Abstract
Post-translational modifications (PTMs) of histones play a role in modifying chromatin structure for DNA-templated processes in the eukaryotic nucleus, such as transcription, replication, recombination and repair; thus, histone PTMs are considered major players in the epigenetic control of these processes. Linking specific histone PTMs to gene expression is an arduous task requiring large amounts of highly purified and natively modified histones to be analyzed by various techniques. We have developed robust and complementary procedures, which use strong protein denaturing conditions and yield highly purified core and linker histones from unsynchronized proliferating, M-phase arrested and butyrate-treated cells, fully preserving their native PTMs without using enzyme inhibitors. Cell hypotonic swelling and lysis, nuclei isolation/washing and chromatin solubilization under mild conditions are bypassed to avoid compromising the integrity of histone native PTMs. As controls for our procedures, we tested the most widely used conventional methodologies and demonstrated that they indeed lead to drastic histone dephosphorylation. Additionally, we have developed methods for preserving acid-labile histone modifications by performing non-acid extractions to obtain highly purified H3 and H4. Importantly, isolation of histones H3, H4 and H2A/H2B is achieved without the use of HPLC. Functional supercoiling assays reveal that both hyper- and hypo-phosphorylated histones can be efficiently assembled into polynucleosomes. Notably, the preservation of fully phosphorylated mitotic histones and their assembly into polynucleosomes should open new avenues to investigate an important but overlooked question: the impact of mitotic phosphorylation in chromatin structure and function.
INTRODUCTION
Histones and their post-translational modifications (PTMs) are intimately involved in chromatin-templated processes (1–3). The availability of fast, reliable and inexpensive methods for obtaining pure histone fractions while preserving their native PTMs is crucial for constructing epigenomic modification maps linked to chromatin function (4–19) and for deciphering the putative epigenetic histone code (2,20,21).
Current histone isolation and fractionation methods rely on mechanical or nonionic-detergent cell lysis under mild, nondenaturing conditions, usually followed by nuclei isolation (and washes) and chromatin solubilization by nucleases, mechanical shearing or sonication (10–19,22–34). These steps are executed singly or in combination in the presence of phosphatase and deacetylase inhibitors to prevent enzymatic hydrolysis of histone biomarkers (4,9–19,34). The extracted histones can be further fractionated by reverse-phase high-performance liquid chromatography (RP-HPLC) (35,36) or analyzed by SDS- or non-SDS polyacrylamide gel electrophoresis (e.g. triton/acetic acid/urea and acetic acid/urea gels). Combinations of various electrophoretic systems can be used to generate more accurate, high-resolution, two-dimensional histone profiles (37).
Despite the progress in the global characterization of phosphorylated, acetylated and methylated histones by mass spectrometry (MS) (4–6,9–19), native histone PTMs may not be fully preserved when using conventional protocols for histone isolation. The turnover of PTMs is catalyzed by a variety of enzymes, most of which lack recognized inhibitors (1,38). Even for the better known super-family of histone deacetylases, no universal inhibitor exists (38). Moreover, for many modifications, the enzymes involved in their turnover remain unknown (38). Additionally, the lengthy operations in the current protocols lead to methionine (Met) and cysteine (Cys) oxidation, even in the presence of reducing agents, making the interpretation of mass spectrometry (MS) data difficult (19).
Further, as noted above, cells are often incubated, prior to or concomitant with cell lysis by non-ionic detergents, in hypotonic solutions to destabilize the cytoskeleton, facilitating the separation of cytoplasm membranes from nuclei (22,34). This severe treatment may induce unwanted protein dephosphorylation (39,40), as well as similar artifactual changes in other PTMs. For example, characterization of the histone H2A-family by top-down MS surprisingly showed no phosphorylation on H2A, and no increase in H2A Ser1 phosphorylation during S- and M-phase (18). This result contradicts experimental evidence showing that bulk H2A is the heaviest phosphorylated histone in proliferating cells (41–44); some H2A iso-species and H4 Ser1 are maximally phosphorylated during S-phase and metaphase (45), and H2AX Ser139 phosphorylation is upregulated during S-phase (46). We have recently shown that RP-HPLC-fractionated histone H2A from unsynchronized mouse carcinoma cells contains ∼4-fold and 6-fold higher phosphate levels than H3 and H4, respectively, and that bulk phosphorylated H2A isoforms were resistant to cAMP-induced global histone dephosphorylation (43).
Another downside of the current methods for histone fractionation is the obligatory use of HPLC for large-scale MS analysis of fast and dynamically fluctuating histone modifications in response to environmental cues (12,15,43). The massive parallel HPLC fractionations are problematic: although HPLC is a powerful technique, it is cumbersome, time consuming, hazardous, expensive and requires highly skilled personnel to operate the instrument. Thus, it may not be available to many labs.
Here, we present novel methods for histone isolation and purification that bypass the use of HPLC and preserve the native PTMs of histones.
MATERIALS AND METHODS
Unless otherwise stated, reagents were of highest quality purchased from Fisher. All experiments were carried out at room temperature, unless otherwise specified.
Purification of histone H3/H4 from intact cells: salt-urea method
The followed method, described herein, allows the separation of histone H3/H4 from histone H2A/H2B and eliminates the majority of contaminating biopolymers (e.g. proteins and DNA).
Discard cell medium and wash adherent, 80%-confluent cells three times with prewarmed (37°C) serum-free growing medium. This step ensures minimum disturbance of the native PTM status and avoids possible interference of serum components with the purification process. With a Pasteur pipette connected to vacuum, maximally aspirate the remaining washing medium.
Salt-urea-cell lysis. We hypothesize that the cell nucleus in concentrated urea solution, containing relatively low concentration of salt (e.g. 10 mM NaCl), would be relatively more stable than the cell cytoplasm. Thus, the stabilized nuclei can be separated from urea-solubilized cytoplasm by low-speed centrifugation. Add 2 ml (per 100 mm dish) of 8 M urea in 10 mM NaCl, 50 mM Tris–HCl, pH 8.0, 2 mM EDTA and 1 mM DTT (lysis buffer). The 8-M urea is intended to inactivate most of the unwanted uncontrolled enzymatic activities, thereby protecting the native histone PTMs. Incubate the cells in lysis buffer, making sure that the buffer spreads homogeneously over the cells (gently rotate the dishes by hand). Incubate for about 10 min, monitoring lysis under a phase contrast microscope; if necessary, increase lysis time. Too prolonged an incubation is not desirable since it may lead to nuclei lysis and release of DNA, making it difficult to handle the samples. Too short of an incubation can result in partial cytoplasm solubilization, nuclear aggregation upon centrifugation and poor H3/H4 purification.
Upon completion of lysis, collect cell lysates into 2 ml microfuge tubes and add increasing concentrations of NaCl (i.e. from a 5 M stock add 100, 200 or 250 mM NaCl final concentration) to individual tubes to optimize the salt concentration required for H3/H4 purification based on cell type. Higher salt concentrations (e.g. 100–250 mM NaCl), in the presence of 8 M urea, would dissociate histone H2A/H2B from the chromatin leaving insoluble DNA-bound H3/H4. Typically, 0.1–0.25 M NaCl yields good recovery and purity of H3/H4 from a variety of cell lines, such as mouse embryonic kidney cells G7, 3T3-NIH, CHO and 1470.2 mouse breast carcinoma cells. Other cell lines (e.g., normal diploid fibroblast 5659 and 5759 cell lines) may require lower salt concentrations (e.g., 0.01, 0.1 or 0.2 M NaCl); concentrations higher than 0.25 M NaCl can result in H3/H4 loss. Spin down nuclei in a micro-centrifuge at 14 000 (14K = 18 000 rcf; Beckman Coulter Inc, Microfuge 18 Centrifuge, rotor F241.5p) r.p.m. for 10–20 min. This step completely lyses the nuclei, collapsing the chromatin and mitotic chromosomes into a gelatinous pellet.
Detach pelleted nuclei from the bottom of the tubes by inverting the tubes and tapping the bottom of tube with fingers. Do not pipette the lysed nuclei to avoid DNA sticking to the pipette tips. If the collapsed chromatin partially sticks to the bottom of the tube, the purity and recovery of H3/H4 will not be affected. Carefully, drag the chromatin pellet into a 50-ml conical tube. With a 200 μl loading tip, aspirate residual cell lysate.
Salt-urea wash of chromatin pellet (see note 1). Add 40 ml of 8-M urea in 50 mM Tris–HCl, pH 8.0, 2 mM EDTA, 1 mM DTT, containing the appropriate concentration of NaCl (i.e. 10, 100, or 250 mM) to the chromatin pellet. Invert the tube trice. Carefully decant supernatant (or drag chromatin pellet into a new tube with fresh washing solution). Aspirate residual solution as in step (iv). Repeat washes until no more protein can be detected in the washing solution by micro-Coomassie protein assay [e.g. by dispensing 10 µl of the wash into 200 µl Coomassie Plus Protein Assay Reagent (Pearce, cat. no. 1856210) and compare with a dispensed 10 µl washing solution used as blank].
