Abstract
Techniques for targeted genetic disruption in Plasmodium, the causative agent of malaria, are currently intractable for those genes that are essential for blood stage development. The ability to use RNA interference (RNAi) to silence gene expression would provide a powerful means to gain valuable insight into the pathogenic blood stages but its functionality in Plasmodium remains controversial. Here we have used various RNA-based gene silencing approaches to test the utility of RNAi in malaria parasites and have undertaken an extensive comparative genomics search using profile hidden Markov models to clarify whether RNAi machinery exists in malaria. These investigative approaches revealed that Plasmodium lacks the enzymology required for RNAi-based ablation of gene expression and indeed no experimental evidence for RNAi was observed. In its absence, the most likely explanations for previously reported RNAi-mediated knockdown are either the general toxicity of introduced RNA (with global down-regulation of gene expression) or a specific antisense effect mechanistically distinct from RNAi, which will need systematic analysis if it is to be of use as a molecular genetic tool for malaria parasites.
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved mechanism found across a range of eukaryotes, where it plays a key role in post-transcriptional gene regulation and in the protection of genomes from intrinsic and extrinsic threats (1). With its exquisite specificity for the target gene as well as its potent and reversible action, RNAi technology has now become a standard technique in the molecular toolbox for reverse genetic experimentation in many systems, providing a quick and easy means to gain valuable insight into gene function, in particular those that are essential to cell viability. The process of RNAi is triggered by the recognition of double-stranded RNA (dsRNA), which is then processed into 21–25 nucleotide sequences by Dicer, a cytoplasmic dsRNA-specific RNaseIII endonuclease (2–5). The processed short interfering RNAs (siRNAs) that are generated associate with an RNA-induced silencing complex (RISC) and unwind in a strand-specific manner (3). The resulting siRNAs are then able to target homologous mRNA for degradation in combination with the RNaseH enzyme Argonaute (Slicer) (6). The first stage of dsRNA processing can be bypassed by introducing sequence-specific siRNAs directly into cells.
The blood stage malaria parasite is responsible for all of the debilitating clinical symptoms of the malaria disease and is therefore a central focus of research to find new therapeutic approaches (7). The malaria parasite is haploid for most of its life cycle, including the pathogenic asexual forms that proliferate in the blood. Current technologies for the genetic manipulation of Plasmodium are limited to this stage of parasite development, through the introduction of exogenous DNA containing drug-resistant markers and subsequent drug selection. It is self-evident, therefore, that a malaria gene encoding a product essential to blood stage asexual growth would be resistant to permanent genetic disruption. Although two methods have recently been developed that allow inducible expression in the most virulent human malaria parasite, Plasmodium falciparum (8,9), neither has successfully been utilized to create full inducible gene knockouts, most likely because of an inherent leakiness in both systems. Given the immense burden of malaria disease and the need for new drug targets, in particular those essential for development of the pathogenic blood stages, the ability to utilize a gene silencing approach such as RNAi would clearly be extremely beneficial.
To date there have been several reports describing the use of RNAi for gene silencing in the blood stages of Plasmodium. These have involved introducing either long dsRNAs by direct electroporation into erythrocytes infected with P. falciparum (10–12) or the addition of siRNAs into the culture medium (13–16). In addition, siRNAs have been injected into mice infected with the rodent malaria parasite P. berghei (17). While these studies suggest that RNAi may be functional in malaria parasites, targeting of specific genes in all of these cases resulted in parasite death or significant growth defects. Since these processes most probably lead to a global down-regulation of expression of many genes, it will be difficult to demonstrate the specific effect of the siRNA on expression of the target gene. For example, based on RNAi targeting of falcipain-1, a protease involved in the degradation pathway for hemoglobin, it has been concluded that this protein is essential for P. falciparum blood-stage growth (14). However, this finding is at odds with the demonstration that disruption of this gene by standard knockout technologies has no apparent effect on blood stage development (18). These observations question the specificity of gene targeting by siRNA's in Plasmodium. Indeed, unpublished observations [cited by Blackman (19)] have suggested that the introduction of long dsRNA or siRNAs directed against P. falciparum SUB-1 or AMA-1 did not lead to silencing of expression of the target genes. In addition, several studies have been unable to detect even distant homologues of either Dicer or Argonaute in the P. falciparum or P. berghei genomes (19–24) and two recent studies failed to isolate P. falciparum micro-RNAs (miRNAs), a class of endogenous regulatory short RNAs that also act through the RNAi pathway (25,26). However, the failure to detect in Plasmodium homologues of the genes involved in RNAi silencing found in other organisms might be due to the unusually high AT-content of Plasmodium genomes which has notoriously caused difficulties in identifying genes where there is poor conservation. Therefore, the question of the presence of a highly divergent RNAi machinery in Plasmodium remains open.
In an attempt to clarify whether RNAi is functional in Plasmodium, we have taken a variety of RNA-based strategies to target genes in P. falciparum and P. berghei that are non-essential to growth or development and analysed their expression. Additionally, we have undertaken an extensive comparative analysis of available Apicomplexan and other protozoan genomes using sensitive profile hidden Markov models of domains (rather than simply sequence homology or PSI/RPS-BLAST based searches) in an attempt to determine whether a primitive RNAi machinery exists in Apicomplexa. Together our data argue that RNAi is absent in malaria parasites and therefore it is unlikely that RNAi-based gene silencing will prove to be a reliable approach to unravel the function of malaria proteins.
