Abstract
Integration of foreign DNA into eukaryotic genomes results frequently in a total or partial loss of gene function, caused by the interruption of indispensable structures of the gene itself. Using T-DNA insertions in Arabidopsis we screened for mutants with deregulated chlorophyll precursor accumulation in etiolated seedlings. A mutant designated rfd1 (red fluorescent in darkness) with increased protochlorophyllide accumulation showed a fluorescent phenotype that was associated with a lack of transcript initiation from the AtRibA1 promoter situated downstream of the integrated T-DNA. Complementation experiments confirmed rfd1 to be a knockout phenotype. Comparison with two SALK insertion lines bearing T-DNA integrations in the 5′UTR of AtRibA1 demonstrated that the insertion event in rfd1 itself does not explain the complete lack of transcript initiation. A 35S tetrameric enhancer sequence present on the rfd1 T-DNA causes the overaccumulation of a large polycistronic transcript originating inside the T-DNA. This 5.5-kb RNA runs over the downstream situated AtRibA1 promoter, which was shown by 5′RACE analyses to be consequently silenced. Hence, a transcription process that starts upstream and overlaps AtRibA1 blocks the initiation at the AtRibA1 promoter in rfd1. This regulatory mechanism has recently been introduced in yeast as transcriptional interference and is described here for the first time in a plant system.
INTRODUCTION
T-DNA insertions have become a common tool in plant mutant analysis and proved extremely useful to unravel functions by forward as well as by reverse genetic approaches. The interruption of gene function by the insertion of T-DNA results in the generation of knockout mutants and enables screens of mutant collections for the identification of unknown genes contributing to specific cellular functions as well as the analysis of the function of known genes (1). Functional knockout or knockdown of gene products are expected if the T-DNA is inserted into the coding region of a gene. However, T-DNA insertions hit introns or up- and downstream regions of a gene of interest as well as intergenic DNA stretches (2). Such insertion events often fail in generating phenotypical changes. Different types of T-DNA constructs have been used to expand the possibilities of T-DNA transformation as a tool in plant biology. Activation tagging and entrapment tagging are the most prominent strategies to explore the genomic neighborhood of the integrated DNA (3,4).
Recent work in yeast (5) demonstrated that transcription of the non-coding upstream region is able to silence the Saccharomyces cerevisiae SER3 gene by preventing the binding of transcriptional activators necessary for recruiting RNA polymerase II. This mechanism was designated ‘transcriptional interference’ to indicate that it is the process of transcription itself that interferes with the initiation of mRNA synthesis from downstream promoters.
Using a forward genetic screen to identify mutants in the regulation of tetrapyrrole biosynthesis, we identified a T-DNA mutant line with a specific recessive phenotype. An enhanced promoter located inside the T-DNA region caused a strong expression extending into the genomic neighborhood of the integration site. This apparently resulted in a drastic perturbation of transcription initiation from the downstream-situated Arabidopsis gene. Our findings suggest transcriptional interference as the mechanism responsible for gene silencing.
MATERIALS AND METHODS
Plant material and growth conditions
An Arabidopsis mutant collection of 30 000 independent lines transformed with plasmid pWA5 (kindly provided by T. Altmann, Gatersleben) was used in the initial screen. Described SALK mutants (2) were obtained from the NASC (Nottingham). Seeds were surface-sterilized by incubation in Meliseptol (Braun, Melsungen) for 3 min. and subsequently rinsed four times with distilled water. Seeds were sown on agar plates containing 0.5× Murashige and Skoog (MS) salts including vitamins (Duchefa, Haarlem) and 0.05% MES, pH 5.7. Plates for growth under etiolated conditions were incubated for 3 h in the light and subsequently wrapped in ligth-tight plastic foil. Following incubation for 2 days at 4°C, the plates were transferred for 5 days to room temperature. For analyses of light-grown rfd1 mutants, surface-sterilized seeds were sown on MS plates containing 2% sucrose.
Microscopy and protochlorophyllide measurement
Etiolated seedlings were inspected using a Leica stereo-microscope with fluorescence unit and a blue excitation filter set (excitation 470/40 nm, barrier 515 nm). For the determination of protochlorophyllide (PChlide) amounts, samples of 20 seedlings were harvested, steamed over a boiling water bath for 2 min and frozen in liquid N2. Homogenization using an eppendorf pistil was done in acetone/0.1N NH4OH (9/1). Pchlide was determined in a Hitachi F4500 fluorescence spectrophotometer using excitation/emission wavelength of 440/632 nm, respectively.
