Abstract
Gene families compose a large proportion of eukaryotic genomes. The rapidly expanding genomic sequence database provides a good opportunity to study gene family evolution and function. However, most gene family identification programs are restricted to searching protein databases where data are often lagging behind the genomic sequence data. Here, we report a user-friendly web-based pipeline, named TARGeT (Tree Analysis of Related Genes and Transposons), which uses either a DNA or amino acid ‘seed’ query to: (i) automatically identify and retrieve gene family homologs from a genomic database, (ii) characterize gene structure and (iii) perform phylogenetic analysis. Due to its high speed, TARGeT is also able to characterize very large gene families, including transposable elements (TEs). We evaluated TARGeT using well-annotated datasets, including the ascorbate peroxidase gene family of rice, maize and sorghum and several TE families in rice. In all cases, TARGeT rapidly recapitulated the known homologs and predicted new ones. We also demonstrated that TARGeT outperforms similar pipelines and has functionality that is not offered elsewhere.
INTRODUCTION
A major discovery of eukaryote genome projects is that unexpectedly large numbers of genes are members of gene families. Gene families comprise 49% of the genes in Caenorhabditis elegans, 41% in Drosophila melanogaster, 38% in Homo sapiens, 65% in Arabidopsis thaliana and 77% in Oryza sativa L. ssp. japonica (1–5). Variation in the sizes of gene families among closely related species indicates that gene duplication and gene family diversification is an ongoing process (6–8).
Duplicate genes arise in several ways including whole-genome duplication (9–11) and segmental duplication (12,13). Segmental duplication events can be further classified into tandem and interspersed (14). A tandem duplication event can result from either homologous (15) or nonhomologous recombination mechanisms (16), while interspersed duplication events are mainly caused by the activity of transposable elements (TEs) (17–20).
Gene family members can be detected by clustering genes based on their similarity (21,22), and new members can be identified through similarity comparison to known members. Many gene family databases have been established, including Pfam (23), TreeFam (24) and PANTHER (25), etc. While these gene family databases are useful recourses, they are not updated at the same rapid pace as that of newly generated genomic sequences. Researchers interested in particular gene families often have to perform their own searches to obtain the most current collection of sequences.
The identification of gene family members using sequence similarity searches is often complicated by the detection of homologs from other gene families. Phylogenetic analysis is a powerful tool to identify homologs of interest and to provide additional information about gene function and evolution. To this end, researchers can perform manual searches using publicly available programs such as BLAT (26), Wise2 (27), BLAST (28), FASTA (29) and HMMER (30), followed by sequence alignment and phylogenetic analysis. However, these procedures can be complicated as they often require extensive manual curation, particularly if homologous regions need to be extracted from genomic sequences. While this is a manageable problem for a small gene family, it can be a tedious and time-consuming process when the target gene family is large. More significantly, the quality of the results often suffers.
In addition to the more traditional gene families, TEs can also be viewed as members of ‘special’ gene families that are able to duplicate themselves by the activity of element-encoded proteins. TEs often constitute the largest component of eukaryotic genomes, and their identification and classification are essential to accurate genome annotation (31,32). However, as with large gene families, the very high copy numbers of some TEs make their retrieval from genomic sequence and characterization an extremely difficult task. The increasing pace of genomic sequencing projects demands a computer-assisted pipeline that can rapidly and accurately identify and characterize gene families.
Several automated pipelines have been developed to ease homolog identification and most are limited to protein or expressed sequence tag (EST) databases. For example, PhyloBLAST (33), Pyphy (34), HoSeql (35), PhyloGena (36) and TRIBE-MCL (37) perform BLASTP searches and retrieve data from protein databases. SimESTs uses TBLASTN to search EST databases (38). Because these programs only compare protein-coding sequences, they will miss any mutational events that occur within noncoding regions.
TARGeT (Tree Analysis of Related Genes and Transposons) is a program to streamline the process of retrieving, annotating and analyzing both gene families and TE families from a genomic database. The core of the TARGeT pipeline is an algorithm called putative homolog identifier (PHI) that uses a series of steps to predict gene structure using BLAST results. From the predicted gene structure, PHI extracts the amino acid sequences of putative homologs for use in subsequent phylogenetic analysis. We have compared TARGeT with two pipelines, FGF and GFScan, which can also be used to retrieve gene families from genomic databases. Results are presented showing that TARGeT significantly outperforms both programs and adds several layers of functionality not present in existing programs. To make it easier for users, especially nonspecialists, TARGeT was implemented as a user-friendly web-based pipeline (http://target.iplantcollaborative.org/). All initial input for TARGeT is organized on a web form and the results are presented in the browser. All results and supporting files are documented and are available for download. TARGeT provides several points where results can be inspected and analyses can be repeated.
METHODS
TARGeT can use either protein or DNA sequence as the query. BLASTN searches are used for DNA queries, while TBLASTN is used for protein queries. The pipeline that uses TBLASTN is the focus of this article because it is more complex and may have wider application. TARGeT uses Muscle (39) to calculate the multiple alignment and TreeBest (24) to generate the phylogenetic tree of the putative homologs with the neighbor-joining method (40). The other functions of TARGeT are carried out by several Perl scripts developed by the authors.
Rice genomic data were obtained from Genbank (41,42) with accession numbers from NC_008394 to NC_008405. Maize genomic data were downloaded from the Maize Genome Sequencing Project (http://www.maizesequence.org; version: Dec. 2008). Sorghum genomic data were from the Sorghum Bicolor Genome Project (http://www.jgi.doe.gov; version: 2008 Sorbi1 assembly).