Acid extraction of H3/H4. H3/H4 can be recovered from the chromatin pellet in diluted acid solutions (0.2 M H2SO4 or HCl), leaving behind the insoluble DNA. Add 150–200 µl of 0.2 M H2SO4 to 100 mm dish pellet. Extract histones for 1 h or overnight on ice with occasional vortexing. Centrifuge the insoluble DNA for 5 min at 14 K. Collect H3/H4 supernatant and precipitate H3/H4 with 35% (final concentration) trichloroacetic acid (TCA, Fisher, cat. no. A322-100), overnight in ice; if desired, keep the insoluble DNA-pellet after the first acid extraction for SDS–PAGE analysis or for re-extraction.
Microcentrifuge the histone pellet at 14 K for at least 30 min; recovery of H3/H4 will be proportional to the time of centrifugation. Discard supernatant (do not use vacuum) and save pellet. In some cases, the histone pellet will be spread over the tube walls, and additional care should be taken to avoid losing it. Following washing and drying, the histone pellet (Note 2), dissolve pellet in H2O or in any other appropriate solvent (70 µl for 100 mm dish) and neutralize aqueous solution before use (Note 2).
Nonacid SP-recovery of H3/H4 from step (v)
Alternatively, H3/H4 can be recovered by sonicating the chromatin pellet in 8-M urea and selective adsorption of H3/H4 onto Sulfopropyl-Sepharose (SP, Sigma-Aldrich cat. no. S1799) beads. The principle of this procedure relies on the ability of high concentration of urea to dissociate H3/H4 from fragmented tetrasomic DNA by unfolding H3/H4. Urea, in turn, has a weak shielding effect on the negative and positive charges on SP and histones, respectively; since urea interacts mainly with hydrophobic moieties. This allows a robust adsorption of H3/H4 to SP. This procedure would preserve acid-sensitive nitrogen phosphorylation that can be subsequently detected in polyacrylamide gels by a neutral Imidazole-Zn reverse staining solution (Note 3) as follows:
Equilibrate chromatin pellet from step (v) (containing H3/H4) by adding 2 ml of 8- M urea, 20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 1 mM DTT (equilibration solution). Repeat this operation four times. For the last two operations, gently rotate the tubes for 5 min at RT to achieve optimum equilibration.
Spin pellet at 14 K for 5 min and discard supernatant.
Add equilibration solution to the chromatin pellet to a final volume of 0.5 ml.
Sonicate pellet at R/T (four times at 20% sonifier power for 30 s) (Branson Digital Sonifier, model CE Converter 102C, micro-tip 3 mm/1/8”). Visually verify for pellet fragmentation by inverting trice the tube. If necessary, sonicate for a longer period.
Nonacid SP-chromatography of sonicated chromatin-containing H3/H4
(v) Wash 1 ml of SP resin in 20 ml double deionized (dd) water. Repeat this operation two more times. Resuspend SP in 1 ml dd-water, mix well, and take 0.3 ml with the aid of a cut 1-ml tip. Centrifuge at 4 K in a microfuge, discard water and add the sonicated chromatin-containing H3 and H4 to 0.15 ml of swollen SP resin.
(vi) Rotate sample at RT for 40 min.
(vii) Spin SP-resin at 4 K for 5 min. Save supernatant for SDS–PAGE analysis.
(viii) Wash SP-bound H3/H4 with 0.5 ml chromatin equilibration solution (at least four times) until no protein can be detected by micro-Coomassie assay. Use 10 μl of 8-M urea as a control blank.
(ix) Wash away urea solution with 25 mM Tris–HCl, pH 8.0, containing 2 mM EDTA.
(x) Elute H3/H4 in batch with 0.4 ml of 1.1 M NaCl, 25 mM Tris–HCl, pH 8.0, 2 mM EDTA. Repeat this operation three more times and collect all fractions.
(xi) Run equal volumes of all samples (input, washes and elution) in a 12.5% or 15% SDS–PAGE and detect the protein bands with a method of choice.
Note 1: A note of caution is warranted here. Urea in solution undergoes slow spontaneous rearrangement to ammonium cyanate (NH4CN) that may react with amino or sulfhydryl groups on proteins (47) and possibly with phosphate groups on DNA (48). Thus, this solution should be made fresh for each purification batch.
Note 2: Whenever histones are acid-precipitated, rinse the histone pellet (to eliminate residual salts that may interfere with MS analysis) twice with 1.5 ml of the same precipitating solution (diluted TCA or perchloric acid) by inverting the tube with the rinsing solution a few times (no pipetting, no vortexing). Rinse acid-washed pellet twice with acetone/0.2% HCl and twice with acetone alone. Aspirate washes with an appropriate pipette (do not use vacuum). After each rinse, centrifuge samples for 5 min at 14 K. Air-dry the histones and dissolve in appropriate solvent (H2O or any buffer) by incubating the pellet for at least 1 h at RT with occasional vortexing. Exercise care not to lose the histones to the tube walls. The ultimate pellet may be acidic; neutralize it right before use with 50 mM final concentration of Tris–HCl, (from 1 M stock) pH 8.0. Check pH by spotting 2 µl of histone solution onto a pH-indicator paper (Whatman, cat. no. 2613991). Storing the isolated histones in acidic solutions prevents Cys and Met oxidation. If desired, the histones can be directly dialyzed in an appropriate buffer, omitting the precipitation step; the buffer should contain low concentration of Tween-20 (0.02–0.1%) to prevent histone binding to surfaces and 2–10 mM DTT to protect oxidation of Met and Cys.
Note 3: Nonacid recovered H3 and H4 (10–20 ng each) can be resolved by SDS–PAGE and visualized using a sensitive neutral imidazole-zinc reverse staining method (49). After excising the H3 and H4 bands, incubate the acrylamide slices for 5 min in 1 ml of 0.5 M EDTA with two changes to eliminate the imidazole–Zn complex that would otherwise prevent the recovery of H3 and H4. The proteins can then be extracted in an appropriate solution containing 0.05–0.1 M EDTA to dissociate the residual imidazole–Zn complex. The extracted H3/H4 can be subjected to MS analysis. Transfer of histones to a membrane should be done in the presence of 0.01–0.03 M EDTA, after the initial removal of the imidazole–Zn precipitate from the SDS gels. If urea samples are intended to be resolved by SDS–PAGE, do not heat the samples since urea solutions cleave protein peptide bonds. Instead, incubate the samples in reducing sample buffer for 10 min at 37°C.
Note 4: Core histones or highly purified H3/H4 in 8 M urea precipitate poorly in TCA or HClO4 solutions, regardless of the histone concentration. To avoid loss during acid precipitation, histones can be first concentrated by re-adsorption onto 0.1–0.2 ml SP-resin as follows. The samples are first diluted to a final concentration of at least 1-M urea in 50 mM Tris–HCl, pH 8.0, 2 mM EDTA, and flowed twice by gravity through 0.1–0.2 ml SP packed in a 0.2-cm diameter disposable column. After washing SP in the same buffer (1 ml) without urea, 2 M NaCl-eluted fractions of 0.1 ml each are collected, pooled, precipitated with 35% TCA and further treated as described in Note 2.
Similarly, if the H3/H4 tetramer in the 2 M NaCl eluate from the SP resin is too diluted for acid precipitation (despite using a 2 M NaCl elution step, which concentrates the histones), the H3/H4 fractions can be diluted to 0.2 M NaCl in the above buffer and concentrated on SP resin as above.
SP-core histone purification and H2A/H2B fractionation from H3/H4 obtained from acid-extracted undisturbed whole cell
Grow cells on three 150-mm dishes to 80–100% confluence and proceed as in salt-urea method [step (i)].
Add 2 ml of 0.1 M H2SO4 (prepared in deionized H2O from ∼18 M H2SO4 concentrated stock) to each dish. Collect cells in three 2 ml Eppendorf tubes by detaching the cells with a plastic cell lifter. Extract bulk histones for 1–2 h (or overnight) on ice with occasional vortexing. Spin cellular debris in a microfuge at 14 K at 4°C, for 5 min. Collect supernatant containing total histones plus contaminating proteins, and pool the three extracts (∼6 ml) in a 14-ml conical tube.
SP-chromatography of acid-extracted whole histones. Neutralize the extract by adding 6 ml of 1 M Tris–HCl, pH 8.0, to the pooled 6 ml 0.1 M H2SO4-extracts (after neutralization the pH of crude extracted histones should be ∼7–8). From concentrated stock solutions, add NaCl, EDTA and DTT to a final concentration of 200 mM NaCl, 2 mM EDTA, 1 mM DTT. Flow neutralized extract (flow rate 0.3–0.4 ml/min; if necessary, the flow rate can be reduced or increased) through 1–2 ml of packed SP fast-flow resin (column: diameter 0.5 cm, height 5 cm) pre-equilibrated with 50 mM Tris–HCl, pH 8.0, 2 mM EDTA (equilibration buffer) plus 200 mM NaCl (use at least 10-fold excess buffer volume over the resin volume). If necessary, collect the SP flow-through (FT) fraction depleted of histones for analysis.
The resin with bound H1 and core histones is washed with 0.5 M NaCl in the above buffer, with at least 10 resin-volumes. This step serves to clean H1 and core histones from contaminants. Check protein contaminants in the wash by dispensing 10 µl of FT into 200 µl Coomassie Plus Protein Assay Reagent (see note 5). If necessary, prolong the wash until no more protein is detected in the wash fractions.