Nucleic acid extraction and PCR
Following the collection of etiolated seedlings under the fluorescence microscope, RNA of seedlings was extracted using Trisure (Bioline GmbH, Luckenwalde) according to the manufacturer's instructions. cDNA synthesis was performed from 1 µg total RNA with random hexamer primers and RiboLock RNase Inhibitor and RevertAid Reverse Transcriptase (Fermentas, St. Leon-Rot). Semi-quantitative RT-PCR used 2–18 ng of reverse transcribed total RNA, Mangotaq DNA polymerase (Bioline GmbH, Luckenwalde) and primer pairs F3 aacgggagtatcagctcgtgacag/R3 cgcagaagcacgttccactaattt (AtRibA1) and tuaF tggttctggattgggttctc/tuaR acagcatgaaatggatacgg (At5g19780, tua5), respectively.
5′ RACE reactions were performed using RNA purified with RNeasy Plant Mini Kit (Qiagen, Hilden) from rfd1 mutants and Arabidopsis wild-type plants grown on agar plates. SALK mutant lines 036891 and 094736 were selfed and individuals homozygous for the T-DNA insertion upstream of AtRibA1 were identified by PCR. Here, as well as for heterozygous rfd1 and rfd4 individuals, RNA was isolated from 3-week-old soil-grown plants and purified as described above. RACE analyses included an initial treatment with calf intestine phosphatase (Invitrogen, Carlsbad) for removal of uncapped 5′ phosphate residues. Decapping using tobacco acid pyrophosphatase (Epicentre Biotechnologies, Madison) and ligation of the RNA oligomer adaptor gugauccaaccgacgcgacaagcuaaugcaagannn to the newly generated 5′ phosphates was performed according to (6). Reverse transcription of adapter ligated RNA employed a mixture of AtRibA1 (tcttcaatggcctcag, gcaggagcagaatcag) or pat-specific (cgatcataggcgtctc, cagaaacccacgtcat) primers. Combinations of adaptor-specific (Ad1 tgatccaaccgacgcgac) and gene-specific primers (RibA1_RACE269 aatggctcttcggtccgatcat, pat_RACE351 ctgtgcctccagggacttca) were used in primary RACE amplifications employing 35 cycles. Secondary RACE PCRs were performed with 25 cycles on 1:100 diluted primary RACEs using nested adaptor primer Ad2 (accgacgcgacaagctaatgc) in combination with RibA1_RACE216 (tccgttggtgaatgagagcaga) and pat_RACE113 (gtacggaagttgaccgtgcttg), respectively. Amplification products were isolated from agarose gels and sequenced using an ABI 377 automatic DNA sequencer (Applied Biosystems).
For DNA analyses by single-seedling PCR, 6-day-old plantlets were homogenized in 100 µl of 200 mM Tris–HCl pH 8.0, 150 mM NaCl, 25 mM EDTA, 0.5% SDS using an eppendorf pistil. Following centrifugation, the supernatant was transferred to a new tube and precipitated using 2.5 vol ethanol and 10 µg glycogen. The pellet was washed with 70% ethanol and dissolved in 20 µl H2O. Larger amounts of DNA were isolated following the method described in (7).
Identification of insertion sites
T-DNA flanking sequences were determined using the protocol of Strizhov et al. (8). PCRs were performed using pWA5 left border primers LB1 tttataataacgctgcggacatctacatttt and LB2 tggtaattactctttcttttcctccatattga. Amplified fragments were sequenced with LBseq tgaccatcatactcattgctga. Insertion of T-DNA in rfd1 was confirmed using oligonucletides F1 tcaagaggattcgtgggaaa and R1 cgttttccttctgattcacc (see Figures 1E and G). For genetic complementation experiments, At5g64300 including promoter sequences was amplified using primers F2 ccggagagatctcgatcaatg and R2 ctgtcatttgagaatccggac (see Figure 1E). The 5-kb product was inserted into binary vector pBinAR following removal its 35S promoter. The resulting plasmid pRIB was used to transform Arabidopsis rfd1 mutants by vacuum infiltration.
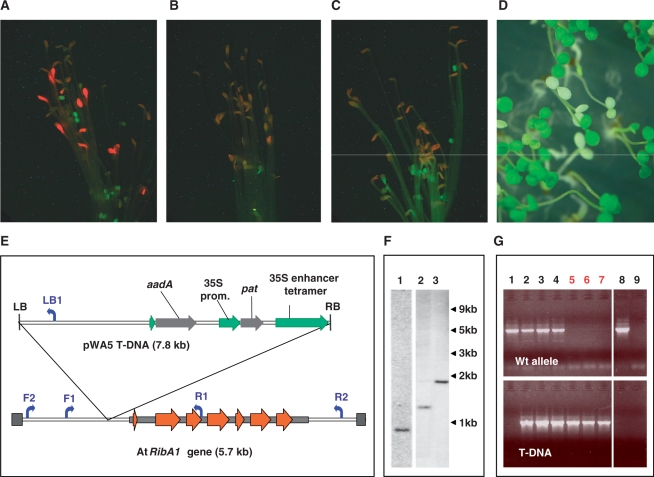
Figure 1.