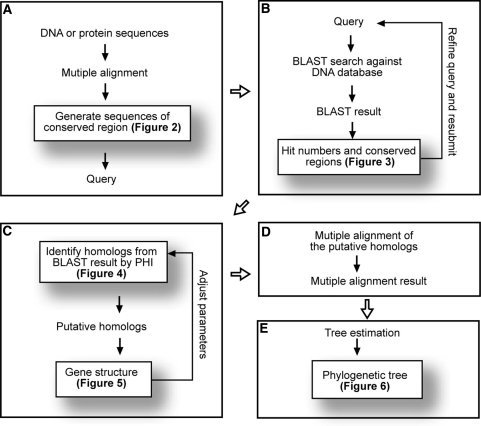
There are five main steps in the TARGeT pipeline with a checkpoint at the end of each: (i) preparation of the query when multiple sequences are to be submitted, (ii) BLAST search (either BLASTN or TBLASTN), (iii) homolog prediction, (iv) multiple alignment and (v) phylogenetic tree estimation (Figure 1). Details of each step are presented in the ‘Results’ section using the ascorbate peroxidase (APx) gene family as an example.
Figure 1.
Map of the five main steps of the TARGeT pipeline. Users are able to inspect the results of each step before going on to the next step. (A) Preparation of the query when more than one sequence is being used. This is an optional step and its output is shown in Figure 2. (B) BLAST search. Results are shown in Figure 3. (C) Homolog identification by PHI. The algorithm is explained in Figure 4 and the result of this step is shown in Figure 5. (D) Multiple alignment. (E) Tree building.
TARGeT can be accessed on a web server, where all data used and generated by TARGeT are entered in a log file. TARGeT output is presented in a single webpage that uses nested tabs to organize the data, images and re-submission forms for each TARGeT run during a session. There is a final tab for each run called Provenance, where the user can view the parameters used by TARGeT in a log file and also download an archive that includes all files and images for offline viewing and analysis. The output includes the XML log file, BLAST results in image and text format, PHI results in image and text format, multiple alignments in FASTA format and the phylogenetic tree in Newick and jpeg formats.
RESULTS
Searching for APx gene family in rice
Rice and Arabidopsis serve as model plant monocot and dicot species, respectively. They diverged from a common ancestor 120–200 million years ago (43) and their genomes are fully sequenced (1,2,44). Thus, they provide excellent opportunities to evaluate the cross-species searching ability of TARGeT. We searched the rice APx gene family using the Arabidopsis APx protein sequence as query and compared the results generated by TARGeT to the published data. The goal of this exercise was to see how well TARGeT would perform at predicting the rice APx family members. We chose APx because it is a small but important gene family that has been well annotated in both Arabidopsis and rice. Based on the literature, there are as many as nine APx family members in Arabidopsis (45) and eight in rice (46) (Table 1). The APx family shares sequence similarity with several other peroxidase families (47) and, as such, is a good dataset to test the ability of TARGeT to discriminate between closely related protein families.
Table 1.
The APx gene family homologs of Arabidopsis, rice, maize and sorghum
|
Arabidopsis |
Rice |
Maize | Sorghum | |||||
|---|---|---|---|---|---|---|---|---|
| Gene namea | Accession no. | TARGeT IDb | Gene namea | Accession no. | Missed rate (%)c | Error rate (%)d | TARGeT IDb | TARGeT IDb |
| APX1 | AT1G07890 | TOAPx_1 | – | Os09g0538600 | – | – | TZAPx_1 | TSAPx_1 |
| APX2 | AT3G09640 | TOAPx_2 | OsAPx4 | Os08g0549100 | 0.41 | 0.41 | TZAPx_2 | TSAPx_2 |
| APX3 | AT4G35000 | TOAPx_3 | OsAPx7 | Os04g0434800 | 0 | 0 | TZAPx_3 | TSAPx_3 |
| APX4 | AT4G09010 | TOAPx_4 | OsAPx6 | Os12g0178100 | 4.03 | 0.37 | TZAPx_4 | TSAPx_4 |
| APX5 | AT4G35970 | TOAPx_5 | OsAPx1 | Os03g0285700 | 1.62 | 0.4 | TZAPx_5 | TSAPx_5 |
| APX6 | AT4G32320 | TOAPx_6 | OsAPx5 | Os12g0178200 | 0 | 1.1 | TZAPx_6 | TSAPx_6 |
| APX7 | AT1G33660 | TOAPx_7 | OsAPx3 | Os04g0223300 | 1.63 | 1.22 | TZAPx_7 | TSAPx_7 |
| SAPX | AT1G77490 | TOAPx_8 | OsAPx8 | Os02g0553200 | 0 | 0 | TZAPx_8 | TSAPx_8 |
| TAPX | AT4G08390 | TOAPx_9 | – | Os08g0522400 | – | – | TZAPx_9 | TSAPx_9 |
| TOAPx_10 | OsAPx2 | Os07g0694700 | 1.22 | 0.41 | TZAPx_10 | |||
| TOAPx_11 | – | Os04g0602100 | – | – | TZAPx_11 | |||
aNames used for previously identified APx genes.
bNames assigned by TARGeT to predicted APx homologs.
cThe ‘missed’ rate is calculated by dividing the number of missed amino acid residues that are at the ends of the sequence by the length of the query.
dThe ‘error’ rate is calculated by dividing the number of the incorrect amino acid assignments by the length of the corresponding region in the previously published rice APx protein sequence.
