Luciferases are widely used to monitor various biological processes. Here, we describe the naturally secreted Gaussia luciferase as a highly sensitive reporter for quantitative assessment of cells in vivo by measuring its levels in blood. The Gluc blood assay complements in vivo bioluminescence imaging which has the ability to localize the signal and provides a multifaceted assessment of cell viability, proliferation and location in experimental disease and therapy models.
Luciferase-mediated bioluminescence imaging has served as a reporting tool for monitoring various biological processes in vitro and in vivo in different fields3, including immunology4 oncology5, virology6, and neuroscience7. After systemic substrate injection, a charge coupled device (CCD) camera can be used to localize the luciferase photon signals in vivo. Recently, we have characterized a naturally secreted luciferase from the marine copepod Gaussia princeps (Gaussia luciferase, Gluc) and found it to be over 2000-fold more sensitive than firefly and Renilla reniformis luciferases and 20,000-fold more sensitive than the secreted alkaline phosphatase (SEAP) when expressed in mammalian cells8,9. Gluc expression levels can be easily quantified in cell-free, conditioned medium by adding its substrate coelenterazine and measuring emitted photons using a luminometer. Since Gluc is secreted from mammalian cells in culture9, we hypothesized that it might also be secreted into the blood of animals harboring cells expressing this reporter.
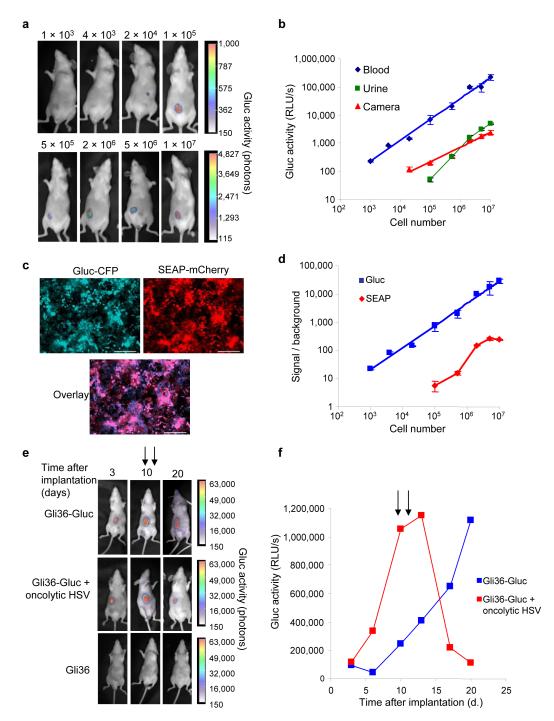
In order to assess the potential of Gluc as a reporter to monitor biological processes by measuring its level in the blood of small animals, we transduced Gli36 human glioma cells with a lentivirus vector encoding Gluc (Gli36-Gluc) and implanted them in different numbers into the flanks of nude mice. We visualized the tumors 3 days post-implantation by in vivo bioluminescence imaging after intravenous (i.v.) injection of the Gluc substrate, coelenterazine (4 mg/kg body weight) and acquiring photon counts using a CCD camera (Fig. 1a). At the same time, we withdrew 5 μl blood samples from these mice and directly aliquoted them into tubes containing EDTA (1 μl 20 mM), after which we measured the Gluc activity by adding coelenterazine (100 μM) and acquiring photon counts using a luminometer (Supplementary Methods online). The Gluc activity in the blood was linear with respect to cell number in a range covering over 5 orders of magnitude, and correlated well with values obtained using the CCD camera (Fig. 1b). Further, Gluc activity could also be detected in the urine, albeit to a lesser extent than in the blood, which indicates it is cleared by the kidneys (Fig. 1b). In addition, there was no detrimental effect of EDTA on Gluc activity measured in blood (Supplementary Fig. 1a online), and no significant differences were detected between the Gluc activity measured in serum or whole blood samples, showing that hemoglobin, which can interfere with luciferase measurement10, did not have a significant effect on Gluc activity under our assay conditions (Supplementary Fig. 1b online). Gluc samples could be stored at 4 °C for several days without significant decay of activity, with Gluc half-life being around 6 days (Supplementary Fig. 2a online). Further, the half-life of Gluc in the circulation in vivo is approximately 20 min (Supplementary Fig. 2b online), suggesting only a minor contribution of Gluc accumulation over time to the total Gluc signal measured in blood samples.
Figure 1. Monitoring of Gluc blood levels with subcutaneous tumor model.
(a) Different numbers of Gli36-Gluc cells were implanted subcutaneously in mice (n = 4) and imaged with a CCD camera three days later. (b) Total relative light units (RLU) per second was calculated for tumors in (a) (red line). Gluc activity was measured in blood (blue line) or urine (green) using the luminometer. Results are presented as mean ± SD with p<0.001 as calculated by student T-test. (c and d) Different numbers of Gli36 cells expressing both Gluc-CFP and SEAP-mCherry (c) were implanted subcutaneously in mice and Gluc (in blood) or SEAP (in serum) activity was assayed 2 days later (d). Results are showing as mean ± SD with p<0.001. Bar, 100 μm. (e and f) Mice were implanted with 1×106 Gli36-Gluc cells subcutaneously and tumor growth was monitored by both in vivo bioluminescence imaging (e) and the Gluc blood assay (f). At day 10 and 13 post-implantation, one set of mice was injected intra-tumorally (arrows) with an oncolytic HSV vector (108 pfu; red line) and another set with PBS (blue line) (n=3/group). The results shown are from one representative mouse from each group.