(A) Phenotype of etiolated rfd1 seedlings. Plants grew 6 days in the dark and were analyzed by fluorescence microscopy as described in ‘Materials and Methods’ section. (B) A. thaliana wild-type seedlings under the conditions described in (A). (C) Etiolated seedlings of line rfd1 in the presence of 5 µM riboflavin. (D) Nine-day-old light-grown seedlings of line rfd1. (E) Scheme of the pWA5 T-DNA insertion upstream of At5g64300 (AtRibA1) in rfd1 DNA. Exons 1–7 are indicated by red arrows, the mRNA sequence by a gray box. pWA5-derived T-DNA is inserted upstream of the AtRibA1 5′ end. The inserted sequence contains a 35S enhancer tetramer next to the right border (RB) sequence. The phosphinotricine resistance marker gene (pat) is controlled by a CaMV 35S promoter (35S prom.). Primers used in (F) are indicated by blue arrows. Since the schemes of AtRibA1 and the inserted T-DNA are scaled differently, sizes of both DNA stretches are given in parentheses. (F) Southern blot analysis of DNA from single heterozygous rfd1 plants cut by restriction endonucleases DraI, BspHI and NdeI (lanes 1–3, respectively). Hybridization was performed using a LB-specific PCR product of pWA5. Sizes of DNA fragments are given on the right. (G) Detection of T-DNA in individual etiolated non-fluorescent (lanes 1–4) or red fluorescent seedlings (5–7) of line rfd1. The Primer pair F1/R1 specifically amplified the wild-type allele (upper panel), whereas primer combination F1/LB1 detected AtRibA1-alleles containing integrated pWA5 T-DNA (lower panel). Arabidopsis wild-type and nontemplate controls are shown in lanes 8 and 9, respectively.
Northern and southern blot analyses
RNA separated on formaldehyde-containing gels was blotted on Hybond N membranes (GE Healthcare, Munich) and hybridized to PCR fragments that were radioactively labeled with α32P-dCTP (Hartmann Analytic, Braunschweig) by Decaprime labeling kit (Fermentas, St. Leon-Rot) using standard protocols (9). Hybridization signals were detected using a Bio-Rad PhosphoImager. Southern blotting was performed according to standard protocols (9) using a 760-bp PCR fragment amplified with primers SB_F caggatatattcaattgtaaatgg and SB_R ggcacctatctcagcgatct from pWA5 plasmid DNA as probe.
RESULTS
Mutant screen and initial analysis of rfd1
The precursor of all tetrapyrroles, 5-aminolevulinic acid (ALA), and its synthesis in dicotyledonous plants is strongly reduced in the dark (10). A screen for mutants affected in control of ALA biosynthesis was performed using a transgenic population of 30 000 independent Arabidopsis T-DNA insertion lines transformed with plasmid pWA5 (kindly provided by T. Altmann, Gatersleben). Mutants that do not sufficiently repress ALA synthesis under etiolated growth conditions accumulate excessive amounts of the chlorophyll precursor Pchlide, which is converted to chlorophyllide in a light-dependent enzymatic step. Several mutants showing a characteristic red fluorescent phenotype under etiolated growth conditions (red fluorescent in darkness, rfd) were identified.
Pigment extraction from fluorescing rfd1 mutant seedlings revealed an ∼8-fold increase in accumulated protochlorophyllide. These highly fluorescent seedlings exhibited normal skotomorphogenic growth with wild-type length hypocotyl and closed cotyledons after 5–7 day's growth on agar plates in the dark (Figure 1A). When etiolated rfd1 mutants were transferred to light, the overaccumulated chlorophyll precursor caused photooxidative damage and the respective seedlings subsequently died. Hence, the phenotype in the dark strongly resembles the flu mutant described by Meskauskine et al. (11).
However, when grown in constant light or dark–light cycles, the mutant showed completely white cotyledons (Figure 1D). True leaves were similarly bleached under normal light conditions although growth under dim light (5–20 µM photons m−2 s−1) results in a slight greening. On sucrose-supplemented media rfd1 developed inflorescences and sets flowers, but no viable seeds were produced.
