Abstract
ALIX recruits ESCRT-III CHMP4 and is involved in membrane remodeling during endosomal receptor sorting, budding of some enveloped viruses and cytokinesis. We show that ALIX dimerizes via the middle domain (ALIX-V) in solution. Structural modeling based on small angle X-ray scattering (SAXS) data reveal an elongated crescent shaped conformation for dimeric ALIX lacking the proline rich domain (ALIXBRO1-V). Mutations at the dimerization interface prevent dimerization and induce an open elongated monomeric conformation of ALIX-V as determined by SAXS modeling. ALIX dimerizes in vivo and dimeric ALIX co-localizes with CHMP4B upon co-expression. We show further that ALIX dimerization affects HIV-1 budding. C-terminally truncated activated CHMP4B retaining the ALIX binding site forms linear, circular and helical filaments in vitro, which can be bridged by ALIX. Our data suggest that dimeric ALIX represents the active form that interacts with ESCRT-III CHMP4 polymers and functions as a scaffolding protein during membrane remodeling processes.
Keywords: ALIX, ESCRT-III, CHMP4, multivesicular body (MVB), budding, HIV, cytokinesis
Introduction
Endosomal sorting processes determine the fate of plasma membrane receptors that are either recycled or delivered to the lysosome for degradation. (Gruenberg and Stenmark, 2004; Hicke, 2001; Katzmann et al., 2002). An intricate network of protein interactions, which includes the sequential recruitment of ESCRT (Endosomal Sorting Complexes Required for Transport) complexes 0, I, II and III and associated molecules, regulates endosomal protein sorting, vesicle formation and budding (Saksena et al., 2007) (Hurley, 2008)(Lata et al., 2009). The accessory protein ALIX interacts with ESCRT-I Tsg101 (Mahul-Mellier et al., 2006; Strack et al., 2003) as well as with all ESCRT-III CHMP4 isoforms (Odorizzi et al., 2003)(Katoh et al., 2003)(Fisher et al., 2007)(McCullough et al., 2008). ESCRT-III proteins polymerize on membranes (Babst et al., 2002)(Lin et al., 2005) by forming filamentous (Ghazi-Tabatabai et al., 2008), circular (Hanson et al., 2008) and helical tubular structures (Lata et al., 2008b) that act during membrane remodeling processes including membrane abscission (Lata et al., 2009). ESCRT-III proteins form inactive closed conformations in the cytosol and undergo polymerization and membrane targeting upon activation (Muziol et al., 2006)(Zamborlini et al., 2006)(Shim et al., 2007)(Lata et al., 2008a)(Lata et al., 2008b). It is most likely that ALIX interacts only with an activated form of CHMP4 via a C-terminal CHMP4 binding motif (McCullough et al., 2008). Thus ALIX function must be linked to the role of ESCRT-III in membrane remodeling processes. In fact, ALIX itself might directly control changes in membrane structures (Matsuo et al., 2004)(Falguieres et al., 2008).
ALIX was originally thought to be a homologue of yeast Bro1 that functions in receptor down regulation (Odorizzi et al., 2003)(Kim et al., 2005), but this role could not be confirmed (Cabezas et al., 2005)(Schmidt et al., 2004)(Doyotte et al., 2008).
ALIX is recruited to the budding process of some enveloped viruses such as HIV-1 and equine infectious anemia virus (EIAV) (von Schwedler et al., 2003)(Strack et al., 2003)(Martin-Serrano et al., 2003). Although EIAV budding is strictly dependent on ALIX (Martin-Serrano et al., 2003)(Strack et al., 2003), the EIAV late domain can be replaced by ESCRT-I Vps28, which provides access to ESCRT-III (Tanzi et al., 2003)(Pineda-Molina et al., 2006). In contrast, ALIX is not sufficient to support HIV-1 budding in the absence of the Tsg101/ESCRT-I function (Garrus et al., 2001)(Martin-Serrano et al., 2001). However, over-expression of ALIX or NEDD4 can rescue HIV-1 late domain mutants (Usami et al., 2007)(Chung et al., 2008)(Usami et al., 2008).