Note 5: H1 is not detected in solution by the currently used Coomassie protein assay. H1 concentration can be determined by SDS–PAGE using any commercially available H1 standard. If H1 is not needed, ignore this step (iv) and go to step (v), in which 0.6 M NaCl is used to wash off contaminants and H1 in a single step. Depending on the cell type, H1 can be practically quantitatively recovered in the following 0.6-M wash, whereas core histones remain bound to SP. Elute H1 at 0.6 M NaCl in the SP equilibration buffer [step (iii)]. Discard the resin FT fraction (0.5–0.7 ml for 1 ml resin). Collect at least five fractions of 1 ml each. For most cell types (but not for 1470.2 cells, see Figure 3B), these fractions contain mainly H1. Additionally, wash the resin with at least 5 resin-volumes of the same buffer and discard the wash. Precipitate H1 in 35% TCA, overnight on ice; process the samples as described in Note 2 for the salt-urea method. Some protein contaminants can co-elute at 0.6 M NaCl and are easily separated from H1 by precipitating them at 5% HClO4. H1 is then precipitated at 20-35% TCA or dialyzed.
(v) Elute bulk core histones in 2 M NaCl in equilibration buffer. Collect eluted histones in fractions equal to the volume of the SP-matrix (e.g. for 1 ml resin, collect 1 ml elution fractions). The majority of core histones and some H1 (for some cell types) elute in the first and second fraction. Some minor contaminants migrating above canonical H3 are observed as well. Precipitate core histones in 35% TCA and process the samples as described in Note 2 for the salt-urea method. Alternatively, dialyze the eluted histones against your buffer of choice. This process yields ∼0.5 mg of purified core histones.
(vi) Differentially precipitate core histones from residual H1 with 4–5% of HClO4 (reagent ACS, 70% in water; Across Organics, cat. no. 424030010), on ice overnight. Pellet precipitated core histones for 30 min at 14 K. If desired, save supernatant and re-precipitate H1 with 35% TCA at 4°C overnight. Pellet H1 for 1 h and proceed as described in Note 2 for the salt-urea method.
(vii) SP-fractionation of H2A/H2B from H3/H4. Alternatively, after washing with 0.6 M NaCl, H2A/H2B can be selectively eluted from SP (keeping H3/H4 bound to the resin) in 0.8 M NaCl, 50 mM Tris–HCl, pH 8.0, 2 mM EDTA, in the first 4-5 fractions of 1 ml each (for three 150-mm confluent dishes). For most cell types, 0.8 M NaCl optimally resolves H2A/H2B from H3/H4. Slight contamination of H2A/H2B with H4, if seen, is most likely due to over-saturation of the SP resin with histones. H3/H4 can be recovered in the first or second 1-ml fractions eluted in 2-M NaCl in the same buffer. Precipitate fractionated H2A/H2B and H3/H4 in 35% TCA overnight on ice. Proceed as described in Note 2 for the salt-urea method. Note that highly purified diluted H3/H4 and H2A/H2B precipitate poorly with TCA or perchloric acid. The fractionated H2A/H2B and H3/H4 can be concentrated by re-adsorption onto SP resin (Note 4).
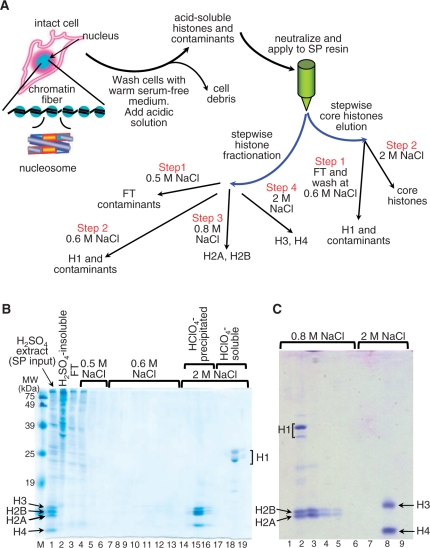
Figure 3.
(A) Flow chart of histone purification through SP, after H2SO4 extraction from whole cells. (B and C) 15% SDS–PAGE analysis of single-step SP-purification of whole core histones (B) and fractionation of H2A/H2B from H3/H4 (C), respectively. (B) Following washes with 0.5 M NaCl (lanes 4–6) and with 0.6 M NaCl (lanes 7–13), histones were eluted at 2 M NaCl and further fractionated by HClO4-precipitation into core histones (lanes 14–16) and linker histones (lanes 17–19). (C) Following the 0.6 M NaCl wash [as in (B)], H2A/H2B were separated from H3/H4 by stepwise elution at 0.8 M NaCl (lanes 2–5), followed by H3/H4 elution at 2 M NaCl (lanes 6–9). Potential problems with TCA precipitation of diluted column fractions can be overcome by another SP run (Note 4).
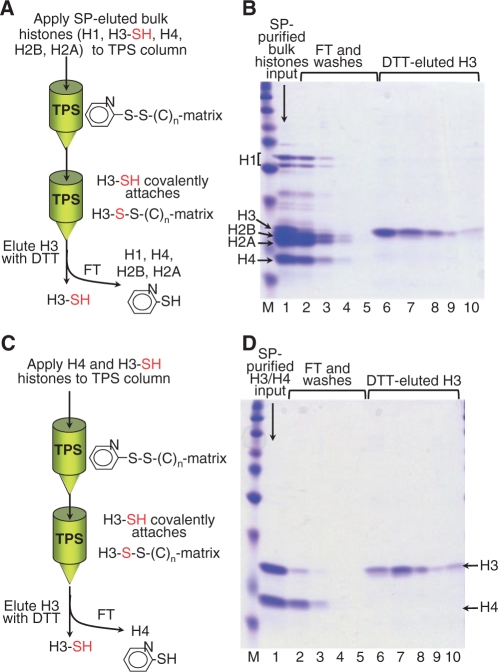
H3 separation from H4 or bulk SP-eluted histones by covalent targeting H3 reduced Cys through thiopropyl-Sepharose-6B
H3 histones in the core particle structure or in H3/H4 tetramers have buried and reduced cysteine residue(s). These residues are exposed in denaturing solvents and can be covalently targeted by activated thiol-residues attached to a solid support [Thiopropyl-Sepharose-6B (TPS; Amersham Biosciences, vendor Fisher, cat. no. NC9631897)]. TPS contains reactive 2-thiopyridyl disulfide groups attached to Sepharose through a chemically stable ether linkage. Thus, reduced Cys-SH groups on H3 can be covalently attached to the matrix through an S-S bond, displacing the resident thiopyridyl groups. None of the other histones has cysteine residues and can thus be recovered in the FT and subsequent washes. In addition, oxidized H3-Cys-SH groups would not react with the TPS. Thus, this type of covalent chromatography acts as an affinity-like step for H3 isolation from its oxidized forms and other histones as well [do not confuse 2,2′-dipyridyl disulfide activated thiopropyl-Sepharose-6B (commonly known as thiopropyl-Sepharose-6B, which is the one used here) with 2,2′-dipyridyl disulfide-activated glutathione-Sepharose-4B, commonly known as activated thiol-Sepharose 4B].
Weigh 1 g of activated TPS resin (1 g = 4 ml) and mix with 50 ml of H2O in a 50-ml conical tube. The matrix swells up fast; gently rotate the tube until crystalline-like additives (compounds preserving the functional properties of the resin) are fully dissolved. Pellet the matrix by low-speed (2-K r.p.m.) centrifugation. Thoroughly batch-wash the matrix five times with 50 ml of H2O each time.
Equilibrate TPS matrix by resuspending it in 50 ml of freshly prepared 8-M urea, 2 mM EDTA, 50 mM Tris–HCl, pH 8.0 (denaturing equilibration buffer, DEB). Decant buffer and repeat equilibration step. Pellet the resin at 4-K r.p.m. (due to high density of the urea solution). Discard supernatant. This step can be done in a column-packed matrix as well.
Resuspend 4 ml of matrix in 4 ml of DEB. With the aid of a cutoff 1 ml pipette tip, take 1 ml of matrix suspension twice and place it into a 2 ml Eppendorf tube. Centrifuge at 3-K r.p.m. and discard the buffer. Dissolve 0.2–0.3 mg of isolated H3/H4 or 0.4–0.6 mg of bulk histones in 0.5 ml of DEB. A note of caution is warranted here: DTT or any other sulfhydryl-containing chemical might interfere with the covalent binding of H3 to TPS, thus, avoid the presence of such compounds in the H3 binding solution. Add the denatured histones to 1 ml TPS and vertically rotate the mix for 2.5 h at RT to achieve homogeneous distribution of the histone solution over the resin. Do not use any other type of device for this operation. The volume of histone solution can be kept low but sufficient to achieve good mixing with the matrix. The rate of the covalent reaction between the TPS and H3-Cys-SH inversely correlates with the reaction volume and decreased H3 concentration. In other words, the more concentrated the histones are, the faster the reaction would take place between H3-Cys-SH group and the matrix. One volume of thiopropyl-Sepharose to 0.5 ml of histone mix would be a desirable mixing ratio.
Collect either H1/H4/H2A/H2B or H4 in the FT and washes. Typically, the unbound histones are recovered in the FT and in one or two column fractions. Histones in urea precipitate poorly even at high histone concentration (0.5–1 mg/ml). Therefore, dialyze the histones against an appropriate buffer. Alternatively, to change the urea buffer and concentrate the histones, these TPS-FTs can be treated as described in Note 4.
