. The extract was either mock–treated or depleted of TSP2 (depl) by IgG affinity chromatography. The level of depletion is demonstrated in the immunoblot shown in the insert below. Standard transcription reactions were carried out with both extracts and with the depleted extract which was reconstituted with PTP-purified and active TRF4/SNAPc/TFIIA (depl + TRF4-P), ∼25% and ∼100% of PTP-purified TSP2 (depl + TSP2-P) and ∼100% of PTP-purified XPD (depl + XPD–P).
The EM structure of T. brucei TFIIH confirms the presence of a full core and the lack of the CAK subcomplex
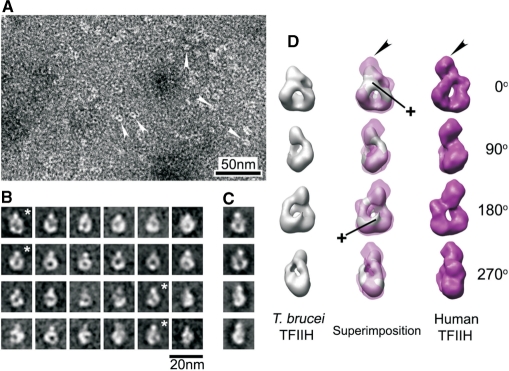
Thus far, our analysis had shown that active T. brucei TFIIH consisted of extremely divergent orthologues of all core subunits, contained two more essential subunits, and lacked a CAK subcomplex. To analyze these characteristics at the structural level, we determined the molecular structure of the XPD–PTP purified complex by EM. A corresponding study of endogenous human TFIIH had shown that the TFIIH core is a flat particle adopting a ring-like shape around a ∼3 nm-wide hole and that the CAK complex forms a ∼5 nm wide bulge protruding from the ring (28). The ring-like shape of the core is conserved because it was also formed by the yeast complex in a crystal (36). Negative staining of the T. brucei complex followed by single particle image processing did not reveal a flat particle but an egg-like shaped particle of 13.3 × 11.2 × 8.2 nm with a ∼3 nm-wide hole (Figure 6A and B). Although the particle length in the vertical axis is shorter than in human TFIIH, the shape around the hole exhibited a remarkable similarity with the ring in projection images of human TFIIH. It should be noted that for this result the class averages of different views of TFIIH were directly obtained from the images and not reference-based (compare Figure 6B and C). The length of the vertical axis in most of the class averages was consistent (Figure 6B), strongly indicating that particles representing different views were rotated around that single axis. Thus, following manual screening, constituent images of a total of 1032 particles from selected class averages were segregated into independent data sets for 3D analysis. The 3D comparative analysis confirmed the observed ring-shaped similarity between the human and the trypanosome structure from 2D analysis and, in fact, revealed a surprisingly high level of congruence (Figure 6D; see movie S7 in the Supplementary Material).
Figure 6.
Single particle EM structure of T. brucei TFIIH. (A) Negative staining of T. brucei TFIIH. White arrowheads indicate individual molecules that were processed. (B) Representative averaged images of ring-like shaped molecules with different orientations. These class averages contain 20–40 images each. (C) Examples of projection images produced from the human TFIIH model (35). Asterisk-marked averages show corresponding views for the projection images of the same row (B and C). (D) Sets of surface views rotated 0, 90, 180 and 270° around vertical axis, taken from 3D reconstruction of T. brucei TFIIH (left panels; gray), superimposed views of the T. brucei TFIIH 3D on human TFIIH 3D (middle panels), and the 3D model of human TFIIH (right panels; magenta). Black arrowheads indicate the position of the bulge protruding from the ring of human TFIIH. Cross signs indicate additional features accommodated in 3D reconstruction of T. brucei TFIIH. The human TFIIH reconstruction (D) and corresponding projections (C) were reduced in size by 10% to match the dimensions of the T. brucei ring structure.
