Abstract
Classic tissue recombination and in vitro lineage tracing studies suggest that condensed metanephric mesenchyme (MM) gives rise to nephronic epithelium of the adult kidney. However, these studies do not distinguish between cap mesenchyme and pre-tubular aggregates comprising the condensed MM, nor do they establish whether these cells have self-renewing capacity. To address these questions, we generated Cited1-CreERT2 BAC transgenic mice, which express tamoxifen-regulated Cre recombinase exclusively in the cap mesenchyme. Fate mapping was performed by crossing these mice with the Rosa26RLacZ reporter line and evaluating the location and cellular characteristics of LacZ positive cells at different time points following tamoxifen injection. These studies confirmed expected results from previous in vitro analysis of MM cell fate, and provide in vivo evidence that the cap mesenchyme does not contribute to collecting duct epithelium in the adult. Furthermore, by exploiting the temporally regulated Cre recombinase, these studies show that nephronic epithelium arising at different stages of nephrogenesis has distinct spatial distribution in the adult kidney, and demonstrate for the first time that the cap mesenchyme includes a population of self-renewing epithelial progenitor cells.
Keywords: Cap mesenchyme, metanephric mesenchyme, kidney development, lineage tracing, Cited1 BAC transgenic mice
Introduction
The process by which the mammalian kidney develops has been the subject of extensive investigation over the past five decades and these studies have elucidated many of the physical and molecular interactions required for proper nephrogenesis (for recent reviews see; Boyle and de Caestecker, 2006; Dressler, 2006; Kopan et al., 2007). However, the precise developmental origins of the nineteen different specialized epithelia comprising the adult nephron, collecting duct epithelium, renal vasculature and interstitium of the adult kidney have yet to be established in vivo.
Development of the mammalian kidney is dependent on reciprocal tissue interaction between the ureteric bud (UB) and the metanephric mesenchyme (MM). Beginning at embryonic day 10.5 (E10.5) in mice the UB emerges from the nephric duct and invades the overlying MM. This process is marked by the patterning of the MM into peripheral, loosely associated stroma, and more condensed elements surrounding the UB tip. The sequence of MM invasion and patterning occurs in an iterative fashion throughout nephrogenesis as the UB branches repeatedly. After E11.5 condensed MM surrounding UB tips is organized into two morphologically and molecularly distinct zones: cap mesenchyme, an organized layer of cells opposed to the dorsal and lateral surfaces of UB tips, and pre-tubular aggregates, which are contiguous with the ventral and lateral aspect of the cap mesenchyme and are committed to differentiate into nephronic epithelium (Sariola, 2002). This distinction between cap mesenchyme and pre-tubular aggregates is important as it is likely that only a proportion of cells within the cap mesenchyme develop into pre-tubular aggregates. This suggests the cap mesenchyme may give rise to other cell types within the adult and/or embryonic kidney, which could include contribution to collecting duct epithelium, endothelium and/or stromal mesenchyme. Furthermore, this raises the possibility that a proportion of cap mesenchyme cells are committed at an early stage to self-renew and repopulate this progenitor pool over the course of nephrogenesis.
Classic tissue recombination experiments have led to the general understanding that nephronic epithelial structures (including proximal and distal tubular as well as glomerular epithelial elements) are derived from the MM, while the collecting system arises from the UB epithelium, which does not contribute to nephronic epithelium (Aubach, 1958; Saxen, 1987). These observations are supported by in vitro lineage tracing studies demonstrating that the MM can give rise to nephronic epithelium (Herzlinger et al., 1992; Qiao et al., 1995). However, the inability to differentially label subpopulations of cells within the MM limits the ability of these techniques to determine the fate of cap mesenchyme versus pre-tubular aggregates and stromal mesenchyme. Furthermore, these in vitro studies are limited in their capacity to track the fate of MM cells at different stages of nephrogenesis, and to determine the spatial and temporal origins of more complex epithelial structures that form at later stages of kidney development. Questions have also been raised by in vitro lineage tracing studies suggesting that the MM may in fact give rise to collecting duct epithelial cells (Qiao et al., 1995).
More recently, in vivo analysis of cell fate during development has been facilitated by the creation of Cre-dependent conditional reporter lines to track the fate of cell populations expressing Cre under the control of tissue-specific promoters. Two previous reports briefly address the fate of MM cells in the developing kidney using Pax3-Cre (Grieshammer et al., 2005) and Rar2b-Cre (Kobayashi et al., 2005). Crossing these mice with the Rosa26RLacZ (R26RLacZ) conditional reporter line demonstrates that the MM gives rise to nephronic epithelium in vivo. However, as Pax3-Cre and Rar2b-Cre are expressed throughout the MM from the earliest stages of metanephric kidney development, these studies do not distinguish between the fates of different cellular compartments within the MM, nor can they address patterns of MM differentiation at different stages of development. In addition, as these lines express active Cre in the MM throughout nephrogenesis, they cannot address questions regarding the capacity of the MM to undergo self-renewal.
Recent studies have identified two distinct transcriptional regulators that are uniquely expressed in the cap mesenchyme, Cited1 and Six2 (Boyle et al., 2007; Self et al., 2006). Six2 mRNA is expressed throughout the cap mesenchyme surrounding the UB tips and branch points (Self et al., 2006), while Cited1 protein (and mRNA) expression is restricted to the more lateral compartment of the cap mesenchyme and unlike Six2 is absent from the UB branch point cleft (Boyle et al., 2007). Neither Six2 nor Cited1 are expressed in pre-tubular aggregates or stromal mesenchyme of the embryonic kidney. We therefore saw the expression domain of Cited1 as a unique opportunity to track the fate of a distinct group of cells within the MM over the course of nephrogenesis. Here, we describe the generation and characterization of Cited1-CreERT2 transgenic mice, which express tamoxifen-activated (and therefore temporally regulated) Cre recombinase in the cap mesenchyme. By crossing these animals with the R26RLacZ reporter line and activating Cre with tamoxifen, we have created the first detailed fate map of cap mesenchyme cells over the course of metanephric kidney development.
Materials and Methods
Generation of BAC transgene and transgenic animals
The Cited1-CreERT2 BAC transgene was created as previously described (Lee et al., 2001). Briefly, a BAC (RP23-204G22, mouse 129/SvJ; Invitrogen) covering the entire Cited1 locus as well as 190 kb of 5′ and 30 kb of 3′ genomic sequence was electroporated into the EL250 E. coli strain, which carries a temperature sensitive λ phage Red recombination system and arabinose inducible Flp recombinase. A targeting vector containing a CreERT2-IRESeGFP-frt TETR frt cassette flanked by 500 bp homology arms corresponding to the Cited1 locus at the transcriptional start site (Fig. 1A) was electroporated into EL250 cells containing the Cited1 BAC and induced to undergo homologous recombination by 42°C heat shock. Homology arms were generated using PCR and designed to remove the Cited1 transcriptional start site and the subsequent 12 bp within the BAC, but do not delete any additional coding sequence to avoid removal of possible intronic regulatory elements within the Cited1 locus (Fig. 1A).