Elution of H3. Add 0.5 ml of 50 mM DTT in DEB to the H3-bound-TPS, mix well and incubate the matrix at 37°C for 30 min. Spin down the resin at 3-K r.p.m. and collect H3 in the supernatant. Repeat this procedure until all H3 is reduced and solubilized. Finally, by SDS–15%–PAGE, check for the absence of H3 on the TPS resin by collecting, previously rinsed in water, 20 µl of resin and heating it at 100°C in 2× reducing sample buffer (do not heat the sample in 8-M of urea, see note 3). Pool the eluted H3 fractions and dialyze against a solution of choice or use the procedure described in step (iv) above for histone concentration.
Cell culture
Cell line 1470.2 (derived from C127 mouse mammary adenocarcinoma cells, a generous gift of Dr. C. L. Smith), G7 embryonic kidney cell line (a gift of Dr. L. A. Parada) and HeLa cells were maintained in high-glucose DMEM, containing 10% fetal bovine serum (Atlanta Biologicals), 2 mM pyruvate, 4 mM glutamine and 10 µg/ml of ampicillin and streptomycin. The growth medium for 1470.2 cells was additionally supplemented with 100 ng/ml of epidermal growth factor (Sigma-Aldrich, cat. no. E1257).
Treatment of cells to obtain hyper-phosphorylated or hyper-acetylated histones
To obtain hyper-phosphorylated histones, HeLa cells (80% confluence) were mitotically arrested by treatment with 1 µg/ml nocodazole (Sigma-Aldrich, cat. no. M1404) for 24 h. Hyper-acetylated histones were obtained by treating HeLa cells with 40 mM sodium butyrate (Alfa Aesar, Lancashire, UK; vendor, Fisher, cat. no. 156-54-7) for 20 h (50,51).
Gel electrophoresis, western transfer and immunoblotting
Cellular extracts were resolved by electrophoresis in sodium dodecylsulfate (SDS)–12.5%- or 15%–polyacrylamide gels. Proteins were visualized by staining of gels with either GelCode Blue Stain Reagent (Pierce; vendor Fisher, cat. no. PI-24590), or Coomassie Blue R-250; or staining of membranes with Ponceau S (Sigma-Aldrich, cat. No. P3504), after western transfer.
Western transfer of proteins onto 0.1 µm pore size nitrocellulose membrane (Whatman, Protran, BA79, Superior Nitrocellulose Membrane; vendor Fisher, cat. No. 09-301-120) was carried out in a Hoeffer Semi-dry transfer unite 77 (GE Healthcare, cat no 80-6211-86) for 40 min at 9 mA per cm2 of membrane in 1× Tris-Glycine buffer containing 0.05% SDS and 7% methanol. Immunoblotting was performed with antibodies against specific H3, H2A, and H4 modifications (Upstate Biotechnology, NY), according to the manufacturer's instructions. The membranes were incubated with peroxidase-conjugated anti-rabbit secondary antibodies (Jackson Immunoresearch), and bound antibodies were detected using a chemiluminescence assay (Pierce; vendor, Fisher, cat. no. PI-32106), after exposure to films. When necessary, membranes were stripped in a solution containing 8–9-M urea/10–20% acetic acid, at 60°C for 1 h or at RT, overnight (43). After re-blocking the membrane with 4% skim milk in TBS, antibody stripping was verified by incubating the stripped membranes with HRP-secondary antibodies and developing the membranes using the chemiluminescence assay.
Supercoiling assay
Nucleosome assembly factors (fraction QS500) were purified from HeLa cells and used for supercoiling reactions as previously described (52). Briefly, 6.5 µg of BlueScript plasmid was relaxed with 15 IU of Topoisomerase I (Promega); 1.25 µg of relaxed DNA was used per reaction. The QS500 fraction (10 µl at 8 µg/µl) was pre-mixed with hyper- or hypo-phosphorylated histones H3/H4 (2 µg per reaction) for 15 min at RT. The relaxed plasmid was added and the deposition reaction was allowed to proceed for 2 h at 37°C, in a final volume of 13 µl. The reaction was stopped with 0.1% SDS (final concentration) and 5 µg proteinase K. The protease digestion was carried out at 37°C for 1 h and the DNA purified by phenol/chloroform/isoamyl alcohol extraction. The DNA topoisomers were resolved, without further manipulation, by 0.9% agarose gel electrophoresis in 1× TAE buffer. Similarly, hyper- and hypo-phosphorylated core histones (4 µg per reaction) were assembled into poly-octasomes and processed as described above.
RESULTS AND DISCUSSION
Salt-urea purification of histones H3/H4 from whole undisturbed cells
In water, hydrophobic clusters responsible for protein folding stabilize intermolecular interactions as well (53–55). Additionally, strong ionic solutes in water render the macromolecular environment even more polarized than pure water, causing ‘salting-out’ effects of interacting hydrophobic surfaces, thus strengthening the hydrophobic interactions among biomolecules (53–55). As a consequence, the interacting hydrophobic surfaces are less accessible to denaturing solutes, such as highly concentrated urea (55) and acids (56) in the presence of salts. On this basis, we hypothesize that the cell nucleus in concentrated urea solution containing salt (e.g. NaCl) would be relatively more stable than the cell cytoplasm. Thus, the stabilized nuclei can be separated from urea-solubilized cytoplasm by low-speed centrifugation. In addition, we exploited the properties of the nucleosome particle to isolate the H3/H4 tetramer from chromatin. In nucleosomes, hydrophobic forces and ionic interactions stabilize the DNA wrapping around the octameric core histones, in which tetrameric H3/H4–DNA interactions are stronger than those between H2A/H2B and DNA (57–59). In addition, salt separation of H2A/H2B from H3/H4 is readily achieved at lower ionic strength, since H2A/H2B have a weaker positive charge than H3/H4.
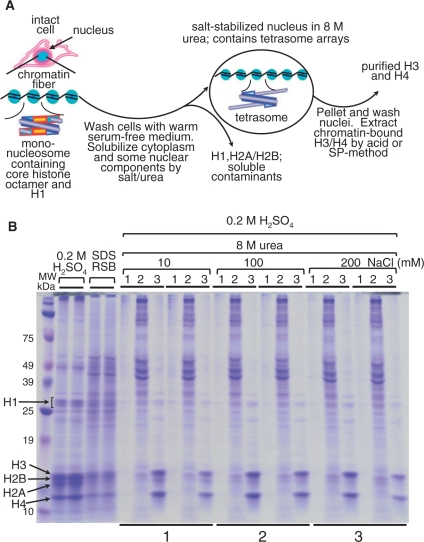
Thus, in contrast with current methods that use mild conditions for cell lysis (9–14,17,22,24–26,28–31,34,60), we first rinse the cells with warm serum-free DMEM medium to ensure minimum metabolic disturbance of the cells (43) and then lyse the cells in 8-M urea containing salt. Therefore, we bypass the traditional nuclei isolation step (see ‘Materials and Methods’ section). We suggest that the precise time for cell lysis, while still preserving the nuclear envelope relatively intact, should be empirically established for each cell type. Of note, nuclear aggregation must be avoided during the initial cell lysis to permit better solubilization of both cytoplasmic and nuclear contaminants; this step is critical for obtaining highly purified H3/H4. The simple and effective salt-urea method yields highly purified H3/H4 with near 100% recovery in a single step (Figure 1A and B, lane 3). This procedure does not require the aide of any binding matrices, such as, for example, hydroxylapatite, previously reported in other methods. Duplicate cell extracts treated at increasing NaCl concentration (10–200 mM NaCl, Figure 1B) clearly show that in the presence of 8-M urea most contaminant proteins are removed (lane 2), including H2A/H2B and part of H1, from bulk and nonfragmented chromatin (lane 3). Curiously, a portion of H1 remains persistently attached to chromatin under these conditions. Since H1 is soluble in 5% HClO4 (29,30), H3/H4 are easily separated from this residual H1 by precipitating the sulfuric acid-extracted histones with 4–5% HClO4 (see Figure 3B, lane 18).
Figure 1.
(A) Flow diagram of salt-urea H3/H4 isolation from whole, undisturbed 1470.2 cells. The salt-urea treatment rapidly dissolves the cell cytoplasm and certain chromatin components, such as H1 and H2A/H2B histones. During this process, the nuclear envelope containing tetrameric H3/H4 histones and DNA (tetrasomes) is relatively well preserved and nuclei do not clump together. This phenomenon is consistently seen using a variety of salts (sodium phosphate, NaCl, KCl, etc.), suggesting that nuclei stabilization is not salt-specific. (B) High yield recovery of H3 and H4 from intact cells by using high urea and salt concentrations. Duplicate samples (bottom underlines 1, 2 and 3) were treated with 8-M urea at increasing concentration of NaCl (10, 100 and 200 mM, as indicated at the top). The histones were sulfuric-acid extracted, TCA precipitated, and resuspended in 80 μl of denaturing reducing sample buffer (RSB) for analysis on 12.5% SDS–PAGE and stained with Coomassie blue. Duplicate histone samples, extracted with H2SO4 and 2× reducing sample buffer (RSB), are shown for comparison (four lanes next to marker lane). The RSB sample contains all cellular proteins and is used as a control for histone yield and purity. With the exception of the RSB samples (which were loaded directly), the extracted histones were TCA precipitated and resuspended in 80 µl of RSB for gel loading. Five microliters of each sample were loaded per well. Indicated at the top of the gel are: lane 1, H2SO4-insoluble cell debris after H3/H4 extraction; lane 2, cell cytoplasm and some nuclear proteins solubilized in 8-M urea at increasing NaCl concentrations (indicated at the top of the gel); lane 3, H2SO4-extracted H3/H4 after salt-urea treatment.