MATERIALS AND METHODS
Generation of RNA interference constructs
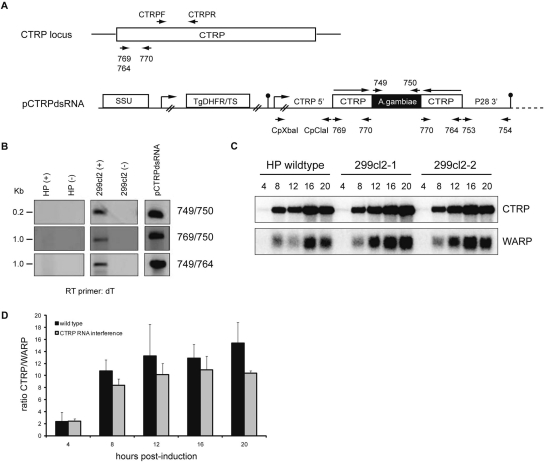
Two dsRNA hairpin structures were created—one based on KAHRP for testing RNAi in P. falciparum and the other based on CTRP for testing RNAi in P. berghei. For the KAHRP hairpin structure, 407 bp of the KAHRP gene (PFB0100c) was PCR-amplified from 3D7 genomic DNA using the oligonucleotides RNAiF and RNAiR or RNAiF(H/X) and RNAiR (Figure 1A, Supplementary Data S1 for oligonucleotide sequences). A luciferase stuffer region of 437 bp was amplified from pDluc⁁D (27) using the oligonucleotides lucF and lucR. The PCR products were cloned sequentially into pHH1 (28) to create plasmid pHH/KRNAi (Figure 1A). For the CTRP hairpin structure, a 2.7 kb promoter fragment upstream of the translation initiation codon of CTRP (PB000233.00.0) was amplified from genomic DNA of the P. berghei line cl5cy11 (HP) with oligonucleotides CpXbaI and CpClaI. For the CTRP inverted repeat, 680 bp of the CTRP ORF was PCR amplified using oligonucleotides 764 and 770 or 769 and 770. A 270 bp stuffer fragment separating the inverted CTRP fragments was amplified from Anopheles gambiae genomic DNA using the oligonucleotides 749 and 750. The P28 3′UTR was amplified using oligonucleotides 753 and 754. These fragments were all cloned sequentially to yield the final plasmid construct pCTRPdsRNA (Figure 2A).
Figure 1.
KAHRP dsRNA is transcribed in P. falciparum but does not silence endogenous KAHRP. (A) Schematic of the two exon structure of the endogenous KAHRP locus and the construct pHH/KRNAi, created to generate dsRNA for silencing endogenous KAHRP. Large arrows above pHH/KRNAi indicate direction of transcription and position of oligonucleotides used for PCR or RT–PCR are indicated by small arrowheads. The human DHFR (hDHFR) selectable marker cassette is under the transcriptional control of the CAM 5′ and HRP2 3′ untranslated regions. Promoter regions are symbolized by right-angled arrows and the 3′ untranslated regions necessary for gene expression by a circle. (B) Transgenic 3D7/KRNAi parasites transcribe dsRNA. Total RNA was isolated from asynchronized parental (3D7) or transgenic (3D7/KRNAi) parasites and cDNA was then generated using oligo d(T) or a luciferase specific primer (lucR). RT–PCRs were performed with the oligonucleotides either in the presence (+) or absence (–) of reverse transcriptase. As controls, genomic DNA (gDNA) isolated from 3D7 or pHH/KRNAi DNA was also used. (C) Northern blot analysis of RNA isolated from 3D7 parental or 3D7/KRNAi transgenic parasites across the asexual life cycle (Exp1) or during the stage of parasite development when KAHRP expression is at its peak (Exp2 and Exp3) using probes directed against luciferase (LUC), PF14_0344 or KAHRP confirms the KAHRP hairpin structure is transcribed yet is not capable of destabilizing KAHRP mRNA. The percentage of relative KAHRP expression was calculated by determining the level of KAHRP transcripts relative to that of PF14_0344 for both 3D7/KRNAi and 3D7, with the latter being normalized to 100%. ER, early ring-stages; R, rings; LR, late rings; ET, early trophozoites; LT, late trophozoites.
Figure 2.
CTRP dsRNA does not silence endogenous CTRP during P. berghei ookinete development. (A) Schematic representation of the endogenous CTRP locus and the construct pCTRPdsRNA used to generate dsRNA for silencing endogenous CTRP. Large arrows above pCTRPdsRNA indicate direction of transcription and position of oligonucleotides used for PCR or RT–PCR are indicated by small arrowheads. The T. gondii DHFR (TgDHFR/TS) selectable marker cassette is under the transcriptional control of the P. berghei DHFR 5′ and DHFR 3′ untranslated regions. Promoter regions are symbolized by right-angled arrows and the 3′ untranslated regions necessary for gene expression by a circle. (B) Transgenic P. berghei 299cl2 transcribe dsRNA. Total RNA was isolated from an in vitro ookinete culture of HP parental and transgenic 299cl2 lines 16-h post-induction and cDNA was then generated using oligo d(T). RT–PCRs were performed with the oligonucleotides shown, either in the presence (+) or absence (–) of reverse transcriptase. As a control pCTRP/dsRNA DNA was also used. (C) Comparison of steady state levels of CTRP and WARP RNA during ookinete development (4–20-h post-induction) by Northern blot analysis. Two independent 299cl2 RNA samples are shown. (D) Ratios of CTRP versus WARP RNA levels in parental HP (two samples) and 299cl2 (three samples). None of the differences are significant (unpaired t-test).
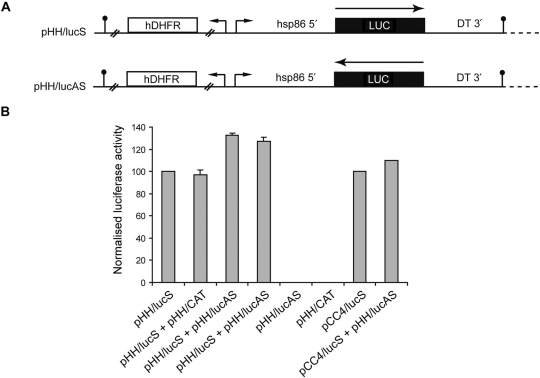
To assess whether antisense mRNA can silence gene expression in P. falciparum, the constructs pHH/lucS and pHH/lucAS were constructed (Figure 3A). The constructs pHH/lucS and pCC4/lucS are based on pHH1 or pCC4, respectively and harbour the luciferase gene in the sense orientation relative to the hsp86 5′ untranslated region, whereas pHH/lucAS harbours the luciferase gene in the antisense orientation. The vector pHH/CAT harbours the chloramphenicol acetyl transferase gene in place of the luciferase gene (29).