DNA blot analyses detected a single T-DNA insertion in the rfd1 mutant line (Figure 1F). Segregation of the mutant phenotype (24.3% white seedlings, Chi square = 9.94, P = 0.0016) as well as of the co-transformed pat (phosphinotricin acetyltransferase) gene (about 70% BASTA-resistent seedlings) on sucrose containing media under normal light conditions confirmed the presence of a single T-DNA insertion. Arabidopsis genomic regions flanking the T-DNA insertion were identified by adapter ligation PCR methods. The right border of pWA5-derived T-DNA is inserted 307-bp upstream of the coding region of At5g64300, which encodes AtRibA1, the first enzyme of the plant riboflavin biosynthesis pathway. Arabidopsis RibA1 specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase that is able to complement Escherichia coli ribA and ribB mutant strains, respectively (12).
Amplification of the Arabidopsis genome region spanning the T-DNA insertion site from individual etiolated seedlings revealed a homozygous state of the T-DNA insertion in those seedlings that exhibited the red fluorescent phenotype (Figure 1G, lanes 5–7). Seedlings that did not over-accumulate Pchlide in the dark were either heterozygous or lacked the pWA5 T-DNA (Figure 1G). Analysis of light-grown seedlings (see Figure 1D) revealed, in line with the findings described for etiolated plants, that white individuals had no wild-type allele of AtRibA1 (data not shown). Hence, seedlings heterozygous for the rfd1 T-DNA insertion did not develop a visible phenotype, neither in the light nor under etiolated conditions (shown for nonfluorescent seedlings in Figure 1G). The term ‘rfd1 mutant’ therefore in the following refers to line rfd1 individuals that are homozygous for the pWA5 T-DNA insertion as indicated by the occurence of the described phenotype(s).
The insertion upstream of the coding region of At5g64300 was shown to be responsible for the mutant phenotype of rfd1 by two different experimental approaches. First, addition of riboflavin suppressed the mutant phenotype in darkness (Figure 1C) and upon light exposure.
Second, the introduction of an 5-kb genomic At5g64300 fragment spanning from F2 to R2 (see Figure 1E) using plasmid pRIB into rfd1 yielded plants homozygous for rfd1 but lacking the described phenotype. Heterozygous rfd1 individuals were transformed using pRIB and F1 transformants were tested for the presence of pWA5 by PCR. F2 offspring was analyzed for the rfd1 phenotype and the presence of integrated pRIB and pWA5 (i.e. rfd1) T-DNA. In total, 29 of 39 (i.e. 74%) analyzed F2 individuals contained integrated pRIB T-DNA. Seven seedlings (18%) displayed the rfd1 mutant phenotype and were shown by PCR to be homozygous for rfd1 but lacking the pRIB fragment. Five of the 39 analyzed seedlings (13%) were homozygous for rfd1 but contained the pRIB insert. All of these five individuals lacked the rfd1 mutant phenotype. The introduction of a wild-type AtRibA1 copy was hence able to complement the rfd1 mutant.
Transcript analyses
Surprisingly, transcript studies by semi-quantitative (sq) RT-PCR revealed a strong increase in transcription of AtRibA1 in rfd1 mutants in comparison to wild-type seedlings. Moreover, enhanced transcript accumulation was observed not only in homozygous, but also in heterozygous rfd1 individuals diplaying no visible phenotype (Figure 2A). To investigate the structure of the over-accumulating transcripts, Northern blot analyses were performed. Due to limiting amounts of etiolated seedling material, heterozyguous rfd1 plants grown on soil were used. The RNA blots revealed a drastically elongated AtRibA1 transcript of about 5.5-kb size in rfd1 plants (Figure 2B). In contrast, wild-type RibA1 mRNA has a size of 2.285 kb (11).
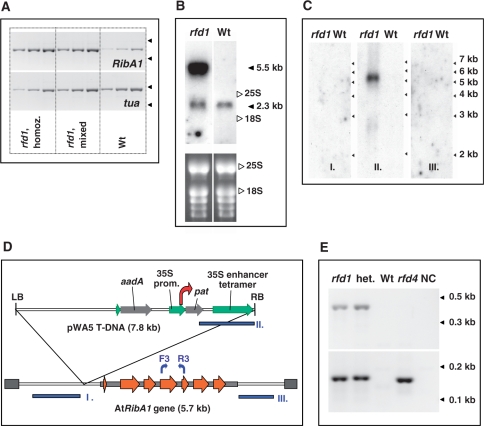
Figure 2.