A flexible feature of this method allows non-acid H3/H4 recovery after DNA fragmentation in the presence of 8 M urea (see ‘Materials and Methods’ section), by selectively capturing H3/H4 by a strong cationic exchange resin (SP in this case, see ‘Materials and Methods’ section). The principle of this procedure relies on the ability of high concentration of urea to dissociate H3/H4 from fragmented tetrasomic DNA by unfolding the histones. Urea, in turn, has a weak shielding effect on the negative and positive charges on SP and histones, respectively; since urea interacts mainly with hydrophobic moieties (55). Indeed, in Figure 2A fragmented DNA is found in the SP flow through and washes (Figure 2A, see Coomassie and ethidium bromide stained gels, lanes 3–6, respectively) while H3/H4 are retained on the SP (Figure 2A, see Coomassie and ethidium bromide stained gels, lanes 7–10). Thus, this procedure would aide in the analysis of the elusive histone nitrogen phosphorylation, which is extremely labile at low pH (61–63). After purification, the H3 and H4 can be resolved by SDS–PAGE and visualized using a neutral and sensitive imidazole-zinc reverse staining method (49) (see Note 3). Additionally, highly phosphorylated H2A and low-molecular-weight protein contaminants that would dramatically interfere with 32P-H3 detection by SDS–PAGE are eliminated from the final H3/H4 preparation without HPLC fractionation (43).
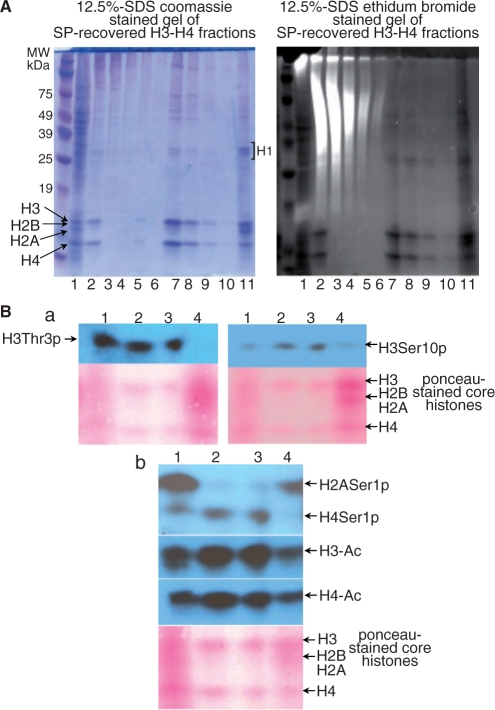
Figure 2.
Nonacid-SP histone H3/H4 recovery from 1470.2 cells after salt-urea treatment. (A) Coomassie-stained 12.5% polyacrylamide gel of SP-recovered H3/H4 fractions (left panel), with the same gel stained with ethidium bromide (right panel). Lane 1, RSB-total cell extract; lane 2, H3/H4 input to SP after salt-urea treatment; lanes 3–6, 8- M urea wash of SP-bound H3/H4; lanes 7–10, H3/H4 fractions eluted with 1.1 M NaCl; lane 11, H2SO4-extracted histones from whole cells. (B, a and b) Comparison by immunoblotting of the preservation of some histone modifications among different histone extraction methods. Lane 1, SDS-RSB-total cell extract; lane 2, input SP of salt-urea purified H3/H4; lane 3, SP-eluted H3/H4; lane 4, core histones extracted by the method described in ref. 34. The anti-histone antibodies used were either specific for particular modifications of particular residues on histones H3, H4 or H2A, or recognized H3 or H4 acetylated at multiple residues, as indicated on the right of each blot. Ponceau S staining was used to assess histone loading.
We next compare the ability of salt-urea-SP procedures (nonacid-SP method) to preserve histone phosphorylation and acetylation (Figure 2B). For this purpose, we compared the preservation of phosphorylated Thr3 and Ser10 on H3 (H3Thr3p and H3Ser10p, Figure 2Ba), and Ser1 on H2A and H4 (H2ASer1p and H4Ser1p, Figure 2Bb); we also examined the total acetylation on H3 and H4 (Figure 2Bb, H3-Ac and H4-Ac, respectively). We used specific antibodies to detect such phosphorylated and acetylated residues. In addition, we compared the levels of such modifications with levels obtained by known published protocols (34) (Figure 2B, a and b, lanes 4). The levels of phosphorylation and acetylation were similar for histones obtained by SDS-reducing sample buffer (SDS-RSB) and urea-salt-SP method [Figure 2Ba, compare lanes 1 (SDS-RSB), 2 (input to SP of salt-urea purified H3/H4 histones) and 3 (H3/H4 eluted from SP)]. Note that since H2A is not recovered using our salt-urea method, the signal for H2ASer1p is almost absent from lanes 2 and 3 in Figure 2Bb, as expected. Surprisingly, phosphorylated Thr3 and Ser10 on H3 (Figure 2Ba, lanes 4) and phosphorylated Ser1 on H4 (Figure 2Bb, lane 4) are dramatically reduced when using conventional published histone extraction protocols (34). Phosphorylated Ser 1 on H2A (H2ASer1p, Figure 2Bb, lane 4) is somewhat diminished as well. However, acetylated levels of H3 and H4 are similar among procedures (Figure 2Bb).
SP-purification of core histones and isolation of H2A/H2B from H3/H4
The salt-urea-SP procedure does not permit the recovery of either whole core histones or isolated H2A/H2B. Thus, we have designed a simple, low-pressure, cation-exchange procedure that yields highly purified core histones (Figure 3A and B, lane 15) or isolated H2A/H2B (Figure 3C, lanes 2–5) and H3/H4 (Figure 3C, lane 8) in a single chromatographic step (see flow chart in Figure 3A). The rationale for using SP for core histone isolation and fractionation of core histone moieties (H2A/H2B and H3/H4) is based on the strong interaction that is expected between highly basic proteins and a strong cation-exchanger such as SP. Histones have the highest isoelectric point among proteins. This unique property renders them positively charged even at high pH, contrasting other proteins that would lose their net positive charge at pH ∼8. The highly basic nature of the histones allows them to bind to SP even at relatively high salt concentration (0.2 M NaCl). In addition, separation of H2A/H2B from H3/H4 is readily achieved at lower ionic strength (0.8 M NaCl), since H2A/H2B have a weaker positive charge than H3/H4.
In the acid-SP procedure (Figure 3A), as in the salt-urea method (Figure 1A), cells are rinsed with warm serum-free DMEM medium to ensure minimum metabolic disturbance of the cells (43), after which histones are directly acid-extracted (pH < 1) (Figure 3A). The acidic environment ensures full protein denaturation and inhibition of the enzymes that may affect the histone native PTMs. Next, the histone solution is neutralized with Tris–HCl, pH 8.0 and made 0.2 M with respect to NaCl to allow histone adsorption, while minimizing binding of contaminants, to SP. We have used this strategy to isolate histone chaperones in complex with other proteins (52). As shown in Figure 3B (lane 3), ∼50% of the contaminating proteins were collected in the SP-FT fraction. After washing with 0.5 and/or 0.6 M NaCl (lanes 4–13), the bound core histones were eluted at 2 M NaCl (Figure 2B, lanes 14–16). Note that core histones are separated from histone H1 by HClO4 precipitation (lanes 14–16 and 17–19, respectively).
As mentioned above, an additional advantage of the acid-SP method is its ability to separate H2A/H2B from H3/H4 (Figure 3C, lanes 2–5 and 8, respectively) without the need for HPLC. This procedure does not require salt gradient elution either: H2A/H2B are eluted at 0.8 M NaCl (Figure 3C, lanes 2–5), and highly purified H3/H4 are eluted at 2 M NaCl (Figure 3C, lane 8). In addition to being more convenient and reproducible, the stepwise elution permits extensive washing of the matrix, ensuring that the histones eluted at higher salt concentrations are not contaminated with proteins from the preceding peak (52).
Preservation of histone modifications by the acid-SP histone purification method
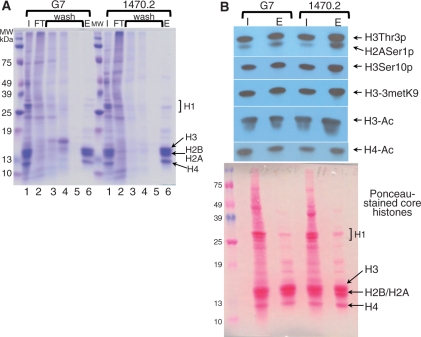
Most important of all, the SP chromatography protects the histone native PTMs by acid-inactivation of unwanted enzymes or by not extracting such enzymes at all (see below and Figure 4). Figure 4 illustrates the preservation of histone phosphorylation, methylation and acetylation in histones purified by the acid-SP method. We have compared the levels of such modifications in input H2SO4-extracted core histones with their SP-purified counterparts from two cell lines (G7 and 1470.2 cells, Figure 4A). Upon acid- extraction, one aliquot was preserved frozen at –20°C as input material for SP (Figure 4A, lanes 1). After SP-core histone purification (Figure 4A, lane 6), histones were resolved by 12.5% SDS–PAGE and transferred onto nitrocellulose membranes. The levels of PTMs were determined with specific antibodies raised against such PTMs (Figure 4B). Input (Figure 4B, lane I) and SP-eluted core histones (Figure 4B, lane E) do not differ significantly in their modification levels; thus the acid-SP histone purification method is suitable for obtaining large quantities of core histones with the preservation of their native PTMs. One modification that may not be protected is the elusive nitrogen phosphorylation, which is sensitive to low pH (61–63). To preserve this particular modification, the nonacid-salt-urea-SP (Figure 2A) should be used.