Figure 3.
Expression of antisense mRNA fails to knock-down transient luciferase activity. (A) Schematic of two constructs, pHH/lucS and pHH/lucAS, designed to generate sense or antisense luciferase mRNA in P. falciparum, respectively. The large arrow indicates the direction of transcription. (B) Luciferase activity observed in parasite extracts transiently transfected with various plasmid combinations. The level of firefly luciferase was normalized to the level of R. reniformis luciferase to control for variation in transfection efficiency across experiments. The firefly luciferase activity observed in parasites transfected with pHH/lucS and pCC4/lucS were set at 100%, with all other luciferase activities observed in other transfected parasites containing pHH/lucS or pCC4/lucS expressed relative to these, respectively. No antisense/sense mediated knockdown of luciferase activity can be seen, irrespective of vector source for luciferase expression.
P. falciparum culturing and transfection
Parasite cultures of P. falciparum 3D7 and D10 were maintained and synchronized as per standard procedures. Stable transfections with pHH/KRNAi and transient trasfections with various combinations of sense and antisense constructs are described in the Supplementary Data.
P. berghei transfection, parasite culture and cloning
Asexual stage P. berghei parasites (HP line) were transfected with pCTRPdsRNA according to Janse et al. (30). Parasites resistant to pyrimethamine selection were cloned by limiting dilution. Three parasite clones (299cl1, 2 and 3) were positive for integration into the C or D SSU (Supplementary Data); clone 2 was chosen for further experiments.
siRNA design and introduction into culture
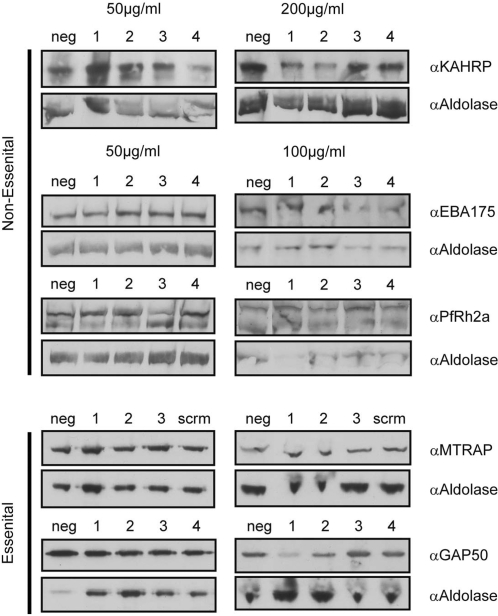
Four independent siRNAs (to avert possible non-functionality of any single RNAi) were designed for three genes that have previously been knocked out in 3D7: KAHRP (31), PfRh2a (32) and EBA175 (33) (see Supplementary Data S2 for sequences). The siRNAs were used at 50–200 µg/ml final concentration as described previously (15,16). 0.5 ml of culture at ring-stages following sorbitol treatment (∼8–12 h) was incubated with each siRNAs in a 24-well culture plate in serum free medium for 30 min with intermittent mixing. Subsequently, Albumax™ (Gibco BRL) was added at a final concentration of 10% and parasites were maintained for 24 h until late trophozoite or schizogony. For immunoblot analysis parasites were harvested at late schizogony (∼40 h post invasion when RfRH2a, EBA175, MTRAP and GAP50 are maximally expressed) or trophozoites (∼20–24 h when KAHRP is maximally expressed), lysed with saponin and resuspended in reducing buffer. Immunoblots were probed with specific antibodies against each target protein [KAHRP (31), PfRh2a (32) and EBA175 (33), MTRAP or GAP50 (34)]. A portion of each parasite culture was retained and allowed to reinvade new erythrocytes. Percentage invasion relative to untreated controls was measured as previously described (35).
RESULTS
Stable expression of a dsRNA hairpin does not effectively silence expression of P. falciparum and P. berghei genes
To determine whether transcription of a gene could be silenced using RNAi in P. falciparum, a double-stranded hairpin construct was made which was designed to specifically target the gene KAHRP (PFB0100c), in a manner similar to that described for Trypanosoma brucei, a protozan pathogen in which RNAi has been validated (36). KAHRP is known to be non-essential to the blood stages as knockout parasites are viable, although they do have a distinct phenotype (31). The construct pHH/KRNAi contains two opposing KAHRP fragments separated by a non-P. falciparum ‘stuffer’ region derived from a portion of the firefly luciferase gene with expression driven by the P. falciparum HSP86 promoter region. This construct was transfected into the 3D7 parasite line pre-selected for the presence of knobs and hence expression of KAHRP, with stable transfectants termed 3D7/KRNAi. Transcription of the dsRNA transgene in P. falciparum was investigated using both RT–PCR and Northern blot analysis (see Supplementary Data for methods). Endogenous KAHRP was successfully amplified using oligonucleotides KF2 and KF3 from both 3D7 and 3D7/KRNAi whilst no product could be detected with cDNA made from a KAHRP knockout line (37) confirming the specificity of the reaction (Figure 1B and data not shown). dsRNA-specific cDNA amplification was, however, only observed in parasites transfected with pHH/KRNAi, indicating that the hairpin structure is indeed transcribed in 3D7/KRNAi (Figure 1B). This was also confirmed by Northern blot analysis of RNA isolated from the two lines and probed with the LUC stuffer region (Figure 1C, top panel). As expected, bands corresponding to LUC transcript were detected in the transfected parasite line 3D7/KRNAi, but not in the wild-type 3D7 line.