We next compared the Gluc blood assay to that of secreted alkaline phosphatase (SEAP), a well-established marker monitored in serum11. We co-infected (>90%) Gli36 cells with two different lentivirus vectors, one carrying the expression cassette for Gluc and cerulean fluorescent protein (CFP) separated by an internal ribosomal entry site (IRES) element, and the other a similar cassette encoding SEAP and mCherry, both driven by the CMV promoter (Fig. 1c). We implanted different numbers of these cells subcutaneously in the flanks of nude mice and assayed the Gluc activity in 5 μL blood samples, as above, and SEAP activity in 5 μL serum as described11. The Gluc blood assay was over 1000-fold more sensitive than the SEAP assay, being able to detect 1000 cells in vivo with signal/background (S/B) of 20, while the SEAP assay could only detect 500,000 cells with S/B of 10. Further the Gluc assay was linear with respect to cell number in a range covering over 5 orders of magnitude, while the SEAP assay fluctuated over 3 orders of magnitude with a plateau at around one million cells which could lead to greater underestimation of cell number. While the Gluc assay can be carried out with equal efficiency in blood or serum samples in few seconds, SEAP activity cannot be measured in blood since hemoglobin inhibits it. Also SEAP assay requires around 2 hrs handling prior to measurement. Gluc activity can be measured in urine, while at no time point was SEAP activity detected in the urine (data not shown). Gluc has a short half-life of 20 min in the blood allowing dynamic events to be monitored, whereas the half-life of SEAP is 3 h11, leading to accumulation over time. Gluc can also be used to localize expressing cells in the animal by in vivo bioluminescence imaging, giving that sufficient cells are present at one location, while SEAP does not have this added advantage.
To determine whether Gluc activity in the blood could be used to monitor tumor growth and therapy in vivo, we implanted nude mice subcutaneously with one million Gli36-Gluc cells. We monitored tumor growth at different time points both with a CCD camera after i.v. injection of coelenterazine, or by assaying 5 μl blood samples for Gluc activity using a luminometer. At day 10 and 13 post-implantation, we injected one set of mice intra-tumorally with an oncolytic HSV virus [hrR3, 108 plaque-forming unit (pfu)]12 and a parallel control set with phosphate buffer saline (PBS). The Gluc signal in the blood from tumors treated with the oncolytic virus decreased dramatically, confirming its anti-tumor activity, while tumors treated with PBS increased logarithmically over time, with correlative changes in tumor volume in the CCD camera images (Fig. 1e and f). These results indicate that the Gluc levels in the blood can serve as a quantitative marker for the number of tumor cells expressing it in vivo, with complementary localization of the signal using a CCD camera.
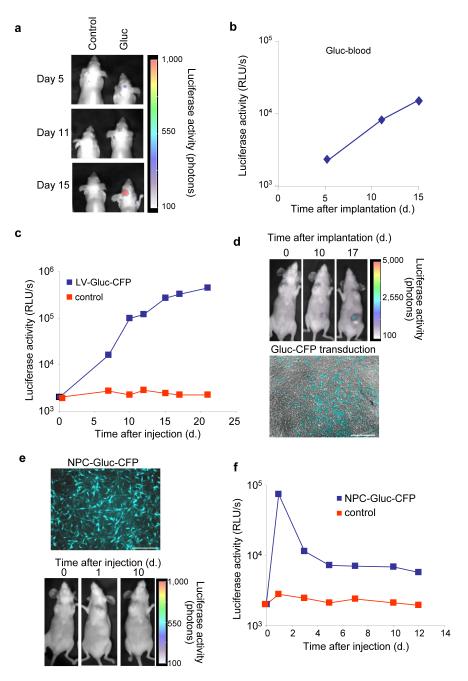
In order to evaluate the usefulness of the Gluc blood assay to monitor tumor growth in deep tissues, we stereotactically injected one hundred thousand Gli36-Gluc cells into the brains of nude mice and monitored tumor volume over time using the CCD camera (Fig. 2a). At the same time, we measured the levels of Gluc activity in the blood which showed increasing signal over time, indicating that Gluc is able to pass out through the brain tumor-barrier (BTB), allowing the easy monitoring of tumor growth in the brain from peripheral blood samples (Fig. 2b). The BTB is more permeable, however, than the blood brain-barrier, so Gluc exit from normal brain into the circulation might be more restricted.
Figure 2. Gluc reporter in the blood as a useful tool to monitor in vivo processes.