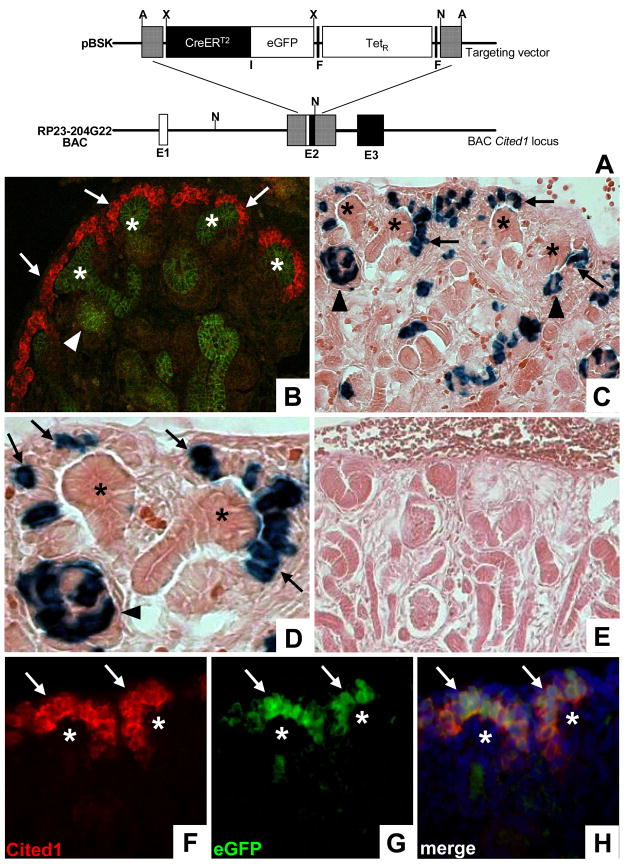
Figure 1. Generation and characterization of Cited1-CreERT2 mice.
A, Schematic of the targeting strategy used to create the BAC Cited1-CreERT2-IRESeGFP transgene. Hatched lines represent regions of homology. Following BAC targeting TetR was removed through Flp mediated recombination E1–Exon 1, F–Frt sites, I–IRES; A–Afl II, N–Not I, X–Xho I. B, Dual immunofluorescence staining of E15 mouse kidneys using anti-Cited1 (red) and anti-ECadherin (green) antibodies demonstrates endogenous Cited1 expression in the cap mesenchyme (arrows), and the absence of Cited1 expression in all ECadherin positive epithelial structures including renal vesicles (arrowhead) and UBs (asterisk). C/D, Low (C) and high (D) power images of the nephrogenic zone of E18 β-Gal stained Cited1-CreERT2/R26RLacZ kidneys following E15 maternal injection of tamoxifen. Recombination is observed within the cap mesenchyme (arrows) and early nephronic epithelia including renal vesicles (arrowheads). No recombination is observed in UBs (asterisks). E, β-Gal stained kidney from E18 Cited1-CreERT2/R26RLacZ embryo whose mother was not injected with tamoxifen. F/G, Anti-Cited1 antibody staining (F) corresponds to eGFP signal (G) within the cap mesenchyme (arrows) in mice carrying the Cited1-CreERT2-IRESeGFP transgene. No expression observed in UBs (asterisk). H, Merged image of (F) and (G) demonstrating overlapping expression of Cited1 and eGFP.
Dual chloramphenicol/tetracycline resistant clones that carried the Cited1 BAC with the predicted insertion were treated with arabinose to activate inducible Flp recombinase, thereby removing the TetR cassette. To remove the flanking LoxP sites in the pBACe3.6 vector backbone which, if present, could result in loss of the transgene upon activation of Cre, two additional modifications of the Cited1 BAC were made sequentially in vitro using homologous recombination. The LoxP511 site was replaced with a kanamycin resistance cassette flanked by 50 bp homology arms complementary to the sequence adjacent to the LoxP511 site (pLoxP511OUTKan). Subsequently, the LoxP site in the pBACe3.6 vector backbone was replaced with a zeocin resistance cassette flanked by 500 bp homology arms complementary to the sequence adjacent to the LoxP site (pLoxPOUTZeo). The details of the pLoxP511OUTKan and pLoxPOUTZeo vectors are available upon request (KKD and EMSS, unpublished data). After targeting procedures were completed, removal of LoxP sites was confirmed by PCR and sequencing across the deleted region with flanking primers. These modifications conferred Kan and Zeo resistance to the Cited1-CreERT2 BAC.
For generation of transgenic animals, Cited1-CreERT2-IRESeGFP BAC DNA was prepared using standard cesium chloride purification and dialyzed into TE pH 7.4 for microinjection using Centriprep-30 columns (Micron 4306) as described (DiLeone et al., 2000; DiLeone et al., 1998). Dilutions of purified BAC DNA were cut with Not I and subjected to pulse-field gel electrophoresis against a standard to estimate concentration. Circular BAC DNA was diluted to 1 ng/μl for pronuclear injection of zygotes derived from FVB mice and injected zygotes were implanted into pseudo-pregnant ICR females.
Mouse lines and tamoxifen injection
Transgenic mice generated using Cited1-CreERT2-IRESeGFP BAC DNA are hereafter referred to as Cited1-CreERT2 mice. R26RLacZ mice were obtained from Jackson Laboratories (Soriano, 1999). For treatment of pregnant females, tamoxifen (Sigma T5648) was dissolved by sonication in 10% EtOH/90% sunflower oil at a concentration of 15 mg/ml. 100 μl (1.5 mg) was injected intraperitoneally (IP) into pregnant R26RLacZ females at times indicated. As tamoxifen injection can compromise the ability of mice to have natural birth, pups were delivered by cesarean section at E19.5–E20 (day of vaginal plug appearance counted as E0.5). After pups were breathing independently they were transferred to foster mothers who had given birth the day before and had begun nursing. The overall success rate of fostering was >80%.