Having established stable expression of KAHRP dsRNA, we next sought to determine whether transcription of the transgene resulted in modulation of the amount of KAHRP transcripts. Northern blots of RNA isolated from 3D7 and 3D7/KRNAi that were hybridized with a probe which recognizes only endogenous KAHRP and not the dsRNA transcript revealed that the presence of the hairpin structure did not affect the levels of KAHRP expression (Figure 1C). To safeguard against asynchrony between the two lines, parasites were harvested at different time points during their life cycle and the transcription levels of KAHRP relative to PF14_0344 (an unrelated gene chosen as a control) in the 3D7/KRNAi line were normalized to levels obtained for 3D7. Accounting for possible asynchrony, and despite the presence of highly abundant dsRNA transcripts present in the 3D7/KRNAi line (Figure 1C, Exp1), no gene silencing effect was observed. Indeed, by the latter time points there was actually more KAHRP expression relative to PF14_0344 in the 3D7/KRNAi line than in the 3D7 wild-type line with overall cumulative transcriptional expression for KAHRP being 86.1% in transfectant versus wild-type. Two repeat experiments (Exp2 and Exp3), this time at the late ring and early trophozoite stages when KAHRP expression is at its peak showed similar results. Together this data strongly suggests that the KAHRP dsRNA was unable to efficiently destabilize the amount of steady state endogenous KAHRP mRNA at least to any levels that could be used to analyse gene function.
Similar results were obtained in our attempts with the rodent malaria parasite P. berghei. A plasmid construct containing a 680 bp inverted CTRP repeat separated by a stuffer fragment from A. gambiae was stably introduced into the C/D-SSU unit on chromosomes 5 and 6 of P. berghei (Figure 2A). The CTRP gene (PB000233.00.0) encodes a surface receptor essential to insect stage parasite development (motility), but is not expressed in blood stage parasites and should therefore be amenable to knockdown or knockout (38). Here, transcription of the dsRNA was placed under the control of the CTRP promoter, ensuring that its expression was transcribed at the same time as the target mRNA. Using three different oligonucleotide combinations it was confirmed that the dsRNA was transcribed in transgenic 299cl2 insect stage ookinetes (Figure 2B, see Supplementary Data for methods). The ability of this dsRNA to specifically destabilize endogenous CTRP mRNA in ookinetes was then investigated by Northern analysis of parental P. berghei HP and transgenic 299cl2 parasites at 4, 8, 12, 16 and 20 h post-induction (hpi) of sexual development using a CTRP probe that recognizes only endogenous mRNA and not the dsRNA (Figure 2C, see Supplementary Data for methods). As expected, transcription of CTRP in all cultures increased steadily and peaked at 16 hpi during ookinete development. When the ratio of expression was calculated relative to WARP (PB000020.03.0), an ookinete specific gene that has a similar temporal expression patterns to CTRP, it was revealed that any down-regulation of CTRP was not significant (Figure 2C and D). Furthermore, at 12 and 13 min post-induction exflagelation centres were counted and the ookinete conversion rates calculated (data not shown). No differences in development were observed between the parental and transgenic 229cl2 parasite lines indicating that there were no significant off-target effects as a result of the transgene expression. This adds further support to our data from P. falciparum that dsRNA is not effective at silencing gene expression in malaria parasites.
Generation of antisense mRNA does not lead to gene silencing in P. falciparum
In the absence of dsRNA-mediated silencing we next sought to determine whether silencing could be achieved using antisense mRNA. P. falciparum parasites were transiently co-transfected with a range of constructs that were designed to generate sense and/or antisense firefly luciferase (F-luc) mRNA (Figure 3A). These parasites were co-transfected with a construct containing the Renilla luciferase (R-luc) gene in order to normalize for the variation in transfection efficiency across different transient transfection experiments by measuring R-luc activity. As can be observed from Figure 3B, F-luc activity could be detected in parasites transfected with either F-luc mRNA sense constructs, pHH/lucS or pCC4/lucS alone, and in parasites co-transfected with both pHH/lucS and pHH/CAT. Furthermore, as expected, no F-luc activity could be detected in parasites transfected with either the antisense construct pHH/lucAS or with pHH/CAT alone. However, the co-transfection of antisense and sense F-luc mRNA constructs together (pHH/lucAS with either pHH/lucS or pCC4/lucS) did not lead to reduction in F-luc activity in two independent transfections (Figure 2B) or in a repeat experiment (data not shown). The inability of antisense mRNA from F-luc to silence luciferase expression from sense mRNA strongly suggests that antisense mRNA is not a general mechanism of gene silencing in malaria parasites.
Introduction of exogenous siRNAs does not lead to gene silencing in P. falciparum
In the absence of evidence that full length RNA constructs can mediate effective gene silencing in P. falciparum, we sought to determine whether we could bypass the need for RNA processing and use exogenously introduced siRNAs to disrupt gene expression. 3D7 parasites were incubated with independent siRNAs (to detect possible experimental variation among siRNAs) at at least two different concentrations (50, 100 or 200 µg/ml). The target genes included the non-essential genes KAHRP, PfRh2b (MAL13P1.176) and EBA-175 (PF07_0128) as well as two putatively essential genes, MTRAP (PF10_0281) and GAP50 (PFI0880c) (34). As a negative control a scrambled siRNA with no homology to the P. falciparum 3D7 genome was used. These experiments were performed following protocols used in those studies that have reported successful siRNA-mediated gene silencing in Plasmodium (13–16). After co-culture of blood stage parasites with the siRNA, parasites were harvested and the targeted protein levels investigated by immunoblot. Although the protein levels of the targeted genes did vary relative to the levels of the PfAldolase (PF14_0425) protein [used as a loading control (34)], no consistent or siRNA-concentration dependent reduction in protein expression was observed for any protein targeted by the siRNA (Figure 4). Furthermore, no reduction in growth rate of the blood stages was seen following the siRNA treatment, with re-invasion levels remaining consistent across all treatments (data not shown). These results strongly suggest that delivery of specific siRNAs to blood stages through the culture medium did not lead to specific gene silencing in P. falciparum.