(A) Semi-quantitative RT-PCR of etiolated seedlings. Homozygous (rfd1, homoz.) as well as nonfluorescent (rfd1, mixed) rfd1 seedlings were collected and compared to A. thaliana wild-type seedlings (Wt). Accumulation of AtRibA1 (RibA1, upper panel) and alpha-tubulin (tua, lower panel) transcripts was investigated using primer pairs F3/R3 (see Figure 2D) and tuaF/tuaR, respectively. Reverse-transcribed total RNA measuring 2 ng, 6 ng and 18 ng (from left to right) were used as template. Arrowheads on the right side of each panel indicate the migration of 0.4-kb and 0.3-kb marker fragments. Note that, due to segregation, the nonfluorescent sample (rfd1, mixed) contained heterozygous rfd1 as well as wild-type seedlings. (B) Northern blot of RNA isolated from heterozygous rfd1 mutants (rfd1) and A. thaliana wild type (Wt) was hybridized to AtRibA1 cDNA. 25S rRNA (3.3 kb) and 18S rRNA (1.7 kb) positions as well as the estimated sizes of specific bands are indicated. The lower panel depicts an ethidium bromide stain of RNA before blotting. (C) Hybridization of RNA blots of heterozygous rfd1 mutants and A. thaliana wild type (Wt) with probes I, II and III, respectively. (D) Localization of DNA fragments I, II and III (blue boxes) used as probes in (C); positions of primers for sq RT-PCR of AtRibA1 (F3, R3) are indicated by blue arrows. The transcription start site of the 5.5-kb T-DNA:AtRibA1 fusion transcript identified in line rfd1 is indicated by a red arrow. (E) 5′ RACE of the pat gene in homozygous (rfd1) or heterozygous (het.) rfd1 mutant plants, compared to A. thaliana wild type (Wt) and an alternative T-DNA insertion line carrying the pWA5 plasmid (rfd4). Primary RACE PCR (upper panel) was performed with 35 cycles using primers Ad1 and pat_RACE351. The secondary, nested amplification used Ad2 and pat_RACE113 and 25 additional cycles (lower panel). A non-template control is shown in lane NC. Fragment lengths are indicated by arrowheads at the right margin.
To characterize the 5.5-kb transcript, three further DNA fragments were used in Northern blot hybridization experiments. Probing with fragments of neighboring genomic regions (probes I/III in Figure 2C and D) did not detect the elongated AtRibA1 transcript. Only a probe specific to the T-DNA right border region detected the 5.5-kb transcript demonstrating that it originated inside the T-DNA region (probe II in Figure 2C and D). According to the size of the transcript and the fact that its AtRibA1 cDNA part was demonstrated to be correctly spliced (data not shown), the 5.5-kb RNA is derived from the 35S promoter driving the pat gene, which initiates transcription at about 3.2-kb upstream of the pWA5 T-DNA right border (red arrow in Figure 2D). To analyze transcript initiation upstream of pat, we used a 5′ RACE strategy based on ligation of a RNA adapter to Arabidopsis total RNA treated with calf intestine phosphatase (CIP) and tobacco acid pyrophosphatase (TAP) (6). The incubation with CIP dephosphorylated the 5′ mRNA ends, which were not protected by a 7-methylguanosine cap. The cap itself was converted in the subsequent TAP treatment step to 5′ monophosphate, which was then ligated to an RNA adapter. Reverse transcription with pat-specific primers was followed by primary RACE amplification (Figure 2E, upper panel). Primary PCRs of homo- as well as heterozygous rfd1 plants were compared with an alternative pWA5 T-DNA containing Arabidpsis mutant (rfd4 in Figure 2E), which does not show enhanced pat gene transcription in northern blot analyses (data not shown). It was confirmed that transcripts initiated upstream of pat overaccumulated in rfd1. Secondary RACE PCRs illustrate that the enhanced transcription initiation originates at or in close proximity to the pat TSS controlled by the 35S promoter in rfd4 (Figure 2E, lower panel). Hence, accumulation of the 5.5-kb T-DNA:AtRibA1 fusion transcript is the result of an enhanced 35S promoter activity in rfd1.
To understand the 5′ structure of the Arabidopsis RibA1 gene and, consequently, the misregulation in rfd1 we inspected transcription initiation sites in wild-type plants. Available EST sequences including the 5′ region of AtRibA1 extended to nucleotide –55 (Acc. Nr. CB254432). Herz et al. (11) described 5′ RACE analyses detecting capped transcripts starting at nt. –284. We performed 5′ RACE reactions as described for pat (see above) using now RibA1-specific primers in reverse transcription and amplification. We identified two major transcript start sites in A. thaliana RNA (Figure 3A, Wt). The majority of transcripts in wild-type plants was initiated at nt. –210 (Transcription Start Site 2 = TSS2) while a minor amount of 5′ RACE amplicons reached to nucleotides –281/–285 (TSS1). The latter positions agreed well with the initiation at nt. –284 described by Herz et al. (12).