Genotyping and quantitative genomic PCR
Founders and subsequent offspring were genotyped using the following primer sets; Cre: 5′ GGC GCG GCA ACA CCA TTT TT; 3′ TCC GGG CTG CCA CGA CCAA. Rosa26RLacZ: 5′A AAA GTC GCT CTG AGT TGT TAT; 5′B GCG AAG AGT TTG TCC TCA ACC; 3′ GGA GCG GGA GAA ATG GAT ATG. Transgene copy number was estimated using TaqMan based real-time PCR using genomic DNA as described (Chandler et al., 2007). Briefly, the chloramphenicol cassette within the BAC vector was amplified using a custom primer set from Applied Biosystems (5′: GCA CAA GTT TTA TCC GGC CTT TAT T; 3′: GTC TTT CAT TGC CAT ACG GAA CTC; and an internal FAM-labeled probe CCG CCT GAT GAA TGC). To estimate copy number, triplicate CT values for the chloramphenicol cassette relative to a genomic DNA control, Jun kinase, were compared to the CT values of known quantities of BAC DNA prepared to estimate copy number equivalents.
β-Gal staining
Kidneys were isolated in cold PBS and fixed in 0.2% glutaraldehyde in PBS with 2mM MgCl2 and 5mM EGTA, or 4% formaldehyde in PBS for dual β-Gal/immunoperoxidase staining. Fixation times were determined empirically and varied from one hour to overnight (O/N) at 4°C according to age. Following fixation, tissues were cryoprotected in a gradient of first 15% (4 hours) and then 30% (O/N) sucrose in PBS at 4°C, embedded in OCT, and sectioned at 10 μm. For staining of frozen sections, slides were thawed and equilibrated in β-Gal wash (0.1M Phosphate buffer pH 7.3 containing 2mM MgCl2, 5mM EGTA, 0.02% NP40, 0.01% Na Deoxycholate) and stained O/N at 37°C in βGal wash containing 1 mg/ml X-Gal and 5mM K ferro- and ferricyanate. Sections were counterstained with eosin, dehydrated and mounted.
Antibody Staining
For immunofluorescence studies kidneys were isolated and fixed for 1 hour with 4% formaldehyde in PBS at 4°C. Following fixation, tissue was rinsed in PBS and cryoprotected in a sucrose gradient, embedded in OCT, and sectioned at 8μm onto charged slides. For staining, slides were thawed, rinsed in PBS and blocked in 10% goat serum (Vector Labs) in PBS for 1 hour. Tissue was incubated with primary antibodies in 10% goat serum in PBS O/N at 4°C, washed 3 times in PBS, and incubated with secondary antibodies for 1 hour at R/T. After washing in PBS, tissue was mounted with Vectashield mounting medium containing DAPI (Vector Labs). When antibodies used for co-labeling were of the same species, sequential sections were used. For immunoperoxidase/β-Gal studies, formaldehyde fixed sections were first β-Gal stained for four hours, rinsed in PBS, then blocked and incubated with primary antibody as described above. Tissues were incubated with goat anti-rabbit HRP secondary antibody (Santa Cruz #SC-2054; 1:100) for 45 min at 37°C, developed with DAB (Sigma #D4168), dehydrated and mounted. Antibodies used were as follows. Primary antibodies; rabbit anti-Cited1 (Neomarkers #RB-9219; 1:250), mouse anti-ECadherin (BD Biosciences #610181; 1:350), mouse anti-Cre (Covance #MMS106P; 1:100), mouse anti-GFP (Molecular Probes Clone Mab3E6 #A11120), rabbit anti-β-Gal (Cappel #55976, 1:5000), mouse anti-Na+/K+ atpase α-1 subunit (Upstate #05-369; 1:250), rat anti-PECAM1 (BD biosciences #553370; 1:250), hamster anti-Podoplanin (Angiobio # 11033; 1:500), rabbit anti-WT1 (Santa Cruz #SC192; 1:100), rabbit anti-Aquaporin 1 (Chemicon # AB3065; 1:250), rabbit anti-Aquaporin 2 (Alpha Diagnostics #AQP21-A; 1:250). Secondary antibodies; goat anti-rabbit Rhodamine X (Jackson #111-295-144; 1:300), horse anti-mouse Fluorescein (Vector Labs #FI-2000; 1:300), horse anti-mouse Texas Red (Vector Labs #TI-2000; 1:300), goat anti-hamster Alexafluor 488 (Molecular Probes #A21110; 1:300) and goat anti-Rat Alexafluor 488 (Molecular Probes #A11006; 1:300). Fluorescence images were acquired using a Nikon Elipse E800 epifluorescence microscope fitted with a SPOT RT slider 2.3.0 digital imaging camera. Color overlays were generated using Adobe Photoshop Version 9.0.2.
Results
Generation and characterization of Cited1-CreERT2 transgenic mice
To create a fate map of the cap mesenchyme we created Cited1-CreERT2 mice using BAC transgenesis. The BAC clone we modified for creation of the Cited1-CreERT2 transgene was selected based on the relative position of the Cited1 locus within its genomic sequence. We used established BAC recombination techniques to insert a CreERT2-IRESeGFP cassette into the BAC Cited1 locus between ~ 190 kb of upstream and ~30 kb of downstream genomic sequence that we can reasonably assume contains the regulatory elements necessary to drive gene expression analogous to endogenous protein (for review on BAC transgenesis see; Giraldo and Montoliu, 2001; Liu et al., 2003; Nishinakamura et al., 2001) (Fig. 1A). This cassette encodes Cre recombinase fused to the ligand binding domain of a modified estrogen receptor that binds tamoxifen with much higher affinity than endogenous estrogens. This system takes advantage of ligand-dependent nuclear import of ERT2, so that Cre translocates to the nucleus (and is thus active) only in the presence of the estrogen analog tamoxifen (Feil et al., 1996; Feil et al., 1997). In addition, the cassette includes an internal ribosomal entry site (IRES) allowing for independent translation of eGFP, which is used as a marker of transgene expression.
Pronuclear injection of ~200 zygotes with the Cited1-CreERT2 BAC transgene resulted in 29 live births, 6 of which were determined by PCR to carry Cre recombinase (data not shown). Four of these animals transmitted the transgene in their germline and were evaluated for recombination potential. For this, we crossed candidate founders with the R26RLacZ conditional reporter (Soriano, 1999), treated pregnant females with a single IP dose of tamoxifen at E15, and evaluated recombination in kidneys at E18 using β-Gal staining. We chose these time points based on the robust expression of Cited1 in the cap mesenchyme at E15 and the abundance of MM present during this period of kidney development (Fig 1B; Boyle et al., 2007). This window also allows time for LacZ expression to be initiated in cells that have undergone recombination. Offspring of three founders clearly exhibited recombination outside of the Cited1 expression domain, including widespread labeling in the UB, which we found was due to ectopic expression of the transgene (data not shown). One line however showed LacZ expression exclusively in the cap mesenchyme and its early derivatives including renal vesicles and S-shaped bodies (Fig. 1C, D), and no recombination was observed in this line in the absence of tamoxifen (Fig. 1E). To demonstrate faithful transgene expression in the cap mesenchyme of Cited1-CreERT2 mice we compared expression of endogenous Cited1 protein and eGFP at E15.5. Direct visualization of eGFP on sections stained with anti-Cited1 antibody revealed that expression of the Cited1-CreERT2 transgene precisely overlaps with expression of endogenous protein (Fig. 1F–H). Staining using an anti-GFP antibody further confirmed proper transgene expression (data not shown). Together, these results indicate that this line of Cited1-CreERT2 mice expresses the transgene throughout the cap mesenchyme, and that this induces relatively efficient recombination at the R26RLacZ locus following a single IP injection of tamoxifen.