On the other hand, the trypanosome structure exhibited distinct characteristics. First, a major difference was found in the bulge. As indicated by measuring the length of the vertical axis from 2D averages, most of the bulge of protein densities was missing in the 3D reconstruction of T. brucei TFIIH (Figure 6D, black arrowheads). Since in the human structure, the bulge is formed by the CAK complex, this finding is in accordance with the absence of this complex in T. brucei TFIIH. Second, there are additional protein densities on both sides of the hole which most likely stem from the additional subunits TSP1 and 2. Most of this extra density is covering the hole on one side of the complex giving it the egg-like shape. It appears that a protrusion reaches through the hole and emerges on the other side of the complex (Figure 6D, cross marks). In summary, the structural data corroborated the presence of a full complement of TFIIH core subunits and the lack of a CAK subcomplex, and they provide a platform for the further analysis of the trypanosome-specific TFIIH subunits.
DISCUSSION
In this study, biochemical characterization of T. brucei TFIIH identified all seven core subunits including the orthologues of p62 (Tb11.01.1200) and TFB5 (Tb10.61.2600). Our identifications are in contrast to a previous bioinformatic analysis in which Tb11.02.2870 and Tb11.03.0815 were proposed to be the corresponding orthologues (21). However, the latter proposition is unlikely to be correct because (i) Tb11.01.1200 and Tb10.61.2600 were co-isolated in three independent TFIIH purifications (21, XPD and TSP2 purifications in this study) whereas Tb11.02.2870 and Tb11.03.0815 did not detectably co-purify in any of these experiments, (ii) Tb11.03.0815 appears to be a dynein light chain sharing 61% identity with human DYNLL1 (data not shown), and (iii) Tb11.02.2870 does not harbor the conserved FW domains characteristic of p62 orthologues, whereas they are present in Tb11.01.1200 and its trypanosomatid counterparts (Figure 2B).
Although we purified an active TFIIH complex which efficiently reconstituted SLRNA transcription in XPD-depleted (18), TSP2-depleted (Figure 5B) and TFIIH-depleted extracts (Figure 5C), the highly conserved subunits of the CAK complex were not detected. This finding was corroborated by the missing bulge in the T. brucei TFIIH EM structure and it is in accordance with the lack of the YSPTSPS motif in the CTD of RPB1 and with a comparative genomics analysis which had predicted that trypanosomatids lack a CDK7 orthologue (37). Although we cannot rule out the possibility that only a very minor, undetected fraction of the purified TFIIH was associated with a CAK complex, the observed transcription reconstitution efficiency of TFIIH-depleted extracts by PTP-purified TFIIH argues against this possibility. Thus, our findings suggest that TFIIH-mediated CTD phosphorylation is not important for transcription initiation of RNA pol II in trypanosomes. On the other hand, the T. brucei CTD is rich in serines and the corresponding RNA pol II subunit RPB1 was shown to be phosphorylated (38). While it is therefore likely that CTD phosphorylation is important for trypanosome RNA pol II transcription initiation, our results predict that the involved kinase(s) are recruited to the PIC in a TFIIH-independent manner.
What are TSP1 and TSP2? In T. brucei, these two proteins are bona fide subunits of TFIIH because the sedimentation analysis did not reveal a ‘complete’ TFIIH core without TSPs. Since eukaryotic TFIIH is recruited into the PIC by the bipartite TFIIE, the TSPs could be the orthologues of the two TFIIE subunits. While trypanosomatid TSP1 and TSP2 sequences could not be meaningfully aligned with TFIIEα, TFIIEβ or archaeal TFE sequences (data not shown), trypanosomatid TSP2s, like all known TFIIEα orthologues, harbor an invariant, internal C2C2 zinc-finger motif. Moreover, the demonstrated interaction of both TFIIE subunits with XPB (39) correlates with our finding that the TSPs become part of TFIIH in conjunction with XPB. Nevertheless, it will require a detailed analysis to determine whether TSP1 and TSP2 are orthologous to the two TFIIE subunits.