Figure 4.
Preservation of histone PTMs by the acid-SP method. (A) Coomassie-stained 12.5% SDS–PAGE gel of samples obtained by SP after H2SO4 protein extraction from whole G7 and 1470.2 cells. Lane 1, input (I) H2SO4-crude extracted histone; lane 2, SP-FT fraction; lanes 3–5, 0.6 M NaCl wash; lane 6, core histones eluted at 2 M NaCl (E). (B) Immunoblotting of SP-input (I) and eluted (E) core histones. The antibodies used are indicated on the right of the blots. Bottom panel is a Ponceau S-stained nitrocellulose membrane of transferred histones.
SP purification of hyper-phosphorylated and hyper-acetylated core histones
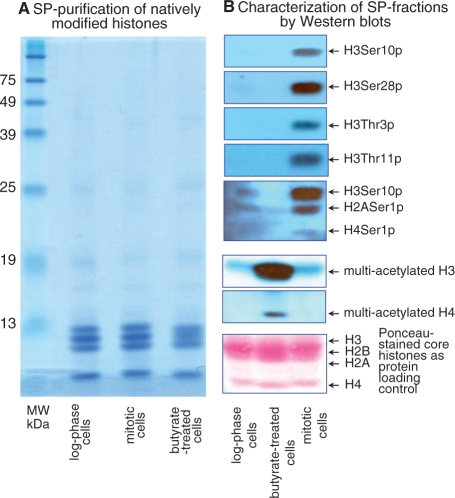
Isolated or in vitro reconstituted polynucleosomes have been extensively used to unveil the role of histone N-termini in chromatin structure (3,64). Acetylation of the N-terminal histone tails has been extensively studied (3,7,8). Much less is known about the effect of histone tail phosphorylation on chromatin structure and function (65). Gene-specific and transient phosphorylation of Ser10 and Ser28 on histone H3 is linked to gene activation (3,8). On the other hand, global and persistent histone hyper-phosphorylation on several H3 residues (Thr3, Ser10, Thr11 and Ser28) and simultaneous phosphorylation of other histones (Ser1 on H2A and on H4) are linked to chromosome condensation (3,8,45). Thus, the most feasible (if not the only) way to obtain fully phosphorylated histones to evaluate their role in chromatin structure and dynamics is by using cell populations synchronized at mitosis. Using mitotically arrested HeLa cells (see ‘Materials and Methods’ section), we evaluated the SP method for its usefulness in purifying and preserving hyper-phosphorylated core histones. We also evaluated the SP method for purifying and preserving hyper-acetylated core histones obtained from HeLa cells treated with 40 mM sodium butyrate (Figure 5) (50,51).
Figure 5.
SP-purification of hyper-phosphorylated and hyper-acetylated core histones from HeLa cells extracted with H2SO4. (A) Coomassie-stained SDS–PAGE of SP-eluted core histones from logarithmically growing HeLa cells (lane log phase cells), metaphase-arrested cells enriched in hyper-phosphorylated core histones (lane mitotic cells) and butyrate-treated cells enriched in hyper-acetylated core histones (lane butyrate-treated cells). (B) Immunoblotting analysis of SP-eluted core histones using antibodies to the most prominent hyper-phosphorylated residues during mitosis (lane mitotic cells), and to globally hyper-acetylated histones H3 and H4 from butyrate-treated HeLa cells (lane butyrate-treated cells) (note that butyrate-treated lane and mitotic cell lane are inverted with respect to the same lanes in the Coomassie-stained gel). The lane log phase cell in (B) is a blotting-control of SP-eluted core histones from logarithmically proliferating HeLa cells; these histones are largely hypo-phosphorylated and hypo-acetylated. The bottom lower panel (in red) is the same membrane stained with Ponceau S, showing transferred core histones, prior to immunoblotting, and serves as a protein loading control.
Coomassie-stained SDS-gel of SP-eluted core histones demonstrates indistinguishable electrophoretic patterns for logarithmic, mitotic, or butyrate-treated cells (Figure 5A). Significantly, immunoblotting analysis with antibodies specific for modifications at specific residues (Figure 5B) clearly shows a dramatic increase in phosphorylation or acetylation of histones purified from metaphase or butyrate-treated cells (compared with logarithmic cells), respectively. Note that no particular enzymatic inhibitors were used during the purification processes. The remarkable degree of preservation of hyper-phosphorylated and acetylated histones is most likely due to inactivation of the respective phosphatases and deacetylases during the acid-extraction step.
H3 separation from H4 or bulk SP-eluted histones by covalent targeting H3 reduced Cys through thiopropyl-Sepharose-6B
Thiol-activated covalent chromatography was originally used to specifically target reduced Cys located within the active cleft of papain (66). The rationale of our method for separating H3 from either bulk histones or H4 exploits the free-sulfhydryl-cysteine residues present on most H3s (67) (but not in other histones) by covalently targeting it with TPS (Figure 6). Bulk histones (Figure 6A and B) or H3/H4 (Figure 6C and D) are denatured in 8-M urea to fully expose the reduced H3 Cys-SH groups to the TPS matrix and to prevent histone–histone interactions during the covalent chromatography (23,27). Nonacid-salt-urea-Sp method, acid-SP and TPS use EDTA to prevent H3-sulfhydryl residues from being oxidized by traces of heavy metals. TPS covalent-bound H3 is recovered by incubating the matrix with 25–50 mM DTT in 8-M urea, 50 mM Tris–HCl, pH 8.0, 2 mM EDTA (Figure 6B and D, lanes 6–10; compare with input histones, lane 1, respectively). We observed that bulk histones (Figure 6B) and highly purified H3 and H4 (Figure 6D) in 8-M urea precipitate poorly, if at all, with TCA or HClO4. Thus, to concentrate the histones and eliminate the urea, the procedure described in Note 4 is recommended. Alternatively, and if convenient, the samples can be dialyzed against an appropriate buffer or water.
Figure 6.
Isolation of H3 from bulk histones (A and B) and from H3/H4 mixtures (C and D) by Thiopropyl Sepharose−6B (TPS) chromatography. TPS contains reactive 2-thiopyridyl disulfide groups attached to Sepharose, allowing H3 capture via its unique reduced Cys. SP-purified core histones (B, lane 1) and SP-purified H3/H4 (D, lane 1) from mouse breast carcinoma 1470.2 cells were incubated with TPS in 8-M urea, 2 mM EDTA, 50 mM Tris–HCl, pH 8.0, to allow maximal exposure of the reduced cysteine on H3s to the immobilized activated thiol groups on the resin. Bulk histones depleted of H3 (B, lanes 2–4) or H4 depleted of H3 (D, lanes 2 and 3) can be recovered in the FT and the washes. H3 is eluted under mild reducing conditions in DTT (B and D, lanes 6–9). Lane 10 contain a 2× SDS–RSB extract of the resin at 100°C, after H3-DTT elution, to verify the effectiveness in H3 elution by DTT. Potential problems with TCA precipitation of diluted column fractions can be overcome by another SP run (Note 4).
Multiple oxidized forms of Cys and Met on H3 have been observed upon lengthy manipulations and storage of histone samples, such as cell lysis, HPLC fractionation and sample lyophilization. The presence of such oxidized forms may compromise the accuracy of MS measurements (19). As the Coomassie-stained gels in Figure 6B and D reveal, the vast majority of H3 has been coupled to TPS (compare the input H3 quantity with that in the FT and the H3 eluates), which suggests a very low level of oxidized SH-groups in these samples. This low oxidation level might be attributed to the very low pH (<1) during the extraction and precipitation procedures (24), the use of 2 mM EDTA throughout the fractionation procedures to chelate heavy metals, and/or the brevity of the entire procedure. One useful property of TPS purification is that it could discriminate between oxidized and reduced thiol groups on H3, thereby allowing capture of only the reduced forms of H3. Of note, our protocols avoid the use of reducing agents, which would interfere with the coupling reaction of H3 to the TPS.