Because BACs contain large segments of genomic DNA (on the order of 200kb) and tend to integrate into the genome as concatamers, stable transmission of transgene copy number from parent to offspring can be variable as a result of linkage disequilibrium, and is often not established until after the F1 generation (DM, personal communication). To characterize copy number transmission in Cited1-CreERT2 mice we utilized a genomic quantitative PCR method (Chandler et al., 2007). Using this method we determined that our founder (F0) carried approximately five copies of the CreERT2 transgene. By crossing this mouse to a wild type female we examined the transmission pattern of the transgene. Analysis of the F1 generation suggested transmission of a single transgene insertion site and showed that Cre-positive F1 animals were estimated to carry between 1 and 5 copies of the transgene. When F1 males carrying an estimated four copies of the transgene were crossed to wild type females, all Cre-positive progeny carried approximately four copies of CreERT2. This four-copy transgene insertion was stably transmitted through the F4 generation. Animals generated from this line are referred to as Cited1-CreERT2 mice and were used for all studies to map cap mesenchyme fate presented in this manuscript.
The cap mesenchyme gives rise to cells in all segments of the adult nephron
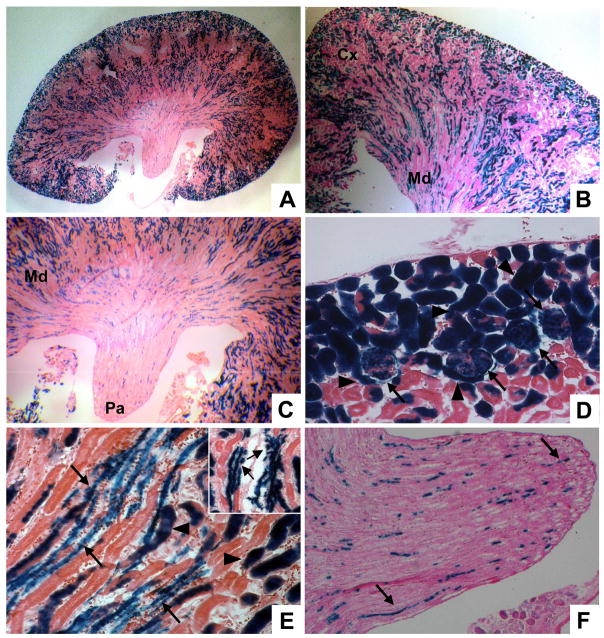
To test the overall lineage potential of the cap mesenchyme, we crossed Cited1-CreERT2 mice to the R26RLacZ line and treated pregnant females with a single IP injection of tamoxifen at E13, when Cited1 is strongly expressed in the cap mesenchyme (Boyle et al., 2007). By waiting until six weeks of age to examine cell lineage (by which time post-natal nephrogenesis is complete) we were able to ascertain the definitive fate potential of Cited1 expressing cap mesenchyme cells. β-Gal staining of adult kidneys revealed widespread LacZ expression in a large proportion of nephrons, including multiple cell lineages in the cortical and medullary regions (Fig. 2A–C). Some of these lineages were evident based on morphology and position, including proximal tubules and cells within glomeruli (Fig. 2D). Cell types were more difficult to distinguish in the renal medulla. Here, more distal nephronic elements and collecting ducts are closely associated with vascular structures as they descend into the papilla (Fig. 2B, C). In these regions, we observed LacZ positive cells incorporated into thinner, more elongated tubular structures (Fig. 2E, F). This morphology and position is consistent with multiple cell types in the adult kidney including collecting ducts, loops of Henle and vascular endothelium.
Figure 2. The cap mesenchyme gives rise to cells in all regions of the adult kidney.
Cited1-CreERT2 mice were crossed to R26RLacZ mice and pregnant females injected with tamoxifen at E13. Lineage was assessed using β-Gal staining in 6 week old mice. A–C, Low power images of the entire kidney (A), corticomedullary region (B) and inner medulla (papillary) region (C) show cap mesenchyme derived cells in all areas of the kidney. Cx-cortex, Md-medulla, Pa-papilla. D, In the cortex, some of these cell types are recognizable by morphology, including proximal tubules (arrowheads) and glomeruli (arrows). E/F, In addition to tubular epithelium (arrowheads), the medulla (E) and papilla (F) contain cap mesenchyme derived cells which have a more elongated phenotype (arrows, inset shows higher power image of these structures).
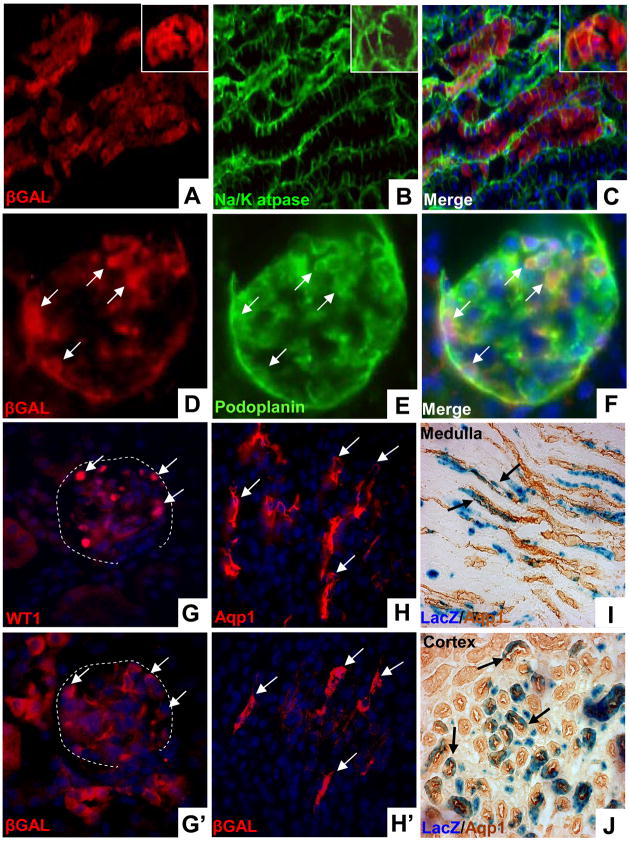
To evaluate this further we utilized an antibody against β-Gal and cell-type specific markers for dual immunofluorescence. β-Gal co-localized with Na+/K+ atpase (α1 subunit), a broadly expressed marker of nephronic epithelia (Fig. 3A–C). In addition, β-gal staining co-localized with podoplanin, indicating cap mesenchyme lineage in the glomerular epithelium (Fig. 3D–F). These findings were confirmed by co-localization of β-Gal with the podocyte specific marker WT1 on sequential sections (Fig. 3G, G′). Aquaporin 1 (Aqp1) is expressed in proximal tubules in the cortex and the thin limb of the loops of Henle in the medulla. Staining of sequential sections with antibodies against Aqp1 and β-Gal revealed that thin limb tubular epithelium in the renal papilla is derived from the cap mesenchyme (Fig. 3H, H′). This lineage is demonstrated with greater resolution using Aqp1 immunoperoxidase staining on sections which have been stained for β-Gal activity (Fig. 3I). These studies also detected LacZ positive cells co-expressing Aqp1 in the cortex, confirming cap mesenchyme lineage in the proximal tubular epithelium (Fig. 3J). Together, these studies show that the cap mesenchyme gives rise to a wide variety of epithelial cell types in the nephron including proximal, distal and glomerular elements.