BLAST search
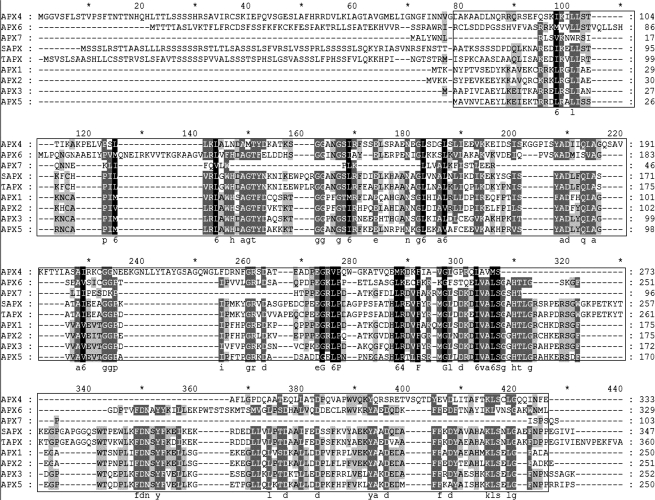
To improve the chances of finding target gene family members, multiple queries can be submitted as long as they are homologs. An optional multiple alignment step is provided for users to select sequences from conserved regions (Figure 1A). As an example, for the APx gene family we selected as query the sequences from the well-aligned (boxed) region in Figure 2.
Figure 2.
Multiple alignment of Arabidopsis APx protein sequences. Sequences in the boxed region were extracted to form the query sequences. APx7 was not included because it aligns poorly.
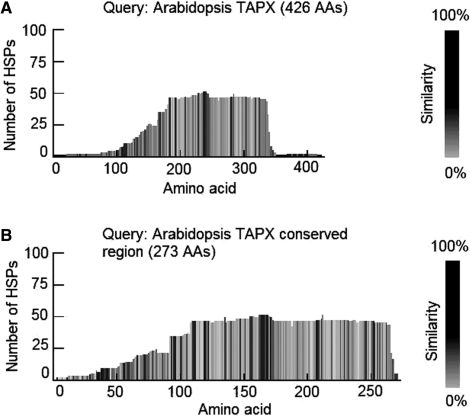
To aid users in viewing the BLAST result, TARGeT produces an image showing a rough estimation of BLAST high scoring pair (HSP) numbers and conserved regions along the length of each query sequence (Figures 1B and 3). This is helpful for a quick overall view especially when the BLAST output is large. In this way, the user can see the information used by TARGeT and, if necessary, modify the query in a subsequent BLAST search. For example, TAPX, which is one of the Arabidopsis APx genes, is 426 amino acids. Using this full-length sequence as the query, low copy regions can be detected at the beginning and at the end (Figure 3A). Readers should note that the number of HSPs (up to 50) is much larger than the number of known APx genes in the rice genome. This inconsistency is largely due to the existence of other gene families that share sequence similarity with the APx gene family. As shown in later steps, true homologs belonging to the APx gene family will be discerned from those of other families. Using the full-length TAPX sequence as the query, only three APx homologs were found in rice (data not shown). However, five APx homologs were found when the sequence from the boxed region (see Figure 2) was used as the new query (Figure 3B).
Figure 3.
TARGeT output provides a rough visualization of the BLAST result. X-axis is the length of the query; Y-axis is the number of BLAST HSPs. The gray gradient shows the similarity which is calculated by dividing the sum of identities and similarities by the number of the aligned amino acids along the HSP. Darker represents higher similarity at that position.
Putative homolog identification
Several factors make it difficult to identify reliable homologs from BLAST output and result in a high false positive rate (48–50). Lack of explicit treatment of frameshifts and introns is also a disadvantage of TBLASTN (51). To solve these problems, we developed a program called PHI, which takes into account the e-value (default 0.01) as well as a second parameter called the minimal match percentage (MMP, defaults to 70%) to find reliable homologs. The two main stages in PHI (grouping and refinement) are explained below.
Grouping
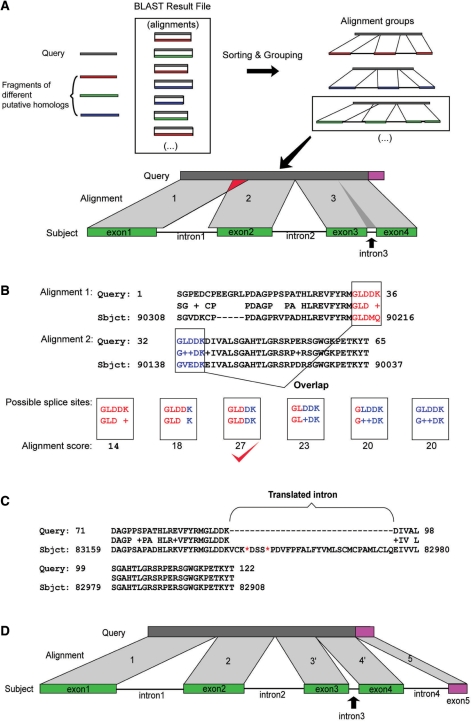
Introns or low-similarity regions can break a complete alignment into smaller HSPs. In addition when a frameshift occurs, TBLASTN produces separate HSPs. To retrieve the intact sequence of each homolog or pseudogene, PHI sorts the HSPs based on position and strand in the genomic sequence. In this step, HSPs that are from the same homolog are grouped together by the sequence position of query and subject (Figure 4A, top part). Two HSPs are assumed to belong to different groups if they are separated by a distance greater than the minimum intron length (a parameter adjustable by the user, defaults to 8000 nt) or if they are on different strands. When there is more than one way to connect the HSPs (which can happen when there are repetitive domains in the query), PHI uses an overall HSP score to determine the correct order. A match percentage is calculated by dividing the sum length of the matches in each group by the length of the query. If this number is greater than the MMP, (see Figure 2) the group is sent to the refinement stage. HSPs that fail to satisfy the MMP are available to interested users as a record file.
Figure 4.