Figure 4.
Exogenous introduction of siRNAs fails to knock-down endogenous protein levels. Immunoblots for five genes targeted by siRNAs specific for four (MTRAP 3 only and scrambled negative control) distinct regions of genes known to be tractable (KAHRP, EBA175 and PfRh2a) or intractable (MTRAP and GAP50) to conventional knockout. Despite incubation at concentrations up to 100–200 µg/ml no consistent reduction in protein level can be seen for any gene.
Comparative genomics suggests malaria parasites lack the core RNAi machinery components Argonaute and Dicer
Previous studies have noted that the RNAi machinery is absent in P. falciparum (19–24), an observation that fits with our inability to find experimental evidence for RNAi in both P. falciparum or P. berghei. To address the possibility that a primitive RNAi machinery might be present, that is highly divergent from—but structurally related to—that of other eukaryotes, we performed a sensitive comparative search of protein sequences from 18 eukaryotic species, including two Plasmodium species and three other apicomplexan and four kinetoplastid protozoan parasites, for key domains involved in RNAi using profile hidden Markov models (HMMs) (see Supplementary Data for methods). Also included in the cohort of analysed genomes were those of species that are known to either perform RNAi (positive controls) or not (negative controls). Domain profile HMMs used included RNase3, double-stranded RNA binding motifs (dsrm) and double-stranded RNA-binding domains (dsRBD) for Dicer orthologues and PAZ and Piwi domains for Argonaute orthologues (2). This method, which has been successfully used to detect Plasmodium proteins that have low sequence similarity to non-malaria proteins (39), greatly improves the chances of finding divergent homologues of Argonaute and other RNAi-related proteins. To further increase the chances of finding such homologues in P. falciparum, a high E-value threshold was used in addition to post hoc analyses of the over-predicted sequences looking for domain co-occurrence and by performing reciprocal domain searches and phylogenetic analysis. Unlike simple homology searches, including the use of PSI-BLAST, this comparative approach provides stronger support for protein absence where domains and domain co-occurrences are not present across entire phylogenetic clades. Furthermore, the inclusion of positive and negative controls allows us to monitor sensitivity and specificity. The results of all domain predictions across the 18 species are shown in Supplementary Data S3. The results of PAZ and Piwi domain profile searches, using both a high E-value threshold (10) and moderately conservative threshold (0.001), are summarized in Table 1, which shows the number of proteins found that contain predicted Piwi or PAZ domains for each species as well as co-occurring predicted Piwi and PAZ domains (E-value ≤10). While sensitive HMM searches with a high E-value threshold predict six putative Piwi and 46 putative PAZ domains in P. falciparum, more conservative searches found none (Table 1). Furthermore, looking for proteins containing both a PAZ and a Piwi domain using a high E-value cut-off (10), we again found none in P. falciparum. This was recapitulated with searches using the sister species P. yoelii. The absence of proteins with a PAZ or Piwi domain as defined by the criteria used in this study supports the idea that Plasmodium does not contain an Argonaute homologue, not even a highly divergent one. As a control, the same search approach that we had applied to Plasmodium confirmed the absence of a true Argonaute in the yeast Saccharomyces cerevisiae, whilst demonstrating its presence in Schizosaccharomyces pombe; results that match the known utility of RNAi in these organisms (24). Similarly, T. brucei is known to be RNAi positive (20), whilst sister species T. cruzi and the related trypanosomatid L. major are unable to perform RNAi and our search strategy supported these observations, detecting Argonaute homologies only in T. brucei (Table 1) (40,41). Of note, we identified in the apicomplexan Theileria annulata a hypothetical protein, TA13085, which contains weak matches to both PAZ and Piwi domains suggesting it may have an active Argonaute. However, the position of TA13085 amongst low scoring apicomplexan domain predictions on the PAZ phylogenetic tree (Supplementary Data S4) may indicate that it is a false positive and its putative function as an Argonaute requires experimental verification. Consistent with the absence of putative Argonaute homologs in Plasmodium, no Dicer/Drosha-like proteins could be detected in Plasmodium when searching using domain profiles consisting of two RNase3 domains or an RNase3 domain and a double stranded RNA binding domain (either dsrm or dsRNA_bind), either individually (using a low E-value threshold) or in combination (using a high cut-off) (Supplementary Data S5). Searches in other species were consistent with their known ability to undertake RNAi.
Table 1.
Number of putative proteins containing predicted Piwi or PAZ domains or both domains across diverse eukaryotic species
| Species |
E-valuesa |
||||
|---|---|---|---|---|---|
| Piwi |
PAZ |
PAZ + Piwib |
|||
| 10 | 10−3 | 10 | 10−3 | 10 | |
| Homo sapiens | 16 | 11 | 15 | 12 | 10 |
| Drosophila melanogaster | 12 | 11 | 16 | 12 | 11 |
| Caenorhabditis elegans | 39 | 35 | 39 | 33 | 31 |
| S. cerevisiae | 10 | 0 | 17 | 0 | 0 |
| S. pombe | 5 | 1 | 12 | 1 | 1 |
| Trichomonas vaginalis | 3 | 2 | 5 | 0 | 2 |
| Giardia intestinalis | 10 | 1 | 9 | 0 | 1 |
| Dictyostelium discoideum | 17 | 5 | 52 | 5 | 6 |
| Enthamoeba histolyca | 12 | 3 | 25 | 2 | 3 |
| T. brucei | 3 | 2 | 6 | 0 | 1 |
| T. cruzi | 2 | 1 | 4 | 0 | 0 |
| Leishmania major | 4 | 1 | 5 | 0 | 0 |
| Leishmania braziliensis | 6 | 1 | 2 | 1 | 1 |
| Toxoplasma gondii | 4 | 1 | 4 | 1c | 1c |
| Cryptosporidium parvum | 24 | 0 | 38 | 0 | 0 |
| T. annulata | 18 | 0 | 23 | 0 | 1 |
| P. falciparum | 6 | 0 | 46 | 0 | 0 |
| P. yoelli | 12 | 0 | 38 | 0 | 0 |
aDomains were predicted using profile HMMs with sensitive (10) and conservative (10−3) E-value thresholds.
