Abstract
A 6-year-old boy had progressive muscle weakness since age 4 and emotional problems diagnosed as Asperger syndrome. His mother and two older siblings are in good health and there is no family history of neuromuscular disorders. Muscle biopsy showed ragged-red and cytochrome c oxidase (COX)-negative fibers. Respiratory chain activities were reduced for all enzymes containing mtDNA-encoded subunits, especially COX. Sequence analysis of the 22 tRNA genes revealed a novel G10406A base substitution, which was heteroplasmic in multiple tissues of the patient by RFLP analysis (muscle, 96%; urinary sediment, 94%; cheek mucosa, 36%; blood, 29%). The mutation was not detected in any accessible tissues from his mother or siblings. It appears that this mutation arose de novo in the proband, probably early in embryogenesis.
Introduction
The mitochondrial encephalomyopathies are a diverse group of disorders often caused by defects in mitochondrial DNA (mtDNA). Hundreds of large-scale mtDNA rearrangements and over 150 mtDNA point mutations have been associated with disease [1]. Patients harboring these mutations usually have multisystem disorders, but phenotypic expression can vary in different families and even in different members of the same family. This complex genotype-phenotype correlation is due to many factors, including the abundance of mutant mDNA and its tissue distribution. Most pathogenic point mutations occur in tRNA genes, are heteroplasmic, and are maternally inherited. Here, we report a patient with weakness and hypotonia, who harbors a previously undescribed and apparently de novo G-to-A mutation at nucleotide position 10406 in the tRNA arginine (tRNAarg) gene.
Case Report
A 6-year-old boy was the third child of non-consanguineous parents. He had normal birth and early development, but showed emotional problems and, at age 4, was given the diagnosis of Asperger syndrome. At the same age, the mother also noted motor problems: the child could not walk long distances, required a walker or a wheelchair for family outings, and needed a handrail to push himself up stairs. The condition did not seem to progress significantly. At age 7, he has occasional dysphagia for solid foods. He attends a regular school and is a good student, but has difficulties with social interactions and does not tolerate changes in his routine. The patient’s mother and older siblings, a sister and a brother, are in good health and there is no family history of neuromuscular disorders. Physical examination is normal. Neurological examination shows poor eye contact and diffusely decreased muscle bulk. His walks with a slight waddle and on his toes because of bilateral ankle equinus contractures, but can run. His strength is decreased (4/5) proximally in his arms and both proximally and distally in his legs. Deep tendon reflexes are normal and plantars are flexor. Sensation is intact.
Serum CK was 134 IU (normal, <190 ) and serum lactate was 62 mg/ml on one occasion (normal, <20 mg/ml) but was normal (14 mg/ml) in a second measurement. Total serum carnitine was normal (50–56μmol/L) but the esterified fraction was increased (23–25 μmol/L), with a relative decrease of free carnitine (28–31 μmol/L). EMG of the left quadriceps muscle was normal. Brain MRI showed mild, nonspecific T2 hyperintensity in the periventricular region.
Methods
Histochemistry and Biochemistry
Histochemical study of muscle using 8-um-thick frozen sections was carried out as described [2]. Biochemical analysis was performed in 10% muscle homogenates as previously described [3].
Molecular analysis
Total DNA was extracted by standard protocol (PUREGENE, Gentra System, Inc, Minneapolis, Minn) following the manufacturer’s instructions. Protocols used for blood, urine, hair roots, and cheek mucosa were as described [4].
Direct sequencing of the 22 tRNA genes of mtDNA was performed in an ABI Prism 310 Genetic Analyzer using Big Dye Terminator Cycle Sequencing Reaction Kits (Perkin-Elmer Applied Biosystems, Foster City, CA).
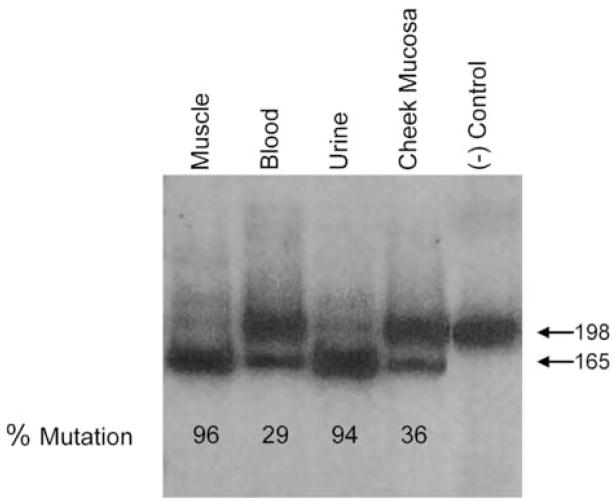
For restriction fragment length (RFLP) analysis, mtDNA was amplified by polymerase chain reaction (PCR) using forward and reverse primers at nt positions 10371–10405 and 10549 –10568, respectively. The forward primer had a mismatch at nt position 10403, which, in combination with the mutant A at position 10406, creates a restriction site for the restriction enzyme Dde1. The 198-bp PCR-amplified product was labeled and digested with DdeI, which cleaves only the fragment containing the G10046A mutation. The digested product was electrophoresed in a 12% non-denaturing acrylamide gel and analyzed in a phosho-imager (Molecular Analyst, BioRad, Hercules, CA) using Image-Quant software (Molecular Dynamics, Sunnyvale, CA) to assess the percentage of the mutation.