The sorting and refinement stages of the PHI program. See the text for details. (A) In the grouping stage, alignments are sorted and grouped. Dark bars are queries and colored bars are homologs. Each group corresponds to one putative homolog. The green group is shown in detail to illustrate potential problems. (B) Two overlapping HSPs together with six possible alternative positions are shown. The separation that produces the highest score in the overlapping region is noted with a red check. (C) An HSP that includes an intron. The intron is detected and cut out by PHI, resulting in two separated HSPs. Red asterisks represent premature stop codons. (D) Figure presentation of the result after the refinement stage. There is no overlap between HSPs 1 and 2. HSP 3 in (C) is separated by the small intron into new HSP 3′ and 4′. An additional exon (5) was found and is shown in pink.
Refinement
After the grouping step, several potential problems often remain in the HSPs of each group. A demonstration figure to illustrate some problems is shown in the lower part of Figure 4A. First, there is an overlap (indicated by a red triangle) between HSP 1 and HSP 2; second, there is an intron (darker area) that has been falsely translated and included in HSP 3; finally, there is a small area in the query (pink region) that has no HSP in the BLAST result, which results in the failure to detect a small exon due to its insignificant e-value. In the refinement stage, the most likely split position is detected within the overlapping region. Introns in the HSPs are removed, and a second round BLAST search is performed to find the missing exons. Several result files will be generated after this step, including the homologous sequences in both DNA and protein FASTA formats.
Resolving the boundary between two overlapping HSPs
In TBLASTN outputs, two successive HSPs often overlap due to coincident similarity beyond the true boundaries, resulting in misalignment between the query and the subject. An example is shown in Figure 4B (boxed regions). In this example, the end of HSP 1 overlaps the beginning of HSP 2 by five amino acids corresponding to amino acids 32–36 of the query (red residues GLDDK), 90212–90216 (red residues GLDMQ) and 90138–90142 (blue residues GVEDK) of the subject. PHI determines the most likely correct boundary by choosing the alignment that has the highest alignment score from all of the possible alignments within the overlapping region. For the example shown in Figure 4B, there are six possible alignments. A score is calculated for each alignment using the BLOSUM62 matrix and any amino acid that aligns to a gap or a stop codon will be penalized 12 points. The third alignment in Figure 4B has the highest score and thus PHI assumes that the true boundary in the subject is between the two aspartic acid residues. After the true boundary is located, additional amino acids will be trimmed off the HSPs (MQ in HSP 1 and GVE in HSP 2). For the rice APx gene family, this step trimmed 21 amino acid residues on average from each homolog.
Identifying small introns
The function of this step is to identify and remove introns that appear as gaps within the HSPs. Any gap in the subject that has a length greater than the minimum intron length parameter (user-adjustable parameter, default 60 nt) is identified as an intron and will be removed resulting in two (smaller) new HSPs (Figure 4C and D). For each rice APx homolog, TARGeT identified, on average, 1.3 introns corresponding to 41.9 falsely translated amino acids.
Identifying small exons
Small exons will be missed by BLAST searches when their alignments do not meet the e-value cut-off. Such small exons may be found by increasing the e-value. However, for a large database, simply increasing the e-value could increase the computational burden of TARGeT, and there is no guarantee that all exons will be identified because the suitable e-value is unknown. To improve the prediction of small exons, PHI can perform a second round BLAST search, using a small database containing only the sequences of putative homologs (including the predicted intronic and flanking regions). Because e-value calculation is dependent in part on the size of the database, short alignments to the original query sequence(s) may now be significant (Figure 4D). For each rice APx homolog, this second round of BLAST identified, on average, 1.6 additional exons and 33.4 amino acids.
Illustration of PHI output
After the refinement stage, an image is generated that provides a view of the predicted gene structure for each putative homolog (Figure 1C and Supplementary Figure 1). Features of this image include the similarity between each putative homolog and its query, the locations of exons, introns, premature stop codons (represented by asterisks in the BLAST output) and frameshifts. Frameshifts are identified by comparing HSPs that are close to each other (less than 5 amino acids by default) and are on the same strand but are in different reading frames. In the demonstration figure, putative pseudogenes may be genes with premature stop codons or frameshifts that are marked with red or blue dots, respectively (Supplementary Figures 7 and 8).
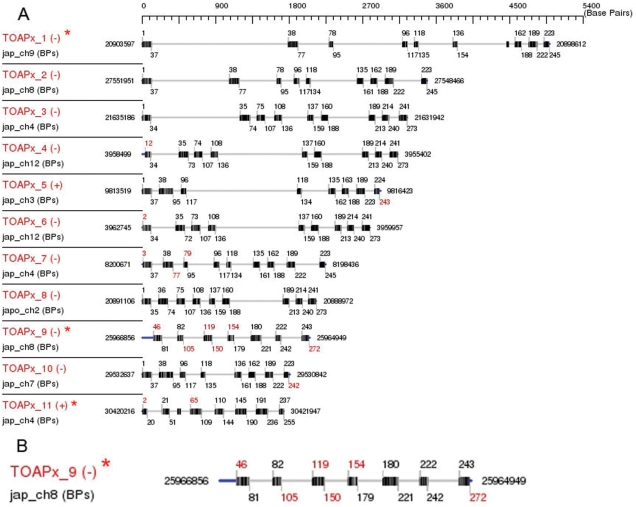
Using default parameters, 46 putative rice APx homologs were identified and clustered into two groups based on their gene structures (Figure 5 and Supplementary Figure 1). There are 11 homologs in the small group, among which, TOAPx_2-8 and TOAPx_10 were found to correspond to known rice APx genes OsAPx1-OsAPx8 (Table 1). For the remaining 35 putative homologs, comparison of their sequences and gene structures revealed that they are not APx homologs but are instead from other peroxidase gene families (data not shown).