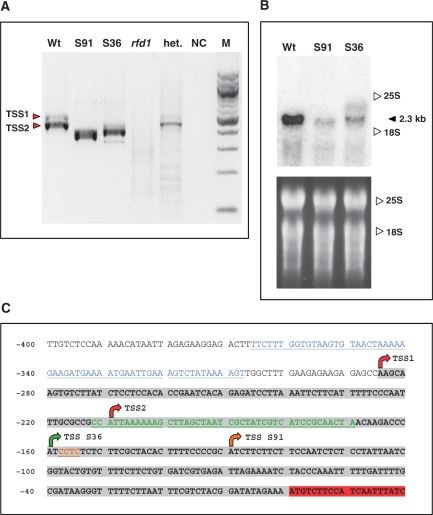
Figure 3.
(A) 5′ RACE reactions to detect AtRibA1 transcript ends in Arabidopsis wild type (Wt), SALK 036891 (S91), SALK 094736 (S36) and homozygous rfd1 seedlings (rfd1). Analysis of a heterozygous rfd1 plant (het.) and a nontemplate control (NC) are included. The DNA marker lane (M) contains a 100-bp DNA ladder (New England Biolabs, Ipswich). Red arrows depict the two major transcription start sites TSS1 and TSS2 detectable in A. thaliana wild type. (B) Northern blot of A. thaliana wild type and SALK lines 036891 and 094736, respectively. The filter was hybridized to an AtRibA1 cDNA fragment. 25S rRNA and 18S rRNA positions are indicated. The lower panel depicts the ethidium bromide stain of RNA before blotting. (C) Overview of the AtRibA1 upstream sequence with transcription start sites mapped by 5′ RACE (arrows) and T-DNA insertion sites. The 5′ UTR (highlighted in gray) in wild-type RNA results from transcription starting at TSS1 and TSS2 (red arrows). Altered transcript 5′ ends in the two SALK lines are designated TSS S36 and TSS S91. Insertions of T-DNA resulted in losses of genomic DNA stretches (colored residues) of different lengths. Insertions of pWA5 T-DNA in rfd1 (blue) and pROK2 in SALK 094736 (green) and 036891 (brown), respectively, are indicated. Nucleotide numbers are given as distances from the AtRibA1 start codon at the left margin.
Publicly available T-DNA mutant collections harbor several lines with insertions in At5g64300. There were no candidates with T-DNA integrated in the coding region, but two insertions are localized in the 5′ UTR. These insertion mutants with indicated T-DNA integrations at nt. –169 (SALK line 094736) and nt. –154 (SALK line 036891), respectively, proved to be useful, since they are situated downstream of the above characterized major TSS of AtRibA1. To characterize the T-DNA integration sites for both SALK lines in detail, the Arabidopsis genomic DNA neighboring the T-DNA border sequences on both sides were amplified and sequenced (Figure 3C). Mutants homozygous for the AtRibA1-specific insertions were produced by selfing and analyzed for the phenotype described for rfd1. While the described insertion sites were confirmed for both lines, neither etiolated nor light-grown mutant seedlings showed visible aberrations of homozygous SALK mutants in comparison to wild-type plants. Northern blots revealed in homozygous individuals of SALK lines 094736 and 036891 an accumulation of AtRibA1 mRNA to significantly lower, but still detectable levels (Figure 3B).
Mapping of AtRibA1 TSS in transgenic lines by 5′ RACE
We analyzed mutants of both SALK lines by 5′ RACE and mapped transcription initiation sites that were situated at nt. –130 (036891) and nt. –160 (094736), respectively (Figure 3A). Hence, the integration of T-DNA downstream of the wild-type AtRibA1 transcription start sites does not abolish transcription of the gene. In fact, the AtRibA1 promoter is rather flexible and able to initiate transcription from downstream sites when disrupted by T-DNA integration events. Surprisingly, in line 036891, we amplified a minor fraction of 5′ RACE products that showed initiation taking place even inside the T-DNA region (–154 + 14 nt of left border sequence of pROK2 T-DNA). Since the left border sequence of pROK2, the vector used to generate the SALK T-DNA lines (2), is not known to encode a promoter, transcription initiation in this T-DNA tagged line seems to be partly determined by promoter structures situated downstream of the actual transcription start site.
Hence, transcript analyses of SALK lines 036891 and 094736 proved that a T-DNA insertion in the 5′ UTR of AtRibA1, i.e. downstream of TSS1 and TSS2, can strongly reduce transcript accumulation, but does not automatically abolish transcript initiation. Furthermore, these T-DNA insertions fail to cause a phenotype resembling that of rfd1.