Both ALIX and ESCRT-III family members are recruited to the midbody during cytokinesis (Carlton and Martin-Serrano, 2007)(Morita et al., 2007) where ALIX competes with Tsg101 for CEP55 interaction (Lee et al., 2008). Although CEP55 itself is not required for virus budding nor any endosome-related processes, virus budding and cytokinesis require ESCRT-III and VPS4 function.
ALIX is composed of three domains, an N-terminal Bro1-like domain (Kim et al., 2005)(Fisher et al., 2007), which harbors the CHMP4B binding site (McCullough et al., 2008), followed by the V-shaped middle domain composed of two three helical bundles that expose the retroviral late domain binding sites (Lee et al., 2007)(Fisher et al., 2007)(Zhai et al., 2008); the third domain is a proline rich domain (PRD) that serves as a platform for multiple interaction partners including ESCRT-I Tsg101 (von Schwedler et al., 2003)(Shibata et al., 2004)(Schmidt et al., 2004)(Chatellard-Causse et al., 2002) and CEP55 (Carlton and Martin-Serrano, 2007)(Morita et al., 2007)(Lee et al., 2008). PRD harbors the Alg-2 binding site (Suzuki et al., 2008), which links ALIX to apoptotic processes (Missotten et al., 1999)(Vito et al., 1999)(Trioulier et al., 2004)(Mahul-Mellier et al., 2006)(Mahul-Mellier et al., 2008). The scaffold function of ALIX is further underlined by its control of the actin cytoskeleton (Cabezas et al., 2005; Schmidt et al., 2003)(Pan et al., 2006). Finally PRD regulates the function of ALIX by keeping it in an auto-inhibited state (Zhou et al., 2008b)(Zhou et al., 2008a).
Here we show that ALIX dimerizes in vitro and in vivo via the V-domain, which helps to explain the dominant negative effect on HIV-1 budding observed for truncated ALIX constructs. We determine a main dimerization interface within the hinge region of the V-domain and present a low resolution model of dimeric ALIX based on SAXS analysis. Electron microscopy data indicate further that dimeric ALIX can bridge CHMP4 filaments. Such a scaffolding function may thus help to position ESCRT-III on membranes and allow recruitment of a variety of effectors via its PRD region.
Results
MALLS and SAXS analysis of ALIX
Both recombinant ALIXBro1-V and ALIXV elute from gel filtration columns in two distinct peaks. Analytical size exclusion chromatography in combination with multi angle laser light scattering (MALLS) revealed monomers for ALIXBro1-V and ALIXV as expected (Fisher et al., 2007)(Lee et al., 2007) as well as dimeric forms with average molecular weights of 158 ± 3 kDa for ALIXBro1-V (Figure 1A) and 76 ± 3 kDa for ALIXV (Figure 1B). The ratio of monomers to dimers purified from E. coli was ~ 5:1 for ALIXBro1-V and ~ 3:1 for ALIXV. Neither ALIXBro1-V nor ALIXV monomers display a concentration dependent equilibrium with their respective dimers. Concentration of ALIXBro1-V and ALIXV monomers up to 20 mg/ml did not produce dimers, nor did dilution of dimers produce monomers as judged by gel filtration chromatography in agreement with analytical ultracentrifugation results (Fisher et al., 2007). Thus no dimer to monomer exchange could be observed.
Figure 1. ALIX forms monomers and dimers in solution.