Figure 3. Cap mesenchyme gives rise to diverse populations of renal epithelial cells.
Cited1-CreERT2/R26RLacZ mice were injected with tamoxifen at E13 and lineage examined at 6 weeks of age. A–C, β-gal positive cells coincide with Na/K atpase expression, a broad marker of nephronic epithelium. Inset shows tubule in cross section. D–F, β-gal (D) and podoplanin (E) expression overlap (F) demonstrating cap mesenchyme lineage in the glomerular epithelial compartment (arrows). G/G′, Staining of sequential sections with anti-WT1 (G, nuclear) and anti-β-Gal (G′, cytoplasmic) shows that the cap mesenchyme gives rise to podocytes (arrows). The glomerular outline is indicated with dashed white lines. H/H′, Staining of sequential sections with anti-Aqp1 (H) and anti-β-Gal (H′) demonstrates that the cap mesenchyme gives rise to thin limb epithelium in the papilla. I/J, Immunoperoxidase staining using anti-Aqp1 on sections which have been β-Gal stained shows cap mesenchyme derived cells in thin limb in the papilla (I) and in proximal tubules in the cortex (J).
The cap mesenchyme does not give rise to collecting duct epithelium or endothelial cells
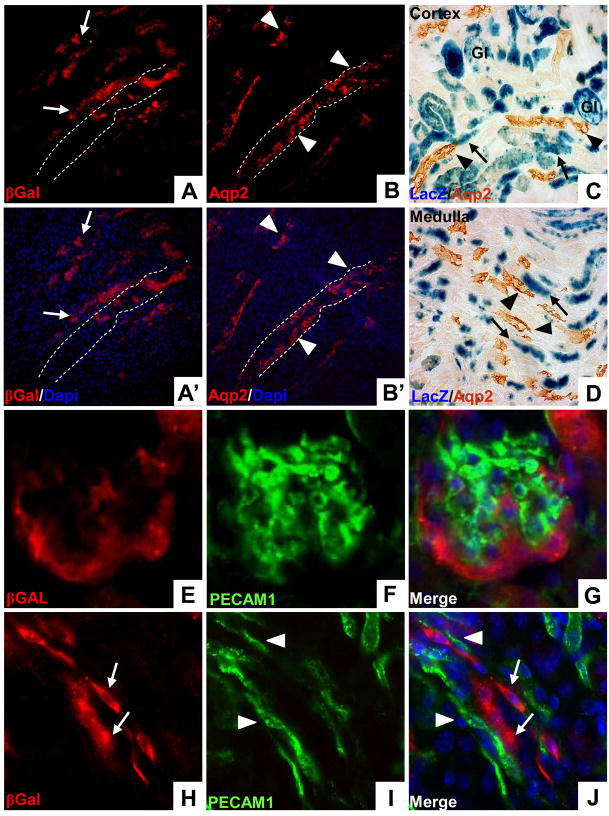
Previous in vitro lineage tracing studies using explanted nephrogenic tissue have suggested that the condensed mesenchyme can give rise to cells within the mature collecting duct epithelium (Quio et. al., 1995). Using the collecting duct specific marker Aquaporin2 (Aqp2) and the β-Gal antibody on sequential sections we asked whether the cap mesenchyme contributes to the collecting system. Using this analysis, we saw no evidence for cap mesenchyme derived cells in medullary collecting ducts (Fig. 4A/A′, B/B′). This was confirmed using immunoperoxidase staining to detect Aqp2 on sections which had been β-Gal stained (Fig 4C, D), again indicating that the cap mesenchyme does not contribute to either cortical or medullary collecting ducts. Given the close proximity of these epithelial elements to vascular structures within the adult kidney we also asked whether the cap mesenchyme gave rise to endothelial cells. For this, we looked for β-Gal positive cells that co-express the endothelial specific marker PECAM-1/CD31. We saw no evidence of overlap between cap-derived cells and endothelium, in either glomeruli (Fig. 4E–G) or in the medulla (Fig. 4H–J). It is likely that the elongated cells we saw in the β-Gal stained renal medullas are nephronic elements (i.e. thin limb epithelium) closely opposed to the associated vasa-recta (Fig. 4J).
Figure 4. Neither collecting duct epithelium or renal endothelium arises from the cap mesenchyme.
Cited1-CreERT2/R26RLacZ mice were injected with tamoxifen at E13 and lineage was examined by staining with anti-β-Galactosidase antibodies at 6 weeks of age. A–B′, Staining of sequential sections with anti-β-Gal (A, A′) and anti-Aqp2 (B, B′) demonstrates that cap mesenchyme derived cells do not populate the collecting duct epithelium. LacZ positive cells (arrows) are closely opposed to, but distinct from, Aqp2 positive collecting ducts (arrowheads). Position of one of the collecting ducts is indicated with dashed white lines, illustrating lack of overlap with β-Gal staining. C/D, Immunoperoxidase staining using anti-Aqp2 on sections which have been β-Gal stained shows that cap mesenchyme derived cells (arrows) do not overlap with either cortical (I, arrowheads) or medullary collecting ducts (J, arrowheads). Gl–glomeruli. E–G, Cap derived cells (E) and PECAM1 positive endothelial cells (F) do not overlap in glomeruli (G). H–J, Elongated cap mesenchyme progeny in the medulla (H, arrows) do not coincide with PECAM1 positive endothelium (I, arrowheads) but instead run alongside their closely associated vasculature (J).
From these studies we conclude that the cap mesenchyme does not give rise to renal endothelium or cells within the collecting system.