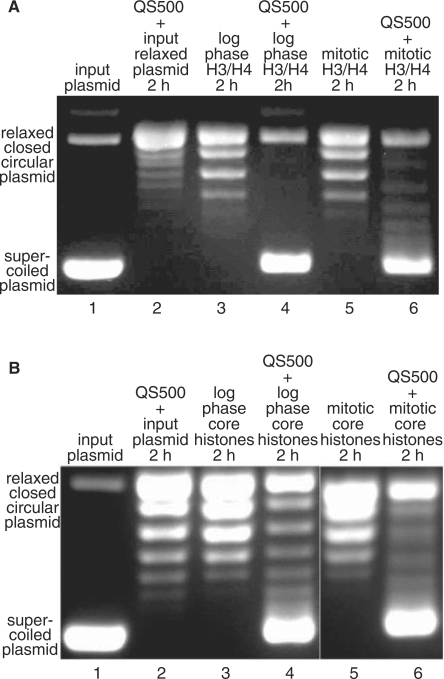
Functionality of purified hyper- and hypo-phosphorylated H3/H4 and core histones: efficient assembly of hyper-phosphorylated mitotic histones into nucleosomes by histone chaperones
Histones denatured by particular chemicals (e.g. 8-M urea, acetonitrile/TFA) can be re-folded simply by replacing the denaturing solution with a physiological one. In effect, structural models of the core particle were obtained with histones solubilized from bacterial inclusion bodies with urea, following their renaturation (58,68). Moreover, nucleosomes reconstituted from RP–HPLC-separated histones are biochemically and biophysically indistinguishable from particles reconstituted with native purified core histones (69). Nevertheless, we assessed the folded state of our purified histones by the ability of a nucleosome assembly fraction containing histone chaperones (52,70) to reconstitute histones into nucleosome-like particles. To that end, we compared the efficiency with which hyper- or hypo-phosphorylated histones are assembled into polytetrasomes and polyoctasomes (nucleosomal arrays containing either [(H3/H4)2]n or [(H3/H4/H2A/H2B)2]n) by acid histone chaperones purified from HeLa cells (52). This is important, since little is known about the ability of hyper-phosphorylated histones to be assembled into nucleosomal-like arrays. Published data have shown that nucleosome assembly protein 1 (Nap1) binds histones H3–H4 by their N-terminal tails (71), therefore we tested if hyper-phosphorylation of mitotic histone tails would interfere with their deposition into nucleosomes by histone chaperones. Therefore, a supercoiling assay was carried out with partially purified histone chaperones from logarithmically proliferating HeLa cells (Q-Sepharose protein fraction eluted at 500 mM NaCl, QS500) (52). The QS500-mediated deposition of folded phosphorylated histones (thereby functional) onto a relaxed closed-circular plasmid in the presence of Topo I results in the formation of tetrasomes and octasomes. Upon removal of the histones, the resulting negatively supercoiled plasmid molecules can be resolved from the more relaxed topoisomers as faster migrating bands in agarose electrophoretic gels. This assay serves as a good assessment of the folded state of isolated histones as unfolded histones would be not able to supercoil relaxed DNA. Figure 7 shows that H3/H4 tetramer (A) and core histone octamer (B) purified from logarithmically grown and M-phase arrested HeLa cells were able to supercoil relaxed DNA in the presence of QS500 fraction (Figure 7A and B, compare relaxed plasmid in lanes 2, 3 and 5 with supercoiled plasmid in lanes 4 and 6). As expected, histone deposition into nucleosomes depends on the presence of both histone chaperones (QS500-fraction) and histones: the QS500 fraction alone or the histones themselves do not have such activity (Figure 7A and B, lanes 2, 3 and 5). This experiment demonstrates that both hypo- and hyper-phosphorylated histones can be efficiently reconstituted into polynucleosomes for further use in chromatin structure and function studies. Hyper-acetylated core histones also supercoiled relaxed plasmid DNA in the presence of QS500 fraction (data not shown).
Figure 7.
Supercoiling assay: purified hyper- and hypo-phosphorylated H3/H4 and core histones are functional and form tetrasomes and octasomes in the presence of chromatin assembly factors. Topo I-relaxed BlueScript plasmid was incubated with the respective histone fractions, which were pre-incubated with Q-Sepharose-isolated nucleosome assembly fraction (QS500) from HeLa cells. The topoisomer distribution in the plasmid population was analyzed by 0.9% agarose gel electrophoresis following proteinase K treatment and phenol/chloroform/isoamyl alcohol extraction. Reconstitution with M-phase hyper-phosphorylated H3/H4 histones (A) or core histones (B). Histones isolated from logarithmically proliferating cells, hence hypophosphorylated, were used in parallel. The components of the reaction mixtures and the histones used are indicated at the top of the figure.
CONCLUSIONS
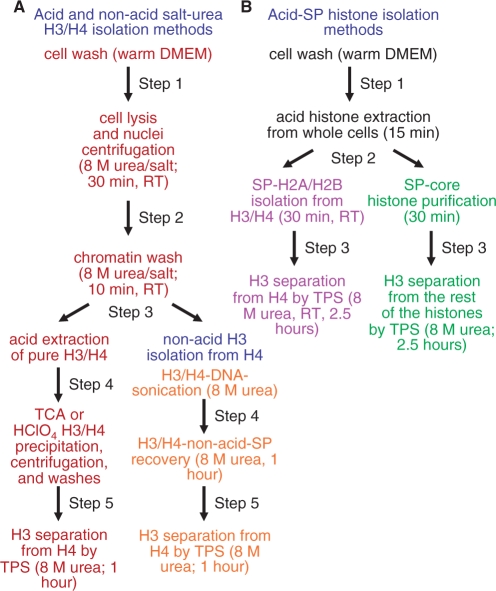
Figure 8 summarizes the main steps of the methods reported here. The essential feature of our methods is the use—at onset of histone isolation—of denaturing solutions such as 8-M urea or sulfuric acid to better preserve certain histone modifications (Figures 2, 4 and 5). Indeed, using conventional methodology that does not use denaturing solvents (34), leads to a drastic histone dephosphorylation (Figure 2). Moreover, the methods reported here are shorter, simpler, less expensive and more flexible. For example, the HPLC is dispensable for obtaining pure H3 and H4. In addition, if one is interested in isolating H3 alone, this can be done through two simple low-pressure chromatographic steps: SP-bulk core histones isolation (Figures 3B and 4A), followed by TPS-H3 separation from bulk histones [Figure 6A and B; Figure 8, chart B, steps (ii) and (iii) outlined in green]. Additionally, highly purified H3 and H4 can be obtained by nonacid extraction methods—but, importantly, under denaturing conditions—which can be valuable for the detection acid-labile modifications [Figures 2 and 8, chart A, steps (i)–(iii), outlined in red; followed by steps (iv) and (v) outlined in orange]. Notably, the preservation of fully phosphorylated mitotic histones (Figure 5) and their assembly into polynucleosomes (Figure 7) would open new avenues to investigate an important but understudied question: the impact of mitotic histone phosphorylation for chromatin structure and function.
Figure 8.
Flow charts of the newly developed histone purification and isolation protocols from mammalian cells. The different colors indicate different strategies for histone purification.
FUNDING
National Cancer Institute (CA122177 to P.R.C.); National Institutes of Health (GM077872 to S.H.L.); and National Science Foundation (0504239 to J.Z.). Funding for open access charge: National Institutes of Health. Grants CA122177 and GM077872.
Conflict of interest statement. Pedro Rodriguez-Collazo, Sanford H. Leuba, and Jordanka Zlatanova are authors of a patent of invention based on this manuscript. The invention is a property of University of Pittsburgh, at Pittsburgh, and has been licensed to Active Motif.