According to the scanning model of ribosome action (13), the over-accumulating 5.5-kb RNA in rfd1 can be assumed to be untranslatable (see ‘Discussion’ section). We therefore inspected rfd1 for the presence of translatable transcripts initiated around the wild-type TSS. The detection by Northern blot technique was infeasible due to limitations in available red fluorescent seedling material. Therefore, etiolated rfd1 seedlings exhibiting the red fluorescent phenotype were analyzed by 5′ RACE. Experiments were performed in parallel with 5′ RACE reactions described above for wild-type plants and SALK lines (Figure 3A). However, in contrast to the latter, no transcription initiation was detectable upstream of AtRibA1 in the homozygous rfd1 seedlings (Figure 3A, lane rfd1). On the contrary, heterozygous rfd1 plants (Figure 3A, het.) yielded RACE products of identical size as observed in wild type. This proves that only homozygous rfd1 individuals are deficient in AtRibA1 mRNA initiated at the wild-type TSS. Hence, a complete lack of properly initiated transcripts in homozygous mutants coincides with the appearance of the characteristic phenotype of rfd1 (see Figure 1A and D).
DISCUSSION
In contrast to the implications of an interruption of an ORF by a T-DNA integration event, insertions in 5′ UTR regions often do not display a ‘knockout’ phenotype. In agreement with this common observation, we demonstrated by using two different SALK lines that an integration event in the 5′ UTR region of AtRibA1 itself was not sufficient to completely abolish transcription (Figure 3A and B). However, the identified rfd1 mutant resembled a complete ‘knockout’. The reversibility of the phenotype by complementation in combination with the presented 5′ RACE analyses proved this assumption.
The integrated pWA5 T-DNA in rfd1 encodes a 35S-enhancer tetramer at the right border sequence neighboring AtRibA1 (Figure 1E). The enhancer did not reinforce the transcription from the AtRibA1 promoter in heterozygous rfd1 plants (Figure 2B and C) or homozygous rfd1 mutant seedlings (see Figure 3A, lane rfd1). Instead, a strong overexpression starting at the T-DNA internal 35S promoter directing pat expression was observed (Figure 2D and E).
The accumulating 5.5-kb transcript comprises 3.2 kb of T-DNA sequence, followed by the AtRibA1 mRNA. By complementation experiments we showed firstly that the phenotype observed in homozygous mutants is abolished by riboflavin (Figure 1C). Second, the introduction of a genomic fragment carrying AtRibA1 under its own promoter was able to complement the phenotype in homozygous rfd1 mutants. This genomic complementation indicated, that the defect was not mediated in trans. Hence, it was not caused by co-suppression of endogenous AtRibA1 mRNA by the overaccumulating 5.5-kb fusion transcript.
The AtRibA1 open reading frame (ORF) is situated at the 3′end of the 5.5-kb transcript accumulating in rfd1. The elongated mRNA contains numerous ORFs starting with the PAT protein coding sequence conferring phosphinotricin resistance followed by at least four ORFs of 115 amino acid each on the 35S enhancer fragment. The AtRibA1 reading frame enconding the riboflavin biosynthetic protein is hence the last on a polycistronic messenger. According to the scanning model of ribosome action (12) translation in eukaryotic mRNAs starts at the most 5′ situated start codon. Re-initiation of translation at a downstream ORF is known to occur only following very short regulatory reading frames in 5′UTRs or when internal ribosome entry sites are present. The latter mechanism has so far only for viral polycistronic RNAs convincingly been described (14). Hence, translation of AtRibA1 from the polycistronic 5.5-kb RNA in rfd1 is not feasible.
We have used a microarray approach to further characterize the rfd1 mutant (Hedtke, unpublished data). Using this technique, transcripts containing AtRibA1 were shown to accumulate more than 8-fold in etiolated homozygous mutants compared to wild-type seedlings.
Since the 35S tetrameric enhancer sequence present in rfd1 has been shown to be able to enforce transcription in promoters over a distance as far as 11 kb (15) we have analyzed the annotated mRNAs residing in a region of 50 kb surrounding the T-DNA insertion site in rfd1. None of the transcripts encoded in this region was found to be significantly enhanced in etiolated rfd1 seedlings. This finding was supported by the fact that the rfd1 phenotype was only observed in homozygous individuals. Hence, the genetic characteristics of the mutant rule out that rfd1 is a simple activation phenotype.
A T-DNA insertion event itself in the promoter region of AtRibA1 does not cause a sufficient reduction of expression to explain the phenotype of rfd1. This was demonstrated using SALK mutant lines 036891 and 094736. In both cases, insertions reside downstream of the T-DNA integration site of rfd1 as well as of the major transcription start sites TSS1 and TSS2 of AtRibA1. However, the integrated T-DNAs in both lines still allow sufficient RNA synthesis starting at aberrant initiation sites (Figure 3A–C) to avoid phenotypic consequences.