It has been shown in vivo and in vitro that mammalian TFIIH is essential for RNA pol I-mediated transcription of ribosomal gene units (40). Similarly, the in vivo analysis of S. cerevisiae temperature-sensitive TFIIH mutants indicated an important role of TFIIH in RNA pol I transcription (40). Trypanosomes harbor a multifunctional RNA pol I which not only transcribes rRNA genes but also the gene units encoding its major cell surface proteins procyclin and variant surface glycoprotein (VSG; 41). While depletion of XPD, TSP2 or TFIIH from extracts inhibited SLRNA transcription, it had no effect on transcription from the class I GPEET procyclin promoter indicating that in trypanosomes TFIIH is not required for RNA pol I transcription in vitro. However, TFIIH may have an important in vivo role in class I transcription because silencing of XPD expression and labeling of nascent RNA in nuclear run-on assays revealed strong perturbations of VSG and procyclin promoter activities in both insect and bloodstream form trypanosomes (21).
Finally, flagellated protists such as trypanosomatids and diplomonads have the most divergent protein sequences among eukaryotes and the apparent lack of GTFs has led to the hypothesis that these organisms, like archaea, have a simplified transcription machinery (6,8), possibly by branching off the main eukaryotic lineage before most of the eukaryote-specific PIC components emerged. Our findings that trypanosomes possess the full complement of core subunits of the eukaryote-specific GTF TFIIH and that the molecular structure of the complex is highly congruent to its human counterpart contradict this interpretation. Together with the recent TFIIA and TFIIB characterizations they strongly indicate that most of the eukaryotic GTF repertoire has evolved before flagellated protists branched off the evolutionary tree and they therefore support a new eukaryotic phylogeny in which flagellated protists appear to be highly evolved and specialized organisms (42). Since the majority of genes in both the Tritryps and G. lamblia remain annotated as hypothetical, it appears premature to classify their molecular machineries as simplified.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (AI059377 to A.G.); Korea Science and Engineering Foundation (C00093 to J.H.L.); American Heart Association postdoctoral fellowship (to H.S.J.). EM was carried out in the Core EM Facility of the UMASS Medical School which is funded by the “Diabetes and Endocrinology Research Center” (grant DK32520). Molecular graphics images were produced using the University of California San Francisco Chimera package whose development was supported by the National Institute of Health [grant P41 RR-01081]. Funding for open access charge: US National Institutes of Health (grant R01- AI059377).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Patrick Schultz (IGBMC, Strasbourg) for providing us with the electron density map of human TFIIH, Mary Ann Gawinowicz (Columbia University) for excellent mass spectrometry, Duncan Sousa (Boston University) for comments on image analysis, and Roger Craig (University of Massachusetts) and Tu N. Nguyen (University of Connecticut Health Center) for critically reading the article.