ACKNOWLEDGEMENTS
Many thanks to Karen Leyden for her comments on the manuscript and to Michelle L. Kienholz for critical reading of the manuscript.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Davie JR. In: Chromatin Structure and Dynamics: State-of-the Art. Zlatanova J, Leuba SH, editors. Amsterdam: Elsevier; 2004. pp. 205–240. [Google Scholar]
- 4.Smith CM. Quantification of acetylation at proximal lysine residues using isotopic labeling and tandem mass spectrometry. Methods. 2005;36:395–403. doi: 10.1016/j.ymeth.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Iizuka M, Smith MM. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 2003;13:154–160. doi: 10.1016/s0959-437x(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 6.Smith CM, Haimberger ZW, Johnson CO, Wolf AJ, Gafken PR, Zhang Z, Parthun MR, Gottschling DE. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl Acad. Sci. USA. 2002;99:16454–16461. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An W. Histone acetylation and methylation: combinatorial players for transcriptional regulation. Subcell. Biochem. 2007;41:351–369. [PubMed] [Google Scholar]
- 8.Bode AM, Dong Z. Inducible covalent posttranslational modification of histone H3. Sci STKE 2005. 2005:re4. doi: 10.1126/stke.2812005re4. [DOI] [PubMed] [Google Scholar]
- 9.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–2574. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 10.Galasinski SC, Resing KA, Ahn NG. Protein mass analysis of histones. Methods. 2003;31:3–11. doi: 10.1016/s1046-2023(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 11.Galasinski SC, Louie DF, Gloor KK, Resing KA, Ahn NG. Global regulation of post-translational modifications on core histones. J. Biol. Chem. 2002;277:2579–2588. doi: 10.1074/jbc.M107894200. [DOI] [PubMed] [Google Scholar]
- 12.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4:1382–1396. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 13.Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- 14.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J. Biol. Chem. 2007;282:27923–27934. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 17.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 18.Boyne MTII, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome. Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 19.Pesavento JJ, Garcia BA, Streeky JA, Kelleher NL, Mizzen CA. Mild performic acid oxidation enhances chromatographic and top down mass spectrometric analyses of histones. Mol. Cell Proteomics. 2007;6:1510–1526. doi: 10.1074/mcp.M600404-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Tordera V, Sendra R, Perez-Ortin JE. The role of histones and their modifications in the informative content of chromatin. Experientia. 1993;49:780–788. doi: 10.1007/BF01923548. [DOI] [PubMed] [Google Scholar]
- 21.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 22.Schnitzler GR. Isolation of histones and nucleosome cores from mammalian cells. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb2105s50. Chapter 21, Unit 21.5. [DOI] [PubMed] [Google Scholar]
- 23.Lorch Y, Kornberg RD. Isolation of the yeast histone octamer. Proc. Natl Acad. Sci. USA. 1994;91:11032–11034. doi: 10.1073/pnas.91.23.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogakou EP, Redon C, Boon C, Johnson K, Bonner WM. Rapid histone extraction for electrophoretic analysis. Biotechniques. 2000;28:38–40, 42, 46. doi: 10.2144/00281bm06. [DOI] [PubMed] [Google Scholar]
- 25.Simon RH, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloom KS, Anderson JN. Fractionation and characterization of chromosomal proteins by the hydroxyapatite dissociation method. J. Biol. Chem. 1978;253:4446–4450. [PubMed] [Google Scholar]
- 27.Bode J, Wagner KG. The use of subunit exchange chromatography for the group specific fractionation of histones. Biochem. Biophys. Res. Commun. 1975;62:868–876. doi: 10.1016/0006-291x(75)90403-9. [DOI] [PubMed] [Google Scholar]
- 28.Bolund LA, Johns EW. The selective extraction of histone fractions from deoxyribonucleoprotein. Eur. J. Biochem. 1973;35:546–553. doi: 10.1111/j.1432-1033.1973.tb02871.x. [DOI] [PubMed] [Google Scholar]
- 29.Johns EW. A method for the selective extraction of histone fractions f2(a)1 and f2(a)2 from calf thymus deoxyribonucleoprotein at pH7. Biochem. J. 1967;105:611–614. doi: 10.1042/bj1050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johns EW, Bulter JA. Studies on histones. 4. The histones of wheat germ. Biochem. J. 1962;84:436–439. doi: 10.1042/bj0840436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johns EW, Forrester S, Riches PL. A method for the large-scale preparation of the two halves of histone fraction F2B. Arch. Biochem. Biophys. 1972;152:287–290. doi: 10.1016/0003-9861(72)90217-2. [DOI] [PubMed] [Google Scholar]
- 32.Murray K. The acid extraction of histones from calf thymus deoxyribonucleoprotein. J. Mol. Biol. 1966;15:409–419. doi: 10.1016/s0022-2836(66)80116-x. [DOI] [PubMed] [Google Scholar]
- 33.Kizer KO, Xiao T, Strahl BD. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods. 2006;40:296–302. doi: 10.1016/j.ymeth.2006.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 35.Gurley LR, Prentice DA, Valdez JG, Spall WD. High-performance liquid chromatography of chromatin histones. J. Chromatogr. 1983;266:609–627. doi: 10.1016/s0021-9673(01)90931-8. [DOI] [PubMed] [Google Scholar]
- 36.Lindner H, Helliger W, Puschendorf B. Histone separation by high-performance liquid chromatography on C4 reverse-phase columns. Anal. Biochem. 1986;158:424–430. doi: 10.1016/0003-2697(86)90570-1. [DOI] [PubMed] [Google Scholar]
- 37.Bonner WM, West MH, Stedman JD. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur. J. Biochem. 1980;109:17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- 38.Holloway AF, Oakford PC. Targeting epigenetic modifiers in cancer. Curr. Med. Chem. 2007;14:2540–2547. doi: 10.2174/092986707782023271. [DOI] [PubMed] [Google Scholar]
- 39.Santell L, Rubin RL, Levin EG. Enhanced phosphorylation and dephosphorylation of a histone-like protein in response to hyperosmotic and hypoosmotic conditions. J. Biol. Chem. 1993;268:21443–21447. [PubMed] [Google Scholar]
- 40.Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 1998;111:3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 41.Ajiro K, Yoda K, Utsumi K, Nishikawa Y. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 1996;271:13197–13201. doi: 10.1074/jbc.271.22.13197. [DOI] [PubMed] [Google Scholar]
- 42.Gurley LR, D'Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Collazo P, Snyder SK, Chiffer RC, Zlatanova J, Leuba SH, Smith CL. cAMP signaling induces rapid loss of histone H3 phosphorylation in mammary adenocarcinoma-derived cell lines. Exp. Cell Res. 2008;314:1–10. doi: 10.1016/j.yexcr.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolby TN, Ajiro K, Borun TW, Gilmour RS, Zweidler A, Cohen L, Miller P, Nieolini C. Physical properties of DNA and chromatin isolated from G1- and S-phase HeLa S-3 cells. Effects of histone H1 phosphorylation and stage-specific nonhistone chromosomal proteins on the molar ellipticity of native and reconstituted nucleoproteins during thermal denaturation. Biochemistry. 1979;18:1333–1344. doi: 10.1021/bi00574a033. [DOI] [PubMed] [Google Scholar]
- 45.Barber CM, Turner FB, Wang Y, Hagstrom K, Taverna SD, Mollah S, Ueberheide B, Meyer BJ, Hunt DF, Cheung P, et al. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma. 2004;112:360–371. doi: 10.1007/s00412-004-0281-9. [DOI] [PubMed] [Google Scholar]
- 46.Halicka HD, Huang X, Traganos F, King MA, Dai W, Darzynkiewicz Z. Histone H2AX phosphorylation after cell irradiation with UV-B: relationship to cell cycle phase and induction of apoptosis. Cell Cycle. 2005;4:339–345. [PubMed] [Google Scholar]
- 47.Spackman DH, Stein WH, Moore S. The disulfide bonds of ribonuclease. J. Biol. Chem. 1960;235:648–659. [PubMed] [Google Scholar]
- 48.Allen CM, Jr, Jones ME. Decomposition of carbamylphosphate in aqueous solutions. Biochemistry. 1964;3:1238–1247. doi: 10.1021/bi00897a010. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Patron C, Castellanos-Serra L, Rodriguez P. Reverse staining of sodium dodecyl sulfate polyacrylamide gels by imidazole-zinc salts: sensitive detection of unmodified proteins. Biotechniques. 1992;12:564–573. [PubMed] [Google Scholar]
- 50.Riggs MG, Whittaker RG, Neumann JR, Ingram VM. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977;268:462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- 51.Vidali G, Boffa LC, Bradbury EM, Allfrey VG. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl Acad. Sci. USA. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez P, Ruiz MT, Price GB, Zannis-Hadjopoulos M. NAP-2 is part of multi-protein complexes in HeLa cells. J. Cell Biochem. 2004;93:398–408. doi: 10.1002/jcb.20163. [DOI] [PubMed] [Google Scholar]
- 53.Zangi R, Hagen M, Berne BJ. Effect of ions on the hydrophobic interaction between two plates. J. Am. Chem. Soc. 2007;129:4678–4686. doi: 10.1021/ja068305m. [DOI] [PubMed] [Google Scholar]
- 54.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 55.Jencks WP. Catalysis in Chemistry and Enzymology. Mineola, NY: Dover Publication; 1987. [Google Scholar]
- 56.Rodriguez P, Fuentes P, Munoz E, Riveros D, Orta D, Alburqerque S, Perez S, Besada V, Herrera L. The streptokinase domain responsible for plasminogen binding. Fibrinolysis. 1994;8:276–285. [Google Scholar]
- 57.Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl Acad. Sci. USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 59.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 60.Tan F, Li G, Chitteti BR, Peng Z. Proteome and phosphoproteome analysis of chromatin associated proteins in rice (Oryza sativa) Proteomics. 2007;7:4511–4527. doi: 10.1002/pmic.200700580. [DOI] [PubMed] [Google Scholar]
- 61.Besant PG, Attwood PV. Mammalian histidine kinases. Biochim. Biophys. Acta. 2005;1754:281–290. doi: 10.1016/j.bbapap.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 62.Matthews HR, Huebner VD. Nuclear protein kinases. Mol. Cell Biochem. 1984;59:81–99. doi: 10.1007/BF00231306. [DOI] [PubMed] [Google Scholar]
- 63.Chen CC, Smith DL, Bruegger BB, Halpern RM, Smith RA. Occurrence and distribution of acid-labile histone phosphates in regenerating rat liver. Biochemistry. 1974;13:3785–3789. doi: 10.1021/bi00715a026. [DOI] [PubMed] [Google Scholar]
- 64.van Holde KE. Chromatin. New York: Springer; 1988. [Google Scholar]
- 65.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Brocklehurst K, Carlsson J, Kierstan MP, Crook EM. Covalent chromatography. Preparation of fully active papain from dried papaya latex. Biochem. J. 1973;133:573–584. doi: 10.1042/bj1330573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marzluff WF, Jr, Sanders LA, Miller DM, McCarty KS. Two chemically and metabolically distinct forms of calf thymus histone F3. J. Biol. Chem. 1972;247:2026–2033. [PubMed] [Google Scholar]
- 68.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 69.Moore SC, Rice P, Iskandar M, Ausio J. Reconstitution of native-like nucleosome core particles from reversed-phase-HPLC-fractionated histones. Biochem. J. 1997;328:409–414. doi: 10.1042/bj3280409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, et al. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 1997;44:253–265. doi: 10.1006/geno.1997.4868. [DOI] [PubMed] [Google Scholar]
- 71.McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]