Figure 5.
TARGeT output of the gene structure of rice APx family members. (A) Exon intron structure of 11 reliable rice APx homologs detected by TARGeT. All 46 putative homologs are in Supplementary Figure 1. (B) A larger figure of TOAPx_9 from (A). Query and subject names are shown on the left. ‘+’ or ‘−’ indicates the strand of the hit. Unmatched query regions at the ends of each homolog are in blue. Black or gray gradient bars represent the exons. Darker represents higher similarity. Numbers flanking each gene structure are positions of the subject, while numbers above and below the exons are the positions of the query. Red numbers indicate discontinuous predicated exons. Putative new APx homologs are indicated by ‘*’.
To assess the accuracy of homolog sequences retrieved by TARGeT, we considered two situations (this is not a step of TARGeT). One situation might occur at the ends of the query-target alignment where the program failed to identify some amino acids at the end. We refer to this as ‘missing’ and can occur when the end of homolog sequences are not as well conserved as the sequences within. By comparing the homolog sequence to the query sequence, the numbers of ‘missing’ amino acids were counted manually. For example, if the query is 100 amino acids and the alignment is from 5 to 97, the missing number of this homolog is 4 + 3 = 7. The ‘missed’ rate is calculated by dividing the number of missed amino acids by the length of the query (7% in the above example). In contrast, we refer to an ‘error’ as a situation where the program incorrectly predicts amino acids within a homolog sequence. By comparing the homolog sequence to the previously published rice APx protein sequence, mismatched amino acids were counted manually as the ‘error’ number of this homolog. The ‘error’ rate is calculated by dividing the number of incorrect amino acid assignments by the length of the corresponding region in the previously published rice APx protein sequence. The missed and error rates may vary for each predicted homolog sequence because they depend on the level of conservation between the homolog and the query sequences. For the rice APx example above, the average missed rate is 1.11% and the average error rate is 0.49% (Table 1).
Multiple alignment and tree estimation
If users are satisfied with the putative homologs found by TARGeT, they can either download the sequences in FASTA format or let TARGeT use the data to generate a phylogenetic tree. Users also have the option to employ other tree estimation methods by downloading the alignment and using the software of their choice. The phylogenetic tree and the figure showing the tree are generated by TreeBest. When there are many homologs, names on the figure will be difficult to read because the figure size cannot be varied. To solve this problem, users can download the newick file and draw their own tree using software such as TreeView (52). We have also provided two more solutions on the server. The first is to use Jalview (53) and the second is to copy the newick format tree file and submit it to PhyloWidget (54), which is a powerful web-based tree viewer.
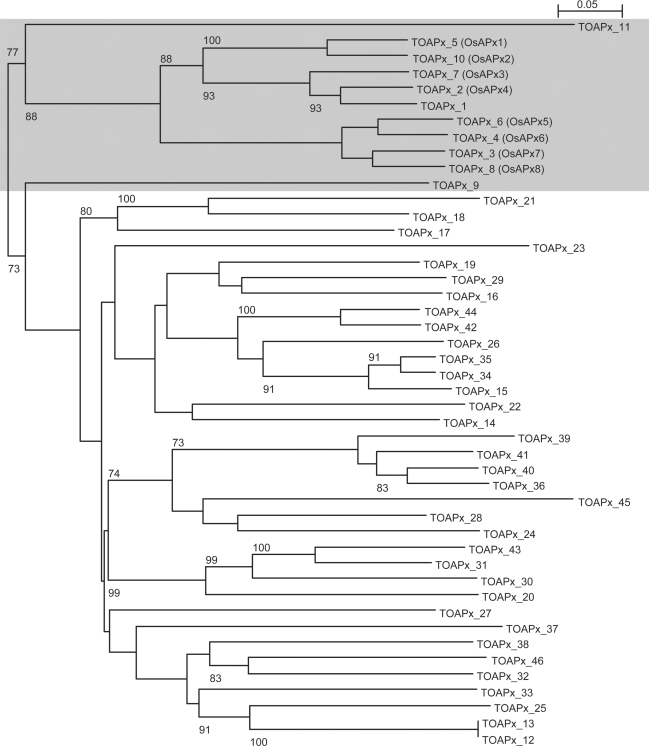
From the TARGeT-generated tree of APx homologs (shaded region in Figure 6), it is clear that the known APx family homologs are separated from the other putative homologs. Consideration of both gene structures (Figure 5 and Supplementary Figure 1) and positions in the phylogenetic tree (Figure 6) led to the identification of three putative new rice APx genes (TOAPx_1, TOAPx_9 and TOAPx_11) that have high similarity to Arabidopsis APx3 (identities = 80%, positives = 92%), APx6 (identities = 62%, positives = 77%) and APx4 (identities = 71%, positives = 82%), respectively. To provide evidence that these are real genes, these sequences were used as queries against the rice cDNA database in Genbank. Each gene matched several cDNAs (data not shown).
Figure 6.
An unrooted phylogenetic tree of all rice APx family members predicted by TARGeT. Previously characterized APx gene names are in brackets. The shaded region contains the true rice APx homologs. Bootstrap values greater than 70 are shown.
Searching for APx gene family members in maize and sorghum
To further evaluate the cross-species search ability of TARGeT, we searched for APx gene families in maize and sorghum, using the same query that was used to search for rice APx genes. The reasons for choosing maize and sorghum are as follows. First, at the time of the final analysis for this study, the available maize and sorghum sequences were incomplete. Maize is being sequenced using a BAC by BAC approach, while sorghum was sequenced using a whole genome shotgun approach. As such, they are more representative of the available genomic databases than the complete rice sequence. Second, search results of maize and sorghum can be compared with the rice and Arabidopsis output. Finally, the APx gene families in maize and sorghum have not as yet been characterized.