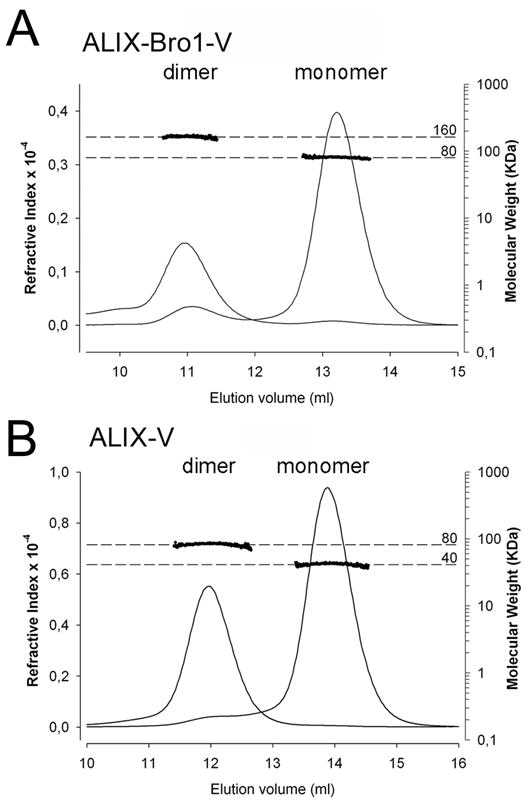
(A) ALIXBro1-V and (B) ALIXV, analyzed by MALLS indicates molecular weights of ~80 and ~160 kDa for ALIXBro1-V and ~40 and ~80 kDa for ALIXV; left y-axis, refractive index; the molecular weight is blotted on the right axis in a logarithmic scale; x-axis shows the elution volume of the peaks.
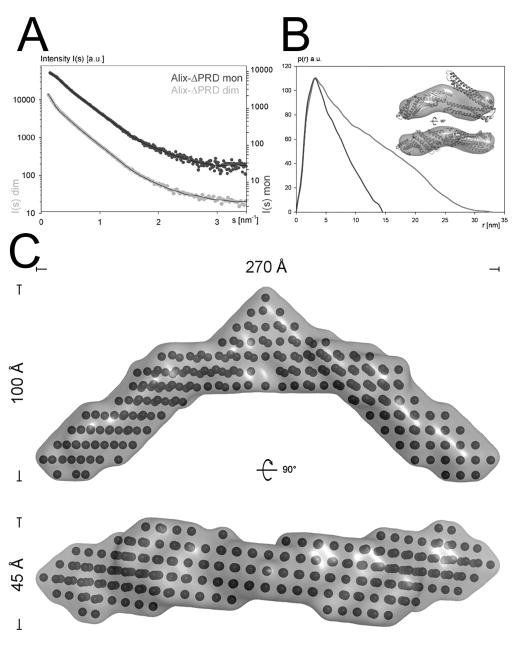
In order to understand the structural basis of ALIX dimerization, both ALIXBro1-V monomers and dimers were analyzed by small angle X-ray scattering. The scattering intensity patterns corresponding to monomeric and dimeric ALIXBro1-V are shown in Figures 2A. The Guinier analysis revealed a radius of gyration (Rg) of 40.6 ± 0.1 Ǻ for monomeric and 81.2 ± 0.8 Ǻ for dimeric ALIXBro1-V. Maximal protein dimensions (Dmax) of ~170 Ǻ for the monomeric ALIXBro1-V and ~300 Ǻ for dimeric ALIXBro1-V were found by the distance distribution function p(r) computed by a Fourier transformation of the scattering intensity (Figure 2B). This thus indicated that dimerization produced an elongated shaped ALIXBro1-V. The shapes of the ALIX conformers were determined ab initio and reconstructed models of ALIXBro1-V fit the corresponding experimental data with the discrepancy χ of 1.9 and 1.2. The ab initio modeling produced an elongated shape with dimensions of ~ 40 × 60 × 145 Ǻ for monomeric ALIXBro1-V (Figure 2B, inset), which is largely consistent with the crystal structure of ALIXBro1-V (Fisher et al., 2007). Comparison of the experimental scattering curve and the one calculated from the ALIXBro1-V crystal structure revealed a discrepancy χ of 2.7. Ab initio modeling of dimeric ALIXBro1-V yielded a crescent-shaped structure with a distance of ~ 270 Ǻ between the ends spanning the concave surface of the dimer (Figure 2C).