Results
Histochemical analysis of the muscle biopsy showed frequent ragged-red fibers (RRF) and cytochrome c oxidase (COX)-deficient fibers. In keeping with the mitochondrial proliferation shown by histochemistry, biochemical analysis revealed markedly increased citrate synthase and succinate dehydrogenase (SDH) activities. In contrast, activities of respiratory chain complexes I+III, II+III, and especially IV (COX) were decreased (Table 1). When normalized to either citrate synthase or SDH, the defects of complexes I, III, and IV were even more apparent.
Table 1.
Mitochondrial enzyme activities in 10% muscle homogenate
| Enzyme | Complex | Patient Activity* | Controls±SD |
|---|---|---|---|
| Cytochrome c oxidase | IV | 0.54 | 2.80±0.52 |
| Succinate cyt. c reductase | II + III | 0.48 | 0.70±0.23 |
| NADH-cyt. c reductase | I + III | 0.51 | 1.02±0.38 |
| NADH-dehydrogenase | I | 18.96 | 35.48±7.07 |
| Citrate synthase | 32.33 | 9.88±2.55 | |
| Succinate dehydrogenase | 4.91 | 1.00±0.53 |
micromoles/min/gm
The presence of RRF and COX-negative fibers and the reduced activities of multiple respiratory chain enzymes suggested a mutation in a mtDNA transfer RNA (tRNA) gene. Direct sequencing of all 22 tRNA genes revealed a novel mutation, G-to-A at nucleotide position 10406 in the tRNA for arginine (tRNAArg). PCR/RFLP analysis showed that this mutation was heteroplasmic in multiple tissues from the patient, with a mutant load of 96% in muscle, 94% in urinary sediment, 36% in cheek mucosa, and 29% in blood (Figure 1). Mutant load in hair follicles varied from 0–7%. This mutation was not detected in any of four accessible tissues (blood, urine, cheek mucosa, and hair follicles) from the patient’s mother, siblings, and – as expected - father (Figure 2).
Figure 1.
A. PCR/RFLP analysis of DNA amplified from multiple tissues from the patient. In the presence of the mutation, a 198 base pair (bp) fragment is digested by Dde1 into two fragments of 165 and 33 bp.
Figure 2.

Pedigree. Mutant load is shown for various tissues from the patient and his family members. Not Detected = No mutation was detected in urine, cheek mucosa, blood, or hair follicles.
Discussion
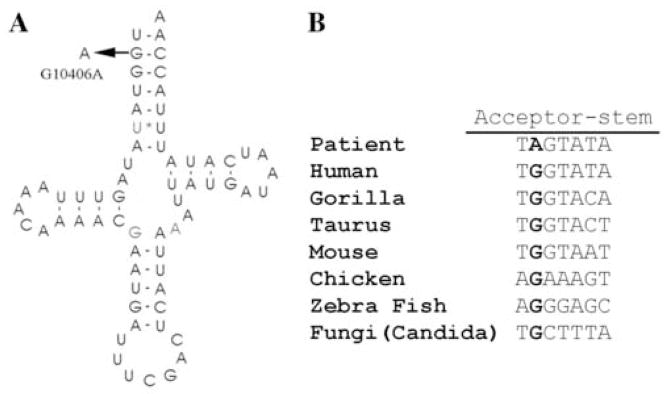
We describe a novel mutation, a G10406A in the tRNAArg gene, which appears to have arisen de novo in the proband, a 6-year-old boy with proximal myopathy and Asperger syndrome. We believe that this mutation is pathogenic for several reasons. First, it is heteroplasmic, and heteroplasmy is a common feature of pathogenic mtDNA mutations. Second, it has not been previously reported and was not detected by us in a group of 100 controls. Third, the point mutation that we encountered in a tRNA gene is consistent with the histochemical observation of COX negative RRF and the biochemical findings of reduced activities for respiratory chain enzymes containing mtDNA-encoded subunits. Fourth, the mutation in the tRNA acceptor stem affects a site that is conserved among species (Figure 3). Although we considered performing single fiber PCR to bolster the evidence of pathogenicity, we decided against it for two reasons: (i) the quality of the frozen sample was not adequate; and (ii) the overall very high level of mutant mtDNAs decreased the value of this assay.