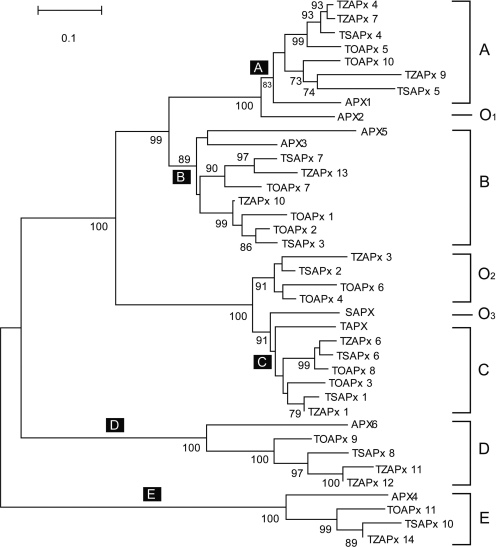
We identified 11 APx homologs in maize and 9 in sorghum (Supplementary Figures 2–5). To get a comprehensive view of the APx family in plants, we produced a phylogenetic tree with MEGA (55) using the published APx data from Arabidopsis and the data predicted by TARGeT for rice, maize and sorghum (Figure 7). APx gene homologs are clustered into five main clades (labeled A–E) with members from all species. These data suggest that ancient duplications preceded species divergence. The putative new rice APx homologs TOAPx_1, TOAPx_9 and TOAPx_11 are in clades B, D and E, respectively. Except for two maize homologs in clade D, there is only one representative for each species in clades D and E. This may be due to the effect of gene dosage balance on these two clades (56–58). In addition to the main clades, there are several putative orphan clades that are missing genes from one or more species. This may be due to either gene lose or insufficient sequence data.
Figure 7.
An unrooted phylogenetic tree of the APx homologs of rice, maize, sorghum and Arabidopsis. This tree was generated with MEGA version4 using the neighbor-jointing method with pairwise deletion and p distance. Five main clades are labeled from A to E. A main clade is defined as a minimal group of homologs that can be found in all species. The remaining homologs are classified into orphan clades O1–O3. Bootstrap values higher than 70 are shown.
Searching DNA TE families in rice
TARGeT is a powerful tool for rapid TE identification, characterization and phylogenetic analysis. We have illustrated this by using TARGeT to search for TEs in the rice genome using as query conserved transposase sequences from five DNA TE superfamilies. The queries were constructed from known TE protein sequences that were downloaded from Repbase (59) and additional sequences annotated as part of another study (data not shown). Here, we focus on the TARGeT results for the Tc1/mariner superfamily because it has been well annotated and characterized in rice.
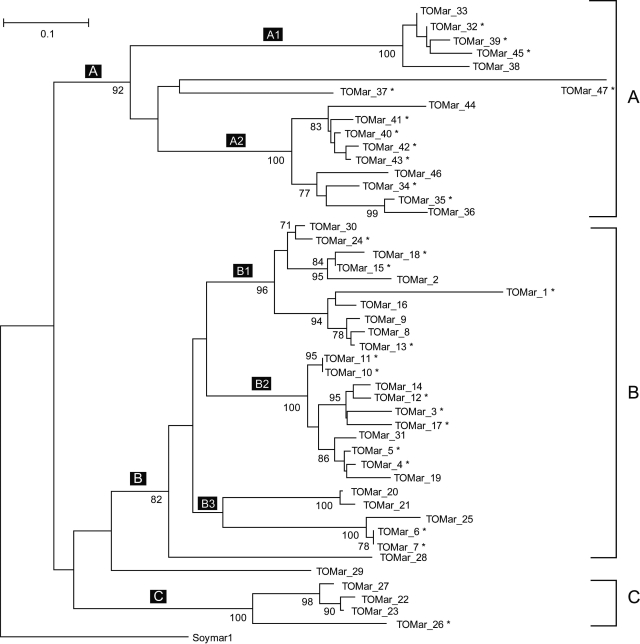
The Tc1/mariner superfamily is widespread in plant and animal genomes (60). A previous study (60) annotated 34 coding mariner-like elements (MLEs) from two partially sequenced rice genomes (14 from the indica database and 20 from the japonica database). Here, we used TARGeT to search the complete japonica database and, in ∼1 min, generated a phylogenetic tree that was consistent with that of Feschotte and Wessler (60). TARGeT successfully retrieved the 20 MLEs reported in the previous study and, in addition, detected 27 new MLEs (Figure 8).
Figure 8.
A rooted phylogenetic tree of predicted rice Tc1/mariner transposases. Three clades (A, B and C) are defined using the phylogenetic tree generated by Feschotte and Wessler (56). Elements denoted by an asterisk are new transposases predicted by TARGeT. Soymar1 was used as an outgroup and the tree was rooted manually using TreeView. Bootstrap values greater than 70 are shown.
Evaluating of the speed of TARGeT
Many factors can affect the speed of TARGeT, such as the number and length of the query sequences, the gene/TE family size, the database size and the number of exons. Other issues that affect the run time include the server hardware and current usage. In addition, because TARGeT is entirely web based, upload and download times vary from user to user. For the gene or TE families that were analyzed in this study, we calculated the average time for each search as an average of 10 independent runs. For example, TARGeT took ∼1.2, 2.5 and 6.8 min to complete the searches of the APx gene family in rice, sorghum and maize, respectively. The search of the rice Tc1/mariner superfamily took ∼1 min to complete.