bIndicates the number of proteins containing both a PAZ and a Piwi domain.
cThe PAZ domain of the TgAGO homolog is upstream of the annotated ATG start codon.
As a final stringent test to rule out the presence of low homology domains, a phylogenetic analysis of PAZ and Piwi domains was undertaken (Supplementary Data S4 and S6). Phylogenetic trees of all predictions (E-value <2) show that proteins containing high scoring Piwi and PAZ domains cluster into groups. Low-scoring domains are however interspersed throughout the tree and have consistently longer branch lengths. This suggests that low scoring hits are either highly divergent homologues or unrelated proteins. The latter conclusion is supported by the fact that the low scoring Piwi domains in these proteins typically do not co-occur with a PAZ domain, arguing that these proteins are not true Argonaute-like proteins. Indeed, as shown in Table 2, the best scoring domain that overlaps each low-scoring Piwi domain in P. falciparum (analogous to the Reciprocal Best Hits), has a highly significant match to an alternative domain which is a much better predictor of the protein's function. Taken together, the failure to detect proteins containing Argonaute- and Dicer-like domains using this sensitive comparative genomics approach strongly suggests that Argonaute-like and other RNAi-related proteins are absent in Plasmodium, supporting earlier studies.
Table 2.
Reciprocal profile HMM search of sequences containing best scoring Piwi domains found in P. falciparum
| Accession | Piwi domain |
Best overlapping domain |
|||||
|---|---|---|---|---|---|---|---|
| Location | Score | E-valuea | Domain name | Location | Score | E-valueb | |
| PF11_0352 | 12–236 | −140.9 | 1.4 | Thioredoxin | 165–275 | 81.6 | 2.6 × 10−21 |
| PFL1370w | 29–258 | −140.9 | 1.4 | Pkinase | 15–298 | 246.8 | 4.7 × 10−71 |
| PF11_0059 | 14–252 | −146.9 | 3.5 | MFS_1 | 17–374 | 5.5 | 0.012 |
| PFI1125c | 10–255 | −149.1 | 4.9 | adh_short | 61–228 | 142.8 | 9.9 × 10−40 |
| PF10_0245 | 456–672 | −151.4 | 6.9 | SIS | 507–636 | 119.1 | 5.1 × 10−15 |
| PFL1545c | 301–555 | −153.7 | 9.7 | Cpn60_TCP1 | 89–615 | 559.5 | 3.4 × 10−165 |
aE-values for Piwi were obtained using hmmsearch.
bE-values of the best overlapping domains were obtained using hmmpfam.
DISCUSSION
Although several studies have reported the use of RNA interference mechanisms for silencing gene expression in malaria parasites, there is still little consensus about whether the genus Plasmodium has an active RNAi pathway. Importantly, the concept has not been supported by a clear demonstration that the Plasmodium genome contains any of the conserved RNAi machinery (e.g. Argonaute, Dicer). Here we combined a novel bioinformatics search of the Plasmodium genome with a variety of biological investigations to establish if RNAi is a valid investigational approach for protein function in Plasmodium with a sound theoretical basis. In previous studies that reported successful targeting of genes with RNAi, the effect of gene silencing was determined by measuring only the general effects on growth and/or metabolism of the treated parasites. In our studies we attempted to measure directly the effect of RNAi on expression of target genes with a clear predicted outcome if successful; non-essential genes (outcome: does-dependent reductions in transcript and protein levels and impaired functionality), essential genes (outcome: dose-dependent parasite death) and transgene reporters (outcome: dose-dependent reduction in luminescence). Stable and abundant expression of dsRNA hairpins targeting transcripts in blood stages of P. falciparum and in ookinetes of P. berghei produced no significant effect on steady state levels of their target mRNA species, precluding this approach for the analysis of the function of these proteins. In order to compensate for the absence of an obvious functional orthologue of Dicer (see below) exogenously produced siRNAs were introduced into the P. falciparum blood stages using a similar approach to previous reports on siRNA silencing of Plasmodium genes (13–16). Again, by measuring the effect on expression of five different proteins, some of which are associated with merozoite invasion of erythrocytes, we did not find consistent and significant reduction in protein expression for any of the transcripts targeted and no reduction in merozoite invasion where relevant. While we have no proof of the efficiency of uptake of siRNAs by P. falciparum, the methodologies used to treat the parasites with siRNA were based on protocols that have previously been reported to give rise to successful siRNA-mediated gene silencing in this and other species. It is possible that other means of transfection such as electroporation of parasites may be required to efficiently introduce siRNAs into the cytoplasm of P. falciparum. However, in P. berghei we used the highly efficient Amaxa method of electroporation (30) to introduce siRNAs that target reporter gene transcripts [green fluorescent protein (GFP) and luciferase] that are expressed from integrated transgenes. Using a variety of electroporation conditions, we were unable to detect significant reduction in the expression of GFP and luciferase in transfected blood stages in multiple independent experiments (data not shown). Although we again have no proof of the efficiency of siRNA delivery into P. berghei by these electroporation conditions, these studies provide no indications that gene silencing by siRNA delivery could be a worthwhile tool to dissect protein function in Plasmodium. Lastly, antisense is an approach that has been proven to be successful in reducing mRNA levels in several other protozoan species (42–47), however, transient transfection of P. falciparum blood stages with plasmids that generated antisense transcripts targeting a luciferase reporter gene did not reduce the expression of luciferase.