Patterns of nephron formation during development
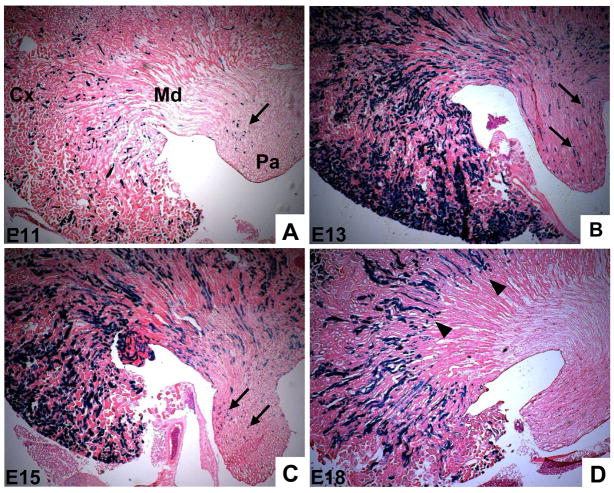
The inducible nature of the Cited1-CreERT2 transgene makes it a useful tool to ask whether the potential of the cap mesenchyme changes over the course of nephrogenesis. To test this we carried out pulse-chase experiments in which we crossed Cited1-CreERT2 mice with the R26RLacZ reporter, treated females with a single dose of tamoxifen at various time points during embryogenesis, and examined lineage patterns at six weeks of age. We observed LacZ positive cells in all parts of the adult kidney following injection at the earliest time point, E11, although recombination was not widespread (Fig. 5A). However, we did observe robust recombination in the heart, where Cited1 is also expressed (Dunwoodie et al., 1998), after an E11 injection of tamoxifen (SB, unpublished data). This makes it unlikely that placental delivery of tamoxifen to the embryo is impaired at E11, and suggests that the relatively low number of nephrons labeled by injecting at this point reflects the low levels of Cited1 in the early condensing MM (Boyle et al., 2007). In contrast to an E11 injection of tamoxifen, treatment at E13 labeled a large proportion of nephrons throughout the adult kidney (Fig 5B). E15 injection resulted in a similar pattern, although fewer total nephrons were labeled (Fig. 5C). This is consistent with the morphometric observation that many primitive nephrons arise prior to E16 (Cebrian et al., 2004). Tamoxifen treatment at later time points, however, resulted in a distinct pattern of LacZ expression. Following injection at E18, LacZ positive cells were observed in the cortex and the outer medulla, but unlike the earlier injection time points, did not reach the papilla (Fig. 5D). These data provide in vivo evidence that nephrons arising at different times during development assume distinct deep and superficial positions in the adult.
Figure 5. Cap mesenchyme derived nephrons assume temporally-dependent deep and superfical positions in the adult kidney.
Cited1-CreERT2 mice were crossed to R26RLacZ reporter mice and injected with 1.5 mg of tamoxifen at times indicated. Mice were sacrificed at 6 weeks of age and lineage assessed with β-Gal staining. A, E11 injections result in recombination in a low percentage of nephrons, but cap derived cells are found in the cortex, medulla, and papilla (arrows). B/C, Injection at E13 (B) and E15 (C) results in recombination in a large proportion of nephrons, some of which extend into the papilla (arrows). D, E18 injection results in recombination in an intermediate number of nephrons, however these structures do not extend into the papilla (arrowheads). Cx–cortex, Md–medulla, Pa–papilla.
The cap mesenchyme includes a self renewing population of epithelial progenitor cells
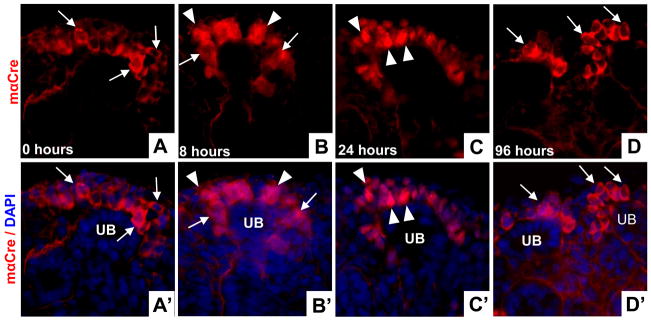
Studies in which we tracked the fate of cap mesenchyme cells labeled at E13 demonstrated that a large proportion of nephronic epithelia in the adult kidney are derived from the relatively small number of cap cells present at this time (Fig. 2). These findings suggest that cap mesenchyme cells not only differentiate into nephronic epithelium, but also give rise to new progenitor cells within the MM that develop and condense around branching UB tips as nephrogenesis proceeds. To determine whether this is the case, we first evaluated the kinetics of tamoxifen-dependent activation of CreERT2 using this model system. We did this to ensure that any labeled cells found in the cap mesenchyme several days after tamoxifen treatment were not the result of persistent CreERT2 nuclear translocation. To evaluate this in Cited1-CreERT2 mice, we examined the sub-cellular localization of Cre over a time course following tamoxifen injection between E14.5 and E16. In uninjected mice, antibody staining revealed that Cre was virtually exclusively localized in the cytoplasm of cap mesenchyme cells (Fig. 6A, A′). Some nuclear translocation of Cre was observed 8 hours after tamoxifen injection (Fig. 6B, B′), but by 24 hours, Cre protein was strongly localized to the nucleus in the majority of cap mesenchyme cells (Fig. 6C, C′). By 96 hours post-injection, Cre protein had redistributed to the cytoplasm and is therefore no longer active (Fig. 6D, D′).
Figure 6. Kinetics of CreERT2 sub-cellular localization following tamoxifen injection.
A–D, CreERT2 localization using anti-Cre antibody staining in E16.5 kidneys; A′–D′ corresponding images with DAPI overlay to illustrate nuclei. Kidneys from uninjected animals (A, A′) show cytoplasmic localization of Cre (arrows). 8 hours after tamoxifen injection (B, B′) Cre is seen in cytoplasmic (arrows) and nuclear compartments (arrowheads). 24 hours after injection (C, C′) Cre is found almost exclusively in nuclei (arrowheads). 96 hours post-injection (D, D′) Cre has redistributed to the cytoplasm in the cap mesenchyme (arrows). UB-ureteric bud.