Comparison of TARGeT with similar programs
Two other pipelines, GFScan (50) and FGF (61), can also retrieve and characterize gene families from genomic databases. GFScan searches for gene family members with the representative genomic DNA motif, while FGF performs TBLASTN search followed by GeneWise and phylogenetic analysis. Here, we briefly compare the features and performance of TARGeT with these two pipelines.
TARGeT versus GFScan
The cross-species searching ability of GFScan was previously tested by using a human query sequence to retrieve the carbonic anhydrases (CA) family from the mouse genome (50). GFScan was able to identify only 5 of the 11 known CA genes along with two putative new CA genes in the available mouse genome sequence. The authors stated that this discrepancy was due to the large difference between the human and mouse motifs. We did a similar search using TARGeT for CA genes in the mouse. Because there is no record of the version of the mouse genomic database used in the GFScan paper, we chose the latest version of the reference data (18 October 2006) from Genbank. A query composed of 14 protein sequences from 14 known human CA genes was constructed. Using default parameters except that the minimal intron length was set to 80 000 nt, TARGeT found 14 out of 16 known CA genes (data in 2008) in mouse, and the remaining two were identified together with a putative new CA homolog after the MMP cut-off was reduced from 0.7 to 0.5.
TARGeT versus FGF
Direct comparison between the results of TARGeT and FGF proved difficult. First, the FGF server is often not available. Second, TARGeT and FGF use different local databases. We ran TARGeT with the queries that were used in the paper describing FGF. Using a peptidylprolyl isomerase Cyp2 gene (AK061894, GI: 115443875) as query to search against the rice database with default parameters, TARGeT found six more putative homologs than FGF (Supplementary Figure 6). We also found one possible mistake in the result of FGF: it identified two overlapping homologs, AK061894_chr06 and AK061894_chr06, while there is no such overlap in the result of TARGeT. Using Hsp90 (GI: 40254816) as the query to search against the human database, both FGF and TARGeT found 15 homologs (Supplementary Figure 7).
DISCUSSION
To date, most gene family search programs can only retrieve homologs from protein sequence databases. More commonly, BLAST has been widely used to search genomic sequence databases. However, manual retrieval of homolog sequences from BLAST outputs requires a great deal of time. This is especially true for large gene or TE families. TARGeT is particularly useful if one wants to quickly retrieve and characterize gene families from DNA databases, especially when a newly sequenced genome is available. TARGeT uses a Perl program named PHI that automatically retrieves homolog sequences from BLAST outputs. In addition, TARGeT can do multiple alignment and phylogenetic analysis with the retrieved homolog sequences. Speed is another major advantage of TARGeT. As demonstrated in this report, TARGeT can routinely retrieve and characterize gene family homologs, including TEs, from plant and animal genome sequences on the order of minutes.
Although TARGeT shares similarity with homology-based TE annotation tools like RepeatMasker (62), there are some important differences. First, instead of showing each fragmented match as RepeatMasker does, TARGeT tries to identify homologs that are long enough for phylogenetic tree estimation. A fragmented TE can be identified as long as the sum length of its fragments satisfies the MMP to the query. As such, using the same query and databases, the number of homologs identified by TARGeT is usually lower than the hit number found by RepeatMasker. Second, when there are no repeat libraries available for a particular species, RepeatMasker gives the user the option of performing a BLASTX search to annotate coding regions of TEs in the submitted sequences. In contrast, TARGeT uses a TBLASTN search to identify coding regions from the whole genomic database. Finally, RepeatMasker lacks most of the functionality that is provided by TARGET including the generation of phylogenetic tree and gene structure figures.
When used to search genomic databases, protein sequence queries can efficiently detect distantly related homologs even when their DNA sequences cannot be aligned. Based on our experience, TBLASTN can detect sequences with identities as low as 25% to the query (data not shown). Comparison of the results of TARGeT, FGF and GFScan show that TARGeT retrieved more homologs. To further improve TARGeT's ability to identify distantly related homologs, we are planning to optimize matrix and BLAST parameters (such as gap penalties).
Using multiple queries can also increase the chances of finding additional gene family homologs. TARGeT can accept multiple queries at one time. Although more than one query may hit one homolog, a unique feature of TARGeT is that it can select the one that has the best match to the homolog.
When there is too much sequence divergence between a homolog sequence and the query, the homolog may not be found by TARGeT. However, TARGeT may still provide a clue for users to find them. For most homologs where HSPs are inadequate to meet the MMP cut-off value, they may still have short matches to the query in conserved regions. In this case, the file containing the BLAST HSPs that do not meet the qualified homolog cut-off would be valuable. Inspecting this file may give users a reason why TARGeT failed to detect some homologs and help users design new queries to find additional homologs.
TARGeT uses two approaches to separate closely related gene families. Because there is no absolute similarity cut-off among genes that are within or between families, closely related gene families may be retrieved, under certain circumstances, with the target gene family. This is often the case when the query is short, such as a domain sequence. An efficient way to separate closely related gene families is using phylogenetic analysis because homologs from the same family tend to cluster on a phylogenetic tree into the same clade (Figures 6–8). However, it may not be obvious which clade represents the homologs of interest. In other situations, the phylogenetic relationships between the homologs may be ambiguous when the root is unknown.
To overcome these limitations, TARGeT displays the gene structure of each homolog and their sequence similarity to the queries. Because different gene families often have distinct gene structures, homologs that have high sequence similarity to the queries and also have similar gene structures can be easily identified as members of the target gene family. For example, the homologs in the shaded clade in Figure 6 have higher sequence similarity to the query sequence than the homologs in the other clade, indicating that they are APx homologs. A determination of whether TOAPx_9 and TOAPx_11 belong to the shaded clade requires the gene structure comparison provided by TARGeT (Figure S1) because the (unrooted) phylogenetic tree alone does not provide sufficient information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (Grant DBI-0607123); Howard Hughes Medical Institute (Grant 52005731 to S.R.W.). Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank The iPlant Collaborative for hosting services on its cyber infrastructure and providing systems administration for the TARGeT web server. In addition, the authors would like to thank Drs Hongyan Shan, Jim Leebens-Mack, Russell Malmberg and Yaowu Yuan for critical reading of the manuscript and for valuable discussions.