Figure 3.
A. Schematic representation of the tRNAArg cloverleaf structure, showing the mutation and the affected base pair. B. Comparison of this region of tRNAArg gene sequences across different species showing that the mutated base pair is highly conserved.
It is interesting to note the tissue distribution of the G10406A mutation in this family (Figure 2). In the proband, the mutant load was high in the urinary sediment (94%) and in muscle (96%), lower in cheek mucosa (36%), and lowest in blood (29%). This pattern is similar to that found for the common A3243G mutation typically associated with MELAS (mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) [4]. To our surprise, we could not detect the mutation in any accessible tissues from the patient’s mother or siblings. To rule out the unlikely scenario of paternal transmission, we analyzed DNA from the father and could not detect the mutation by PCR/RFLP. It appears that this mutation arose de novo in the proband, probably early in embryogenesis because it is present in multiple tissues.
Mutations in tRNAArg have been reported twice before. A T10410C mutation was found in a patient with “Alpers disease”, who, however, was not described clinically [5]. When mutant mtDNA was transferred to cells depleted of mtDNA (rho0 cells) to create cybrid cell lines, it was observed that the T10410C mutation alone did not produce a biochemical phenotype, leading to the conclusion that this was a polymorphism, not a pathogenic mutation.
The second mutation, A10438G, was found in an 8 year-old Finnish boy with moderate mental retardation, broad-based gait, weak facial muscles, and decreased visual acuity [6]. The blood lactate/pyruvate ratio was increased. Muscle histochemistry showed a predominance of type 1 fibers, but no ragged-red fibers, and electron microscopy revealed increased amount of lipid droplets and giant mitochondria. Biochemical analysis showed decreased complex IV activity. This mutation was heteroplasmic, with 88% mutant mtDNA in muscle and 73% in blood from the patient, 19% in the mother’s blood, and 17% in the grandmother’s blood. There are no similarities between this mutation and the one we are reporting. Not only were the clinical presentations of the two patients different but the positions of the two mutations in the cloverleaf structure of human tRNAs are also different. The mutation we are reporting, G10406A, is in the acceptor stem whereas the T10438C mutation is in the nucleotide flanking the anticodon. In addition, the T10438C mutation was present (as expected) in maternal relatives of the patient, whereas the 10406 mutation appears to have arisen de novo. It is noteworthy that our patient was diagnosed with Asperger syndrome because several patients with autistic spectrum disorders carried mtDNA mutations or mtDNA depletion [7].
This case underscores the importance of sequencing all 22 mtDNA genes in cases where morphology and biochemistry suggest a tRNA mutation, even when there is no evidence of maternal inheritance.
Acknowledgments
This work has been supported by NIH grant HD32062, a grant from the Muscular Dystrophy Association, and the Marriott Mitochondrial Disorders Clinical Research Fund (MMDCRF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schon EA. Appendix. In: DiMauro S, Hirano M, Schon EA, editors. Mitochondrial Medicine. London: Taylor & Francis; 2006. pp. 329–35. [Google Scholar]
- 2.Tanji K, Bonilla E. Optical imaging techniques (histochemical, immunohistochemical, and in situ hybridization staining methods) to visualize mitochondria. Meth Cell Biol. 2001;65:311–32. doi: 10.1016/s0091-679x(01)65019-2. [DOI] [PubMed] [Google Scholar]
- 3.DiMauro S, Servidei S, Zeviani M, et al. Cytochrome c oxidase deficiency in Leigh syndrome. Ann Neurol. 1987;22:498–506. doi: 10.1002/ana.410220409. [DOI] [PubMed] [Google Scholar]
- 4.Shanske S, Pancrudo J, Kaufmann P, et al. Varying loads of the mitochondrial DNA A3243G mutation in different tissues: Implications for diagnosis. Am J Med Genet. 2004;130A:134–7. doi: 10.1002/ajmg.a.30220. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi JI, Ohta S, Kagawa Y, et al. Functional and morphological abnormalities of mitochondria in human cells containing mitochondrial DNA with pathogenic point mutations in tRNA genes. JBiolChem. 1994;269:19060–6. [PubMed] [Google Scholar]
- 6.Uusimaa J, Finnila S, Remes AM, et al. Molecular epidemiology of childhood mitochondrial encephalomyopathies in a Finnish population: sequence analysis of entire mtDNA of 17 children reveals heteroplasmic mutations in tRNAArg, tRNAGlu, and tRNALeu(UUR) genes. Pediatrics. 2004;114:443–50. doi: 10.1542/peds.114.2.443. [DOI] [PubMed] [Google Scholar]
- 7.Pons R, Andreu AL, Checcarelli N, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. Pediatr. 2004;144:81–5. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]