(a and b) 1×105 Gli36-Gluc cells were implanted in the brains of nude mice (n=3/group) and tumor growth was monitored by in vivo bioluminescence imaging (a) or by measuring Gluc activity using the luminometer (b). (c and d) 1×106 Gli36 cells were implanted subcutaneously and tumors were either injected with LV-Gluc-CFP or PBS, 3 days later. Viral delivery was monitored over time by measuring Gluc activity in blood samples (c), by in vivo bioluminescence imaging using the CCD camera (d, upper panel) and by monitoring CFP expression in tumor sections (d, lower panel). (e and f) One millions C17.2 NPCs expressing Gluc and CFP (e, upper panel) or PBS were injected i.v. in nude mice. Gluc activity was monitored over time using the CCD camera (e, lower panel) and in blood samples using the luminometer (f). Data shown are from a representative mouse from each set. Scale bar, 100 μm.
To determine the usefulness of the Gluc blood assay to monitor gene transfer in vivo, we implanted Gli36 tumors subcutaneously in nude mice and injected them with either a lentivirus vector carrying the expression cassette for Gluc and CFP or with PBS. A clear increase in the Gluc blood values was observed after viral transduction, indicating that stable gene transfer had occurred with inheritance to daughter cells (Fig. 2c). The Gluc signal was localized to the tumor by in vivo bioluminescence imaging and gene transfer was confirmed by analysis of tumor sections for CFP fluorescence (Fig. 2d). These results show that the Gluc blood assay can also be used to monitor gene transfer and proliferative fate of transduced tumor cells and thus serve as an index of gene therapy. This type of analysis could be extended for quantitative assessment of replication of virus vectors with applications in virology and vaccination.
In order to evaluate whether the Gluc blood assay could be used to monitor circulating cells in vivo, we injected nude mice systemically (i.v.) with one million C17.2 neuronal precursor cells (NPCs) expressing Gluc and CFP (Fig 2e) and monitored Gluc activity in 20 μl blood over time. Mice injected with NPC-Gluc cells showed an initial high Gluc value which decreased after 3 days, indicating that a significant number of the NPCs survived the injection procedure. By four days post-injection the level of Gluc stabilized, indicating that the surviving cells were maintained but did not proliferate (Fig. 2f). We were not able to localize a Gluc signal by CCD camera imaging anywhere in the body, suggesting that the NPC cells did not concentrate in any one tissue (Fig. 2e). These results show that the Gluc blood assay can be used to monitor circulating cell viability in vivo and, as such, could be extended to monitor stem cell transplantation.
Here, we describe a novel use for the Gaussia luciferase (Gluc) reporter as a tool for quantitative assessment of different biological processes in small animals by measuring its level in the blood. The Gluc blood assay was shown to be useful in monitoring tumor growth and therapy, gene transduction as well as circulating cell survival and can be readily extended to many other applications involving luciferase-mediated bioluminescence. The Gluc blood assay provides a sensitive and quantitative assessment of numbers of transduced cells in vivo, complementing in vivo bioluminescence imaging which has the unique ability to localize the signal, and thereby greatly facilitating non-invasive monitoring of numerous biological processes.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Cancer Institute P50 CA86355-04 (RW and XOB), 1K99CA126839-01 (BAT) and the Brain Tumor Society (XOB and BAT). We thank M. Sena-Esteves for help with vector generation and useful discussions, M. Whalen, F. Swirski and M. Pittet for advice, as well as C. Maguire and N. Lewandrowski for technical help.
References
- 1.Contag CH, Ross BD. It’s not just about anatomy: in vivo bioluminescence imaging as an eyepiece into biology. J Magn Reson Imaging. 2002;16:378–387. doi: 10.1002/jmri.10178. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross S, Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Negrin RS, Contag CH. In vivo imaging using bioluminescence: a tool for probing graft-versus-host disease. Nat Rev Immunol. 2006;6:484–490. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 5.Adams JY, et al. Visualization of advanced human prostate cancer lesions in living mice by a targeted gene transfer vector and optical imaging. Nat Med. 2002;8:891–897. doi: 10.1038/nm743. [DOI] [PubMed] [Google Scholar]
- 6.Luker GD, et al. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J Virol. 2002;76:12149–12161. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J, Lin AH, Masliah E, Wyss-Coray T. Bioluminescence imaging of Smad signaling in living mice shows correlation with excitotoxic neurodegeneration. Proc Natl Acad Sci U S A. 2006;103:18326–18331. doi: 10.1073/pnas.0605077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2:e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Colin M, et al. Haemoglobin interferes with the ex vivo luciferase luminescence assay: consequence for detection of luciferase reporter gene expression in vivo. Gene therapy. 2000;7:1333–1336. doi: 10.1038/sj.gt.3301248. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu N, Kasai A, Hayakawa K, Yao J, Kitamura M. Real-time detection and continuous monitoring of ER stress in vitro and in vivo by ES-TRAP: evidence for systemic, transient ER stress during endotoxemia. Nucleic Acids Res. 2006;34:e93. doi: 10.1093/nar/gkl515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramm CM, et al. Herpes vector-mediated delivery of marker genes to disseminated central nervous system tumors. Hum Gene Ther. 1996;7:291–300. doi: 10.1089/hum.1996.7.3-291. [DOI] [PubMed] [Google Scholar]
- 13.Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.