Figure 2. Small angle X-ray scattering analysis of ALIXBro1-V.
(A) Experimental scattering intensity patterns obtained for monomeric (dark grey) and dimeric (light grey) ALIXBro1-V are shown as a function of resolution and after averaging and subtraction of solvent scattering. The scattering intensity patterns calculated from the ALIXBro1-V monomer and dimer SAXS model with the lowest χ value are shown as black lines.
(B) P(r) function of both monomeric (dark grey) and dimeric (light grey) ALIXBro1-V (both curves have been adjusted to the same height). The ab initio model envelope of monomeric ALIXBro1-V is shown together with the manually docked ALIXBro1-V structure (Fisher et al., 2007) as inset.
(C) Ab initio modeling of dimeric ALIXBro1-V reveals a crescent shape model spanning ~ 270 Å; two orientations of the bead model including the molecular envelope are shown.
Mapping of the dimer interface
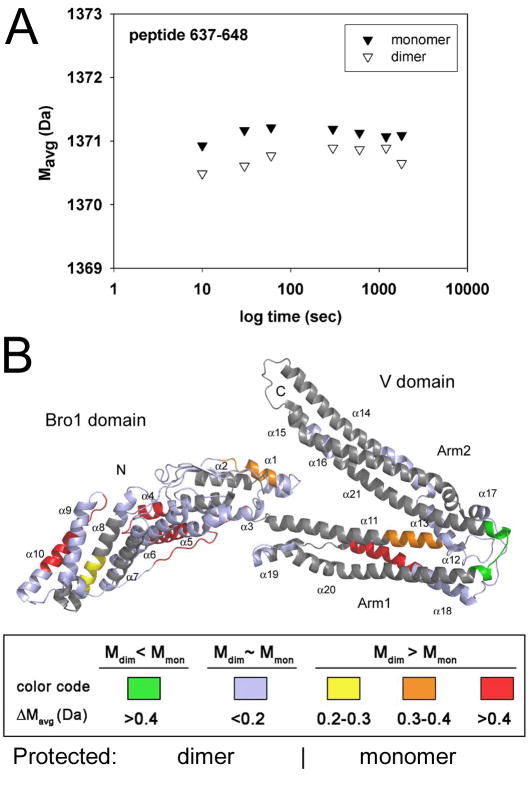
In order to analyze the mode of dimerization, D2O labeling of ALIXBro1-V coupled to peptide mapping by mass spectrometry analyses were applied. The resulting peptides were mapped onto the structure of monomeric ALIXBro1-V and scored depending on an increase or decrease in D2O labeling. The most pronounced decrease in D2O labeling was observed for a peptide (aa 637–648) corresponding to a potential hinge region between the two arms of ALIXV (Fisher et al., 2007)(Lee et al., 2007), suggesting that this region is occluded in dimeric ALIX (Figure 3A). The labeling method also revealed higher accessibilities to D2O labeling in the dimer as compared to the monomer (Figure S1); this includes one region between the two arms of ALIXV (helices α11 and α18), several short helical segments within ALIXBro1 (helices α4, α5, α8 and α10) and the long extended conformation spanning the concave surface of ALIXBro1 connecting α10 andα11 (Figures 3B and S1). The data thus indicate a dimerization interface within ALIXV and potential conformational flexibility within ALIXBro1.
Figure 3. Mapping of the conformational flexibility of ALIXBro1-V.
Hydrogen/deuterium (H/D) exchange labeling coupled to mass spectrometry analysis was used to determine conformational differences between monomeric and dimeric ALIXBro1-V. (A) Example of local H/D exchange kinetic for the peptide sequence 637–648. The average mass (Mavg) is plotted against D2O incubation time. This reveals a difference in H/D exchange within this region where dimeric ALIXBro1-V shows less deuterium exchange compared to monomeric ALIXBro1-V over a period of 30 min.