The analysis of the AtRibA1 promoter in wild-type plants by 5′ RACE analyses identified transcription start sites at positions –285 and –210. A genome-wide identification of Arabidopsis TSS tags containing cap signatures by Yamamoto and Obokata (16) revealed AtRibA1 initiation sites at positions –285 and –76. Although we could not identify the TSS at nt –76 described by Yamamoto and Obokata in our 5′RACE reactions, obviously several motifs present in the AtRibA1 upstream region have the potential to serve as alternative promoters leading to various TSS. This is illustrated by the alternative transcription start sites detected in the two analysed SALK lines (see Figure 3C).
That transcription of AtRibA1 in the SALK lines can be explained by an introduction or generation of new promoter structures as a possible consequence of the insertion of pROK2 is unlikely for two reasons. First, in both cases transcription starts at a different distance from the inserted LB pROK2 sequence. Second, there are no pulished hints neither on pROK2 left border internal promoters nor on a frequent generation of random promoters due to pROK2 T-DNA insertion events.
Most recently, a mechanism designated transcriptional interference was described in S. cerevisiae. The induction of the intergenic transcript SRG1 is able to silence the downstream situated, overlapping SER3 gene (5). Further investigation revealed that the transcription of noncoding SRG1 is serine-dependent and thereby adds an additional level of regulation on the expression of the serine biosynthetic gene SER3 (17). The regulatory mechanism of transcriptional interference hence represents an activation event that directly represses a (downstream situated) gene.
Our data on transcription of AtRibA1 in rfd1 strongly suggest a transcriptional interference mechanism to be responsible for the lack of transcript initiation at the gene's own promoter in homozygous mutant individuals. First, the investigation of the two different SALK mutants proved that AtRibA1 promoter structure is highly flexible and tolerates T-DNA insertion events that are situated even closer to the translational start than the integration present in rfd1. Hence, the physical interruption of the AtRibA1 upstream region by the insertion of pWA5 T-DNA is highly improbable to be responsible for the observed phenotype. Second, by complementing the mutant with a genomic fragment containing AtRibA1 we demonstrated that RibA1 silencing in homozygous rfd1 mutants is not caused by an induction of mRNA degradation, i. e. by co-supression. This would be a trans effect that also would silence the introduced complementing genomic fragment, which is not observed.
Recent work on transcription in eukaryotic organisms revealed an unforeseen amount of RNA synthesis in non-protein-coding regions (18). Also, plant genomes produce numerous such RNAs, including large (i.e. >40 nt) transcripts that are designated as mRNA-like non-coding (mlnc) RNAs (19).
The mechanism shown here to silence the transcription of AtRibA1 in rfd1 mutants demonstrates that transcriptional interference can be used also in plant systems to negatively regulate gene expression. Therefore, among the noncoding RNAs described in Arabidopsis so far a number of transcriptional interferers might be expected.
FUNDING
The Deutsche Forschungsgemeinschaft in subproject B9 of the collaborative research center SFB 429. Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Thomas Altmann (Gatersleben) for the Arabidopsis insertion mutant population which was the basis of the performed mutant screen.
REFERENCES
- 1.Bouché N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 2.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 3.Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springer PS. Gene traps: tools for plant development and genomics. Plant Cell. 2000;12:1007–1020. doi: 10.1105/tpc.12.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 6.Kühn K, Weihe A, Börner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Chory J. Preparation of DNA from Arabidopsis. Methods Mol. Biol. 1998;82:55–60. doi: 10.1385/0-89603-391-0:55. [DOI] [PubMed] [Google Scholar]
- 8.Strizhov N, Li Y, Rosso MG, Viehoever P, Dekker KA, Weisshaar B. High-throughput generation of sequence indexes from T-DNA mutagenized Arabidopsis thaliana lines. Biotechniques. 2003;35:1164–1168. doi: 10.2144/03356st01. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 10.Beale SI. Biosynthesis of the tetrapyrrole pigment precursor, d-aminolevulinic acid, from glutamate. Plant Physiol. 1990;93:1273–1279. doi: 10.1104/pp.93.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herz S, Eberhardt S, Bacher A. Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry. 2000;53:723–731. doi: 10.1016/s0031-9422(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145:722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto YY, Obokata J. ppdb: a plant promoter database. Nucleic Acids Res. 2008;36(Database issue):D977–D981. doi: 10.1093/nar/gkm785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Rymarquis LA, Kastenmayer JP, Hüttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]