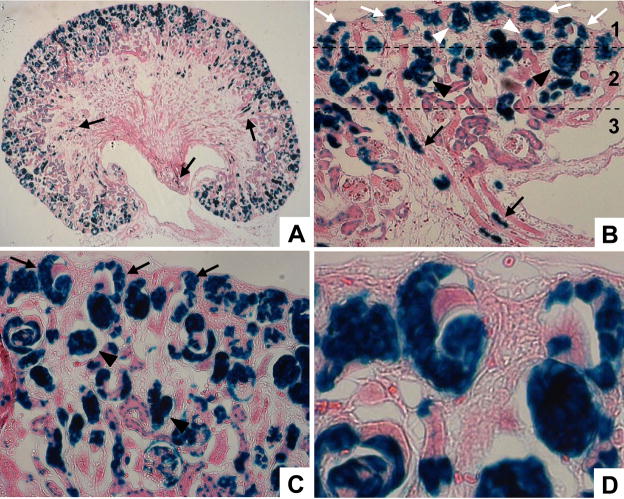
Having established that tamoxifen-dependent activation of the CreERT2 transgene was complete no later than 4 days following a single IP injection of tamoxifen, we evaluated cap mesenchyme lineage in Cited1-CreERT2/R26RLacZ mice at E19.5 following tamoxifen treatment at E13. By this time several generations of nephrons have emerged from the MM, yet the nephrogenic zone remains robust due to the repetitive nature of branching and induction during kidney development. β-Gal staining of kidneys from these mice revealed that most of the cap mesenchyme derived cells are located in the outer cortex, with only a few labeled cells in the embryonic medulla (Fig. 7A). Looking closely at the nephrogenic region and early corticomedullary boundary, we observed three general zones of LacZ positive cells (Fig. 7B). The deepest region (zone 3, Fig. 7B) contained labeled cells that have gone through the stepwise process of epithelial differentiation and are extending into the medulla. These presumably were the first cells to differentiate from the cap following tamoxifen injection. The intermediate zone (zone 2, Fig. 7B) was characterized by primitive nephrons at various stages of differentiation, such as comma- and S-shaped bodies. The outermost zone (zone 1, Fig. 7B) included LacZ positive cells in the early stages of differentiation, the renal vesicles and pre-tubular aggregates. Strikingly, we also saw that a high percentage of cap mesenchyme cells were still labeled at E19.5 (Fig. 6B–D), demonstrating that these are progeny of cap cells originally labeled at E13. Given the observation that cap mesenchyme labeled at E13 gives rise to a large number of primitive nephrons by E19.5, and that the UB has branched extensively over this time period (accompanied by the corresponding increase of cap mesenchyme), these findings demonstrate that the cap mesenchyme is repopulated by an intrinsic, self-renewing population of cells.
Figure 7. The cap mesenchyme gives rise to successive generations of renal progenitor cells.
Pregnant Cited1-CreERT2/R26RLacZ mice were injected with tamoxifen at E13 and lineage was examined at E19.5 using β-Gal staining. A, Low power image of whole kidney shows cells of cap mesenchyme origin have begun to move into distal locations (arrows). B, Nephrogenic zone and primitive corticomedullary junction; cap derived cells are seen in the medulla (arrows, zone 3), inner nephrogenic region (zone 2) associated with comma- and S-shaped bodies (black arrowheads), and outer nephrogenic region (zone 1) containing cap mesenchyme (white arrows) and renal vesicles (white arrowheads). C, Outer nephrogenic zone demonstrating cap mesenchyme lineage in primitive nephrons (arrowheads) and retention of labeled cells in the cap mesenchyme (arrows). D. High power image of the branched UB tip. Persistent labeling of cells in the cap mesenchyme at E19.5 indicates that these are progeny of cap cells originally labeled at E13.
Discussion
In these studies, we describe the generation of Cited1-CreERT2 BAC transgenic mice which express tamoxifen-activated Cre recombinase in a discrete subpopulation of cells within the condensed MM known as the cap mesenchyme. These mice provide a unique reagent that induces efficient Cre-dependent recombination restricted to the cap mesenchyme following a single IP injection of tamoxifen, and incorporates a functional IRES-eGFP marker that could be used for enrichment of Cited1 positive cells in future studies. We have exploited this transgenic system to generate the first detailed in vivo fate map of cap mesenchyme cells over the course of nephrogenesis. By crossing Cited1-CreERT2 BAC transgenic mice with the R26RLacZ conditional reporter line, and activating Cre with tamoxifen, we were able to track the fate of cap mesenchyme derived cells at different stages of nephrogenesis. Using this approach our studies confirm what was predicted from the results of previous in vitro studies indicating that the cap mesenchyme gives rise to a wide variety of nephronic epithelial cell types. They also clarify conflicting in vitro data regarding the mixed ontogeny of the collecting duct epithelium by demonstrating definitively that the cap mesenchyme does not contribute to collecting duct epithelium within the adult kidney. We also demonstrate that the positioning of nephrons within the adult kidney depends on the stage at which they develop from the cap mesenchyme, and importantly, provide the first substantive evidence that these cells contain a population of self-renewing epithelial progenitor cells.
We chose to use Cited1 promoter elements to drive Cre expression given its unique and restricted expression domain in the cap mesenchyme. Previous studies from our laboratory have shown that Cited1 expression increases in the condensed MM from E12.5 and persists in the cap mesenchyme throughout nephrogenesis, but is absent in the adult kidney (Boyle et al., 2007). Cited1 expression is distinct from that of Six2, which is expressed strongly in the MM prior to UB invasion and in early condensations, and extends outside of the Cited1 expression domain to overlie UB branch points (Self et al., 2006). These findings indicate that Cited1 is expressed in a subpopulation of Six2 positive cap mesenchyme, and will be important for future comparison with lineage tracing studies using other Cre lines expressed in the MM.
Initial characterization of this transgenic line demonstrated Cre-mediated recombination at the Rosa26RLacZ locus in approximately 40–50% of cap mesenchyme cells 72 hours after a single injection of tamoxifen. In addition, there was widespread β-gal expression in nephronic epithelia throughout the adult kidney after a single IP injection at E13. As Cre-dependent recombination is a stochastic event (Nagy, 2000), these findings suggest that with repeated injection of tamoxifen, this transgenic line will likely induce highly efficient recombination in the acute setting and will provide a powerful tool to study the effect gene deletions within the cap mesenchyme. Further characterization of this transgenic line indicated that Cre-dependent recombination reflects the expected pattern of Cited1 expression over the course of nephrogenesis (Boyle et al., 2007). For example, we have previously shown that Cited1 expression at E11 is weak and restricted to a few cells in the MM. Correspondingly, when we injecteCited1-CreERT2/R26RLacZ mice at E11 and examined cap mesenchyme lineage in the adult, we observed a low percentage of total nephrons labeled. This was not due to inefficient placental transfer of tamoxifen as we saw efficient Cre-dependent recombination within the developing myocardium at the same time point. In contrast, when we treated mice with tamoxifen at E13, by which time Cited1 expression in the cap mesenchyme is robust (Boyle et. al, 2007), we saw recombination in a large proportion of adult nephronic epithelium. Furthermore, the declining percentage of labeled epithelium in the adult following injection at E15 and E18, respectively, is consistent with the fact that many primitive nephrons arise prior to these time points.