(B) The H/D exchange results have been plotted onto the structure of ALIXBro1-V (Fisher et al., 2007) and the differences in deuteration between monomer and dimer (ΔMavg) are scored according to the following color code: green regions are more protected from H/D exchange in dimeric ALIXBro1-V while regions shown in red < orange < yellow are less accessible to H/D exchange within monomeric ALIXBro1-V. Regions in blue show similar accessibility between monomeric and dimeric ALIXBro1-V and regions in grey were not covered by peptide mapping of the deuterated forms.
Generation of an elongated “open” monomeric ALIXV conformation
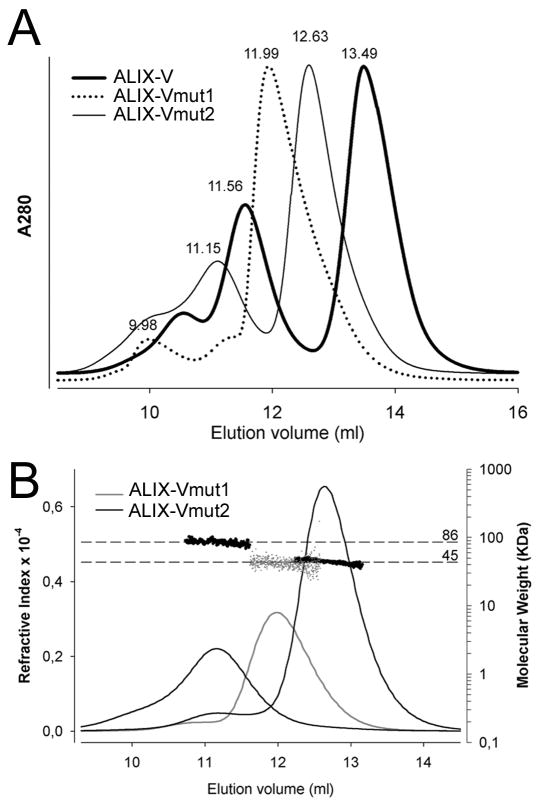
In order to analyze the role of the ALIXV hinge region in dimerization, exposed residues within the segment 638-KMKQSNNE-645, that revealed differences in deuterium incorporation (Figure 3A) were changed to residues 638-EAAQSYKK-645 (ALIXVmut1) and to residues 638-KMKQSYKK-645 (ALIXVmut2). Size exclusion chromatography (SEC) analyses showed that ALIXVmut1 eluted as a single peak at ~12 ml from a Superdex 200 column, a behavior which is different from the elution profile of ALIXV, which forms monomers and dimers (peaks at ~11.6 and ~13.5 ml) (Figure 4A). SEC of ALIX-Vmut2 produced two new peaks eluting at ~11.2 and at 12.6 ml (Figure 4A). Applying MALLS, the single peak of ALIXVmut1 was determined to correspond to a molecular weight of ~ 42 kDa and the two peaks obtained for ALIXVmut2 were determined to correspond to monomeric (~42 kDa) and dimeric forms (~86 kDa) of ALIXV (Figure 4B). Although ALIXVmut2 can still form dimers, the elution positions determined by SEC indicate that both monomers and dimers exhibit a larger hydrodynamic radius as compared to wild type ALIXV.
Figure 4. Mutagenesis of the ALIXV dimer hinge region generates elongated ALIXV monomers.
(A) SEC reveals that ALIXVmut1 produces only one peak as compared to wild type ALIXV and ALIXVmut2.
(B) MALLS analyses show that ALIXVmut1 is monomeric and ALIXVmut2 exists in monomer and dimer conformations. Both refractive index and molecular weight analyses (logarithmic scale) are plotted against the elution volume. The molecular weight distribution across each peak is indicated by dots.