REFERENCES
- 1.Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 2.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 3.Li WH, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 4.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 6.Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyckegaard EM, Clark AG. Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86:1944–1948. doi: 10.1073/pnas.86.6.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neitz M, Neitz J. Numbers and ratios of visual pigment genes for normal red-green color vision. Science. 1995;267:1013–1016. doi: 10.1126/science.7863325. [DOI] [PubMed] [Google Scholar]
- 9.Wendel JF. Genome evolution in polyploids. Plant Mol. Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 10.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 11.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung J, Wilson MD, Zhang J, Khaja R, MacDonald JR, Heng HH, Koop BF, Scherer SW. Recent segmental and gene duplications in the mouse genome. Genome Biol. 2003;4:R47. doi: 10.1186/gb-2003-4-8-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 14.Hurles M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2004;2:E206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 18.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 19.Tchenio T, Segal-Bendirdjian E, Heidmann T. Generation of processed pseudogenes in murine cells. EMBO J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanin EF. Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 21.Dayhoff MO. The origin and evolution of protein superfamilies. Fed. Proc. 1976;35:2132–2138. [PubMed] [Google Scholar]
- 22.Heger A, Holm L. Towards a covering set of protein family profiles. Prog. Biophys. Mol. Biol. 2000;73:321–337. doi: 10.1016/s0079-6107(00)00013-4. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 30.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 31.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 32.Meyers BC, Tingey SV, Morgante M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brinkman FS, Wan I, Hancock RE, Rose AM, Jones SJ. PhyloBLAST: facilitating phylogenetic analysis of BLAST results. Bioinformatics. 2001;17:385–387. doi: 10.1093/bioinformatics/17.4.385. [DOI] [PubMed] [Google Scholar]
- 34.Sicheritz-Ponten T, Andersson SG. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 2001;29:545–552. doi: 10.1093/nar/29.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arigon AM, Perriere G, Gouy M. HoSeqI: automated homologous sequence identification in gene family databases. Bioinformatics. 2006;22:1786–1787. doi: 10.1093/bioinformatics/btl179. [DOI] [PubMed] [Google Scholar]
- 36.Hanekamp K, Bohnebeck U, Beszteri B, Valentin K. PhyloGena–a user-friendly system for automated phylogenetic annotation of unknown sequences. Bioinformatics. 2007;23:793–801. doi: 10.1093/bioinformatics/btm016. [DOI] [PubMed] [Google Scholar]
- 37.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank RL, Mane A, Ercal F. An automated method for rapid identification of putative gene family members in plants. BMC Bioinformatics. 2006;7(Suppl. 2):S19. doi: 10.1186/1471-2105-7-S2-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Karsch-Mizrachi I, Ouellette BF. The GenBank sequence database. Methods Biochem. Anal. 2001;43:45–63. [PubMed] [Google Scholar]
- 42.Burks C, Fickett JW, Goad WB, Kanehisa M, Lewitter FI, Rindone WP, Swindell CD, Tung CS, Bilofsky HS. The GenBank nucleic acid sequence database. Comput. Appl. Biosci. 1985;1:225–233. [PubMed] [Google Scholar]
- 43.Salse J, Piégu B, Cooke R, Delseny M. Synteny between Arabidopsis thaliana and rice at the genome level: a tool to identify conservation in the ongoing rice genome sequencing project. Nucl. Acids Res. 2002;30:2316–2328. doi: 10.1093/nar/30.11.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 45.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J. Mol. Evol. 2004;59:761–770. doi: 10.1007/s00239-004-2666-z. [DOI] [PubMed] [Google Scholar]
- 47.Passardi F, Theiler G, Zamocky M, Cosio C, Rouhier N, Teixera F, Margis-Pinheiro M, Ioannidis V, Penel C, Falquet L, et al. PeroxiBase: the peroxidase database. Phytochemistry. 2007;68:1605–1611. doi: 10.1016/j.phytochem.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Frickey T, Lupas AN. PhyloGenie: automated phylome generation and analysis. Nucleic Acids Res. 2004;32:5231–5238. doi: 10.1093/nar/gkh867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 50.Xuan Z, McCombie WR, Zhang MQ. GFScan: a gene family search tool at genomic DNA level. Genome Res. 2002;12:1142–1149. doi: 10.1101/gr.220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 53.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordan GE, Piel WH. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–1642. doi: 10.1093/bioinformatics/btn235. [DOI] [PubMed] [Google Scholar]
- 55.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 56.Qian W, Zhang J. Gene dosage and gene duplicability. Genetics. 2008;179:2319–2324. doi: 10.1534/genetics.108.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang H, Plazonic KR, Chen J, Li WH, Fernandez A. Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genet. 2008;4:e11. doi: 10.1371/journal.pgen.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 59.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 60.Feschotte C, Wessler SR. Mariner-like transposases are widespread and diverse in flowering plants. Proc. Natl. Acad. Sci. USA. 2002;99:280–285. doi: 10.1073/pnas.022626699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng H, Shi J, Fang X, Li Y, Vang S, Fan W, Wang J, Zhang Z, Wang W, Kristiansen K. FGF: a web tool for Fishing Gene Family in a whole genome database. Nucleic Acids Res. 2007;35:W121–W125. doi: 10.1093/nar/gkm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 2004. http://www.repeatmasker.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.