These fate mapping studies provide in vivo evidence that the cap mesenchyme gives rise to a wide variety of nephronic epithelial cell types populating the cortex and medulla of the adult kidney, confirming results predicted from classical tissue recombination experiments as well as in vitro lineage tracing studies. However, technical limitations of in vitro explant studies do not allow for the differential labeling and fate mapping specifically of cap mesenchyme vs. pre-tubular aggregates and stromal mesenchyme within the MM. Our studies, therefore, provide direct evidence that the cap mesenhcyme is the primary source of the epithelial progenitor cells that will comprise the mature nephron. In addition, these studies show, in vivo, that the cap mesenchyme does not contribute to collecting duct epithelium.
The transient nature of tamoxifen-induced activation of CreERT2 provides an opportunity to track the fate and spatial distribution of cells derived from the cap mesenchyme at different stages of nephrogenesis. This enabled us to address one of the key unanswered questions in this field: Is the cap mesenchyme repopulated over the course of nephrogenesis intrinsically or through the migration of extrinsic cells? Our studies show that a high proportion of cap mesenchyme cells still express LacZ 6.5 days after a single IP injection of tamoxifen. It is unlikely this is due to persistent tamoxifen-induced activation of CreERT2 as we demonstrated that tamoxifen-induced nuclear localization of CreERT2 was maximal 24 hours following injection with no detectable nuclear Cre after 96 hours. Furthermore, previous studies have characterized the kinetics of Cre activation in more detail, indicating that most of the Cre returns to the cytoplasm 48 hours after a single tamoxifen injection (Hayashi and McMahon, 2002; Nakamura et al., 2006). On this basis, if exogenous cells were being recruited to repopulate this niche, we would expect that a high percentage of cap mesenchyme cells present at the time of injection would not be labeled 6 days later, as the originally tagged cells would have been depleted by induction and differentiation and progressively replaced by unlabeled cells. These findings provide the first strong in vivo evidence that an intrinsic, self-renewing population of progenitor cells reconstitutes the cap mesenchyme over the course of nephrogenesis.
By tracking the fate of nephronic epithelium labeled early vs. late in nephrogenesis, our studies also provide in vivo evidence that deep nephrons extending into the renal papilla arise only during the early phase of nephrogenesis and that nephrons arising at later stages of development are restricted to the cortex and outer medulla. As deep and superficial nephrons contain all of the same cell types, this does not represent a shift in cell fate per se, but provides in vivo evidence that the positioning of nephrons within the adult kidney is temporally regulated. There are several possible explanations for this phenomenon. It is possible that nephrons arising later are physically ‘blocked’ from extending into the medulla by early nephrons that have already migrated into the papilla as corticomedullary patterning begins at ~E16 (Cebrian et al., 2004). Given the fact that formation of the renal pelvis is poorly understood, a more intriguing explanation would be the differential expression of an unidentified chemotactic factor that directs migration of distal elements into the papilla. Perhaps this factor is down-regulated once an appropriate number of nephrons have arrived, leaving tubules arising later to occupy superficial positions. In any case, the temporally dependent organization of nephronic elements observed in our studies supports what has been predicted by classical anatomical studies of the adult kidney indicating that nephrons are organized into deep and superficial structures (Kriz and Koepsell, 1974).
In summary, we describe the generation and characterization of Cited1-CreERT2 BAC transgenic mice, which express tamoxifen-regulated Cre recombinase exclusively in the cap mesenchyme. By crossing these mice with a Cre reporter line, we have used these mice to evaluate the fate of cap mesenchyme cells over the course nephrogenesis. These studies confirm the expected findings that cap mesenchyme gives rise to diverse nephronic epithelia in the adult kidney, and provide the first evidence that the cap mesenchyme contains a population of self-renewing epithelial progenitor cells.
Acknowledgments
We thank Chris Wright and Alan Perantoni for advice and critical review of the manuscript, David Frank for technical advice and Kevin Tompkins for assistance with pronuclear injections. Work was supported by supported by: NIH R01 DK61558 and P50 DK39261 (MDC), NIH T32 HD007502 (SB) and NIH R21 DK064251 (MSS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aubach RGC. Inductive interactions of embryonic tissues after dissociation and re-aggregation. Exp Cell Res. 1958;15:384–387. doi: 10.1016/0014-4827(58)90039-9. [DOI] [PubMed] [Google Scholar]
- Boyle S, de Caestecker M. Role of transcriptional networks in coordinating early events during kidney development. Am J Physiol Renal Physiol. 2006;291:F1–8. doi: 10.1152/ajprenal.00447.2005. [DOI] [PubMed] [Google Scholar]
- Boyle S, Shioda T, Perantoni AO, de Caestecker M. Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn. 2007;236:2321–2330. doi: 10.1002/dvdy.21242. [DOI] [PubMed] [Google Scholar]
- Cebrian C, Borodo K, Charles N, Herzlinger DA. Morphometric index of the developing murine kidney. Dev Dyn. 2004;231:601–8. doi: 10.1002/dvdy.20143. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007;27:2934–51. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone RJ, Marcus GA, Johnson MD, Kingsley DM. Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci U S A. 2000;97:1612–7. doi: 10.1073/pnas.97.4.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone RJ, Russell LB, Kingsley DM. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–8. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–29. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Rodriguez TA, Beddington RS. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–90. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–7. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–57. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–18. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Herzlinger D, Koseki C, Mikawa T, al-Awqati Q. Metanephric mesenchyme contains multipotent stem cells whose fate is restricted after induction. Development. 1992;114:565–72. doi: 10.1242/dev.114.3.565. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–23. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- Kopan R, Cheng HT, Surendran K. Molecular Insights into Segmentation along the Proximal-Distal Axis of the Nephron. J Am Soc Nephrol. 2007 doi: 10.1681/ASN.2007040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz W, Koepsell H. The structural organization of the mouse kidney. Z Anat Entwicklungsgesch. 1974;144:137–63. doi: 10.1007/BF00519771. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–12. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128:3105–15. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- Qiao J, Cohen D, Herzlinger D. The metanephric blastema differentiates into collecting system and nephron epithelia in vitro. Development. 1995;121:3207–14. doi: 10.1242/dev.121.10.3207. [DOI] [PubMed] [Google Scholar]
- Sariola H. Nephron induction revisited: from caps to condensates. Curr Opin Nephrol Hypertens. 2002;11:17–21. doi: 10.1097/00041552-200201000-00003. [DOI] [PubMed] [Google Scholar]
- Saxen L. Organogenesis of the kidney. Cambridge University Press, Cambridge [Cambridgeshire]; New York: 1987. [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. Embo J. 2006;25:5214–28. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]