REFERENCES
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol. Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 4.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl Acad. Sci. USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Sci. USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 7.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The Genome of the Kinetoplastid Parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das A, Banday M, Bellofatto V. RNA polymerase transcription machinery in trypanosomes. Eukaryot. Cell. 2008;7:429–434. doi: 10.1128/EC.00297-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilinger G, Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Günzl A, Ullu E, Dörner M, Fragoso SP, Hoffmann KF, Milner JD, Morita Y, Nguu EK, Vanacova S, Wünsch S, et al. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 1997;85:67–76. doi: 10.1016/s0166-6851(96)02816-2. [DOI] [PubMed] [Google Scholar]
- 11.Hartree D, Bellofatto V. Essential components of the mini-exon gene promoter in the trypanosomatid Leptomonas seymouri. Mol. Biochem. Parasitol. 1995;71:27–39. doi: 10.1016/0166-6851(95)00034-x. [DOI] [PubMed] [Google Scholar]
- 12.Yu MC, Sturm NR, Saito RM, Roberts TG, Campbell DA. Single nucleotide resolution of promoter activity and protein binding for the Leishmania tarentolae spliced leader RNA gene. Mol. Biochem. Parasitol. 1998;94:265–281. doi: 10.1016/s0166-6851(98)00083-8. [DOI] [PubMed] [Google Scholar]
- 13.Luo H, Gilinger G, Mukherjee D, Bellofatto V. Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem. 1999;274:31947–31954. doi: 10.1074/jbc.274.45.31947. [DOI] [PubMed] [Google Scholar]
- 14.Schimanski B, Nguyen TN, Günzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol. Cell Biol. 2005;25:7314–7322. doi: 10.1128/MCB.25.16.7314-7322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimanski B, Brandenburg J, Nguyen TN, Caimano MJ, Günzl A. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2006;34:1676–1684. doi: 10.1093/nar/gkl090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palenchar JB, Liu W, Palenchar PM, Bellofatto V. A divergent transcription factor TFIIB in trypanosomes is required for RNA polymerase II-dependent spliced leader RNA transcription and cell viability. Eukaryot. Cell. 2006;5:293–300. doi: 10.1128/EC.5.2.293-300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Nguyen TN, Schimanski B, Günzl A. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot. Cell. 2007;6:641–649. doi: 10.1128/EC.00411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet. 2003;19:578–584. doi: 10.1016/j.tig.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat. Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 21.Lecordier L, Devaux S, Uzureau P, Dierick JF, Walgraffe D, Poelvoorde P, Pays E, Vanhamme L. Characterization of a TFIIH homologue from Trypanosoma brucei. Mol. Microbiol. 2007;64:1164–1181. doi: 10.1111/j.1365-2958.2007.05725.x. [DOI] [PubMed] [Google Scholar]
- 22.Schimanski B, Nguyen TN, Günzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA. 2000;6:1069–1076. doi: 10.1017/s1355838200000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Laufer G, Schaaf G, Bollgönn S, Günzl A. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell Biol. 1999;19:5466–5473. doi: 10.1128/mcb.19.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laufer G, Günzl A. In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 2001;113:55–65. doi: 10.1016/s0166-6851(00)00380-7. [DOI] [PubMed] [Google Scholar]
- 27.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly JM. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 29.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 32.Ruan JP, Arhin GK, Ullu E, Tschudi C. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol. Cell Biol. 2004;24:9610–9618. doi: 10.1128/MCB.24.21.9610-9618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lustig Y, Sheiner L, Vagima Y, Goldshmidt H, Das A, Bellofatto V, Michaeli S. Spliced-leader RNA silencing: a novel stress-induced mechanism in Trypanosoma brucei. EMBO Rep. 2007;8:408–413. doi: 10.1038/sj.embor.7400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brandenburg J, Schimanski B, Nogoceke E, Nguyen TN, Padovan JC, Chait BT, Cross GA, Günzl A. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007;26:4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TN, Schimanski B, Günzl A. Active RNA polymerase I of Trypanosoma brucei harbors a novel subunit essential for transcription. Mol. Cell Biol. 2007;27:6254–6263. doi: 10.1128/MCB.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 37.Guo Z, Stiller JW. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics. 2004;5:69. doi: 10.1186/1471-2164-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman AB, Agabian N. Trypanosoma brucei RNA polymerase II is phosphorylated in the absence of carboxyl-terminal domain heptapeptide repeats. J. Biol. Chem. 1994;269:4754–4760. [PubMed] [Google Scholar]
- 39.Maxon ME, Goodrich JA, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 40.Iben S, Tschochner H, Bier M, Hoogstraten D, Hozak P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 41.Günzl A, Bruderer T, Laufer G, Schimanski B, Tu L-C, Chung H-M, Lee MG-S. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dacks JB, Walker G, Field MC. Implications of the new eukaryotic systematics for parasitologists. Parasitol. Int. 2008;57:97–104. doi: 10.1016/j.parint.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.