Taken together, our studies do not support the observations of previous studies in which RNAi has been used to silence genes. In one study, smaller molecular weight RNA species were detected after introducing exogenous dsRNA in the blood stages of P. falciparum and the presence of these small RNAs interpreted as evidence of digestion of the mature full length mRNA target by Dicer (14). Our inability to achieve gene silencing with stably produced dsRNA at different life cycle stages of the parasite and the absence of proteins with Dicer domains in Plasmodium (see below) might suggest that the small RNAs simply reflect degradation of the introduced dsRNA. In previous studies of siRNA-gene silencing (13–16), a general defect in growth and/or metabolism of the parasites has been reported. These defects might also be explained not as the result of specific gene targeting but as a result of non-specific inhibitory effects. For example, the very high concentrations of siRNAs used in these studies (over 100 µg/ml) may cause non-specific growth inhibition of P. falciparum and consequently a global down-regulation of gene expression. One suggestion is that observed RNA-mediated silencing effects in these studies could have been due to an antisense phenomenon (48), whereby the antisense transcripts annealed to the mRNA, preventing processes such as translation by ribosomes. Indeed, antisense transcripts have been extensively detected in the transcriptomes of P. falciparum although their functional significance remains obscure (49–51). In our hands, however, we found no evidence for antisense mediated silencing of expression when we targeted the expression of the reporter protein luciferase with antisense mRNA.
The lack of experimental evidence in our studies for the presence of an active RNAi machinery in Plasmodium is given credence by the current and previous failures to bioinformatically identify Dicer and Argonaute orthologues in P. falciparum and P. yoelli. PAZ and Piwi domains have been found in all functional Argonaute proteins identified to date (2), and while low scoring hits to these domains were observed in several P. falciparum proteins, the two domains were never found to co-occur within a single protein. Furthermore, all the putative Piwi domain sequences that were identified had a more significant match to an alternative domain in a reciprocal search. Importantly, our bioinformatics analysis approach when applied to other organisms that employ RNAi were in keeping with those of previous studies (20,24). In general the lack of Dicer and Argonaute homologies holds true for all apicomplexan genomes examined. Besides the Theileria protein that was noted, there is one further anomaly: the T. gondii genome harbours a protein with a strong match to Argonaute, although the PAZ domain in T. gondii lies upstream of the putative start codon (52). This is interesting from an evolutionary perspective as it may present evidence for the loss of the RNAi machinery in protozoans.
In other organisms RNAi plays a key role in biological processes such as the control of endogenous gene expression, the silencing of transposable elements and repetitive genes or as a defence against viral attack (53). The Plasmodium genome does not apparently possess retrotransposons or viral pathogens, and thus there may be no selective advantage to the malaria parasite in either retaining (or gaining) an RNAi machinery (54). Furthermore, native miRNAs, which are produced by the cleavage of endogenous dsRNAs by Dicer to regulate gene expression by inhibiting protein translation have not been identified in P. falciparum infected red blood cells nor in purified parasites (25,26).
In conclusion, our studies did not reveal any functional or bioinformatics evidence for RNAi activity in Plasmodium, indicating the absence of the RNAi machinery and the inadvisability of the application of RNAi to investigate gene function in Plasmodium. Although our findings also indicate that gene regulation via antisense inhibiting protein translation is non-functional in P. falciparum, the abundant native antisense transcripts found in this organism leave open the question that antisense regulation of gene activity may be gene-specific (55) and thus further exploration of this technology in malaria parasites may be warranted.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council (NHMRC) of Australia; BIOMALPAR Network of Excellence Grant and The Functional Genomics Initiative of the Wellcome Trust; Career Development Award from the NHMRC (to J.B.). A.F.C. is an International Research Scholar of the Howard Hughes Medical Institute. Funding for open access charge: National Health and Medical Research Council Project Grant 516740.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Australian Red Cross Blood Bank for the provision of human blood and serum and Fiona Angrisano for technical assistance.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM. Dicing and splicing. The core machinery of the RNA interference pathway. Febs Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 3.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Ketting RF. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Gene Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl Acad. Sci. USA. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 8.Meissner M, Krejany E, Gilson P, de Koning-Ward T, Soldati D, Crabb B. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood-stages using Toxoplasma gondii transactivators. Proc. Natl Acad. Sci. USA. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong C, Goldberg D. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat. Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 10.McRobert L, McConkey GA. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 2002;119:273–278. doi: 10.1016/s0166-6851(01)00429-7. [DOI] [PubMed] [Google Scholar]
- 11.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 2005;34:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 12.Tuteja R, Pradhan A, Sharma S. Plasmodium falciparum signal peptidase is regulated by phosphorylation and required for intra-erythrocytic growth. Mol. Biochem. Parasitol. 2008;157:137–147. doi: 10.1016/j.molbiopara.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Kumar R, Adams B, Oldenburg A, Musiyenko A, Barik S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: demonstration of its essential role using RNA interference. Malar. J. 2002;145:1245–1254. doi: 10.1186/1475-2875-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malhotra P, Dasaradhi PVN, Chauhan V. Double-stranded RNA mediated gene silencing of the cysteine proteases falcipain 1 and 2 in Plasmodium falciparum. Mol. Micro. 2002;45:1245–1254. doi: 10.1046/j.1365-2958.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- 15.Dasaradhi PV, Mohmmed A, Kumar A, Hossain MJ, Bhatnagar RK, Chauhan VS, Malhotra P. A role of falcipain-2, principal cysteine proteases of Plasmodium falciparum in merozoite egression. Biochem. Biophys. Res. Commun. 2005;336:1062–1068. doi: 10.1016/j.bbrc.2005.08.213. [DOI] [PubMed] [Google Scholar]