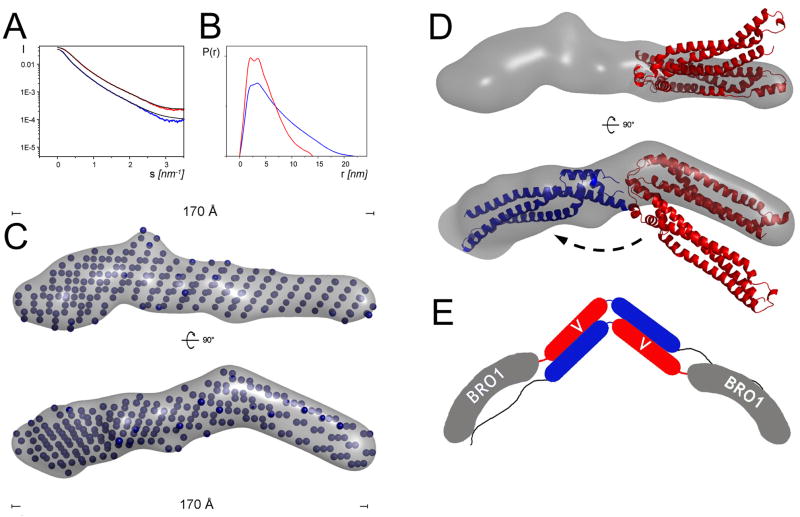
Monomeric ALIXVmut1 was further analyzed by small angle X-ray scattering and compared to wild type ALIXV. The Guinier analysis of the scattering curves (Figure 5A) revealed a radius of gyration (Rg) of 53.6 Ǻ, and maximal protein dimensions (Dmax) of ~190Ǻ were calculated by the distance distribution function p(r) for ALIXVmut1 (Figure 5B). Wild type ALIXV SAXS analysis produced a smaller Rg of 33.4 Ǻ and a smaller Dmax of ~ 125 Ǻ (Figure 5B) confirming that the mutations resulted in an elongated conformation. Comparison of the theoretical scattering curve derived from the ALIXV crystal structure (pdb 2ojq) and the experimental data (Figure 5A) indicated a high discrepancy χ > 5, suggesting that crystallization locked the V-domain in the observed conformation.
Figure 5. SAXS model of ALIXVmut1.
(A) X-ray scattering curves of wild type ALIXV (red) and ALIXVmut1 (blue) is shown after averaging and subtraction of solvent scattering. The inset shows the corresponding Guinier plots. The theoretical scattering profiles calculated from the ab initio models with the lowest χ values are shown as black lines.
(B) P(r) functions of both wild type and mutant ALIXVmut1. (C) Ab initio calculated model of ALIXVmut1 reveals an elongated 170Å long rod-like structure; two orientations of the bead models plus envelopes (grey) rotated by 90° are shown. (D) The three helical bundle structure (red ribbon) of one arm of ALIXV fits into the molecular envelope of ALIXVmut1 and the second arm (blue) would fit after a rotation of ~ 140° along the ALIXV hinge region.
(E) Model for dimeric ALIX. Dimerzation requires that ALIXV opens and allows anti-parallel interaction of the ALIX V-domains arms.
The shape of the ALIXVmut1 was determined ab initio and resulted in a 170 Å long conformation (Figure 5C). The reconstructed average model of ALIXVmut1 fit the corresponding experimental data with the discrepancy χ of 1.56 (Figure 5A). The model suggests an open conformation of ALIXV, whereas one arm of ALIXV needs to rotate ~ 140° to fit the elongated envelope (Figure 5D). The monomeric conformation of ALIXVmut1 indicates that the mutations not only prevent dimerization but may also stabilize an open ALIXV conformation. Together with the SAXS data on dimeric ALIXBro1-V, these results provide support for a dimerization model, whereas opening of ALIXV conformation (as observed in the crystal structures) precedes dimerization. This might entail an anti-parallel association of the ALIXV arms producing an arrangement that positions the Bro1 domains at the extreme ends of the arch-shaped ALIXBro1-V dimer model (Figure 5E).
ALIXBro1-V expression in mammalian cells induces dimerization