- 16.Sunil S, Hossain MJ, Ramasamy G, Malhotra P. Transient silencing of Plasmodium falciparum Tudor Staphylococcal Nuclease suggests an essential role for the protein. Biochem. Biophys. Res. Commun. 2008;372:373–378. doi: 10.1016/j.bbrc.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 17.Mohmmed A, Dasaradhi PV, Bhatnagar RK, Chauhan VS, Malhotra P. In vivo gene silencing in Plasmodium berghei – a mouse malaria model. Biochem. Biophys. Res. Commun. 2003;309:506–511. doi: 10.1016/j.bbrc.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 18.Sijwali PS, Rosenthal PJ. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 2004;101:4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackman MJ. RNAi in protozoan parasites: what hope for the Apicomplexa? Protist. 2003;154:177–180. doi: 10.1078/143446103322166482. [DOI] [PubMed] [Google Scholar]
- 20.Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 21.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Nelson KE, Bowman S, Paulsen IT, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall M, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 24.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. Febs Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 26.Xue X, Zhang Q, Huang Y, Feng L, Pan W. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar. J. 2008;10:47. doi: 10.1186/1475-2875-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Koning-Ward TF, Speranca MA, Waters AP, Janse CJ. Analysis of stage specificity of promoters in Plasmodium berghei using luciferase as a reporter. Mol. Biochem. Parasitol. 1999;100:141–146. doi: 10.1016/s0166-6851(99)00042-0. [DOI] [PubMed] [Google Scholar]
- 28.Reed MB, Caruana SR, Batchelor AH, Thompson JK, Crabb BS, Cowman AF. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc. Natl Acad. Sci. USA. 2000;97:7509–7514. doi: 10.1073/pnas.97.13.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- 30.Janse C, Franke-Fayard B, Waters AP. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 2006;1:614–623. doi: 10.1038/nprot.2006.53. [DOI] [PubMed] [Google Scholar]
- 31.Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 32.Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22:1047–1057. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc. Natl Acad. Sci. USA. 2003;100:4796–4801. doi: 10.1073/pnas.0730883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger TW, Green JL, Holder AA, Cowman AF. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem. 2006;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- 35.Baum J, Maier AG, Good RT, Simpson KM, Cowman AF. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Pathog. 2005;1:e37. doi: 10.1371/journal.ppat.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngô H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted gene disruption shows that knobs enable malaria-infected cells to cytoadhere under physiological shear stress. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 38.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE. CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 1999;18:6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat. Rev. Microbiol. 2006;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- 40.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 41.DaRocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Chen DQ, Kolli BK, Yadava N, Lu HG, Gilman-Sachs A, Peterson DA, Chang KP. Episomal expression of specific sense and antisense mRNAs in Leishmania amazonensis: modulation of gp63 level in promastigotes and their infection of macrophages in vitro. Infect. Immun. 2000;68:80–86. doi: 10.1128/iai.68.1.80-86.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drozdz M, Quijada L, Clayton CE. RNA interference in trypanosomes transfected with sense and antisense plasmids. Mol. Biochem. Parasitol. 2002;121:149–152. doi: 10.1016/s0166-6851(02)00018-x. [DOI] [PubMed] [Google Scholar]
- 44.Somanna A, Mundodi V, Gedamu L. Functional analysis of cathepsin B-like cysteine proteases from Leishmania donovani complex. Evidence for the activation of latent transforming growth factor beta. J. Biol. Chem. 2002;277:25305–25312. doi: 10.1074/jbc.M203034200. [DOI] [PubMed] [Google Scholar]
- 45.Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl Acad. Sci. USA. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touz MC, Conrad JT, Nash TE. A novel palmitoyl acyl transferase controls surface protein palmitoylation and cytotoxicity in Giardia lamblia. Mol. Micro. 2005;58:999–1011. doi: 10.1111/j.1365-2958.2005.04891.x. [DOI] [PubMed] [Google Scholar]
- 47.Mundodi V, Kucknoor AS, Alderete JF. Antisense RNA decreases AP33 gene expression and cytoadherence by T. vaginalis. BMC. Microbiol. 2007;7:64. doi: 10.1186/1471-2180-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Militello KT, Refour P, Comeaux CA, Duraisingh MT. Antisense RNA and RNAi in protozoan parasites: Working hard or hardly working? Mol. Biochem. Parasitol. 2008;157:117–126. doi: 10.1016/j.molbiopara.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Militello KT, Patel V, Chessler AD, Fisher JK, Kasper JM, Gunasekera A, Wirth DF. RNA polymerase II synthesizes antisense RNA in Plasmodium falciparum. RNA. 2005;11:365–370. doi: 10.1261/rna.7940705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 2004;136:35–42. doi: 10.1016/j.molbiopara.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell. 2001;12:3114–3125. doi: 10.1091/mbc.12.10.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al Riyahi A, Al-Anouti F, Al-Rayes M, Ananvoranich S. Single argonaute protein from Toxoplasma gondii is involved in the double-stranded RNA induced gene silencing. Int. J. Parasitol. 2006;36:1003–1014. doi: 10.1016/j.ijpara.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 54.Wickstead B, Ersfeld K, Gull K. Repetitive elements in genomes of parasitic protozoa. Microbiol. Mol. Biol. Rev. 2003;67:360–375. doi: 10.1128/MMBR.67.3.360-375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardiner DL, Holt DC, Thomas EA, Kemp DJ, Trenholme KR. Inhibition of Plasmodium falciparum clag9 gene function by antisense RNA. Mol. Biochem. Parasitol. 2000;110:33–41. doi: 10.1016/s0166-6851(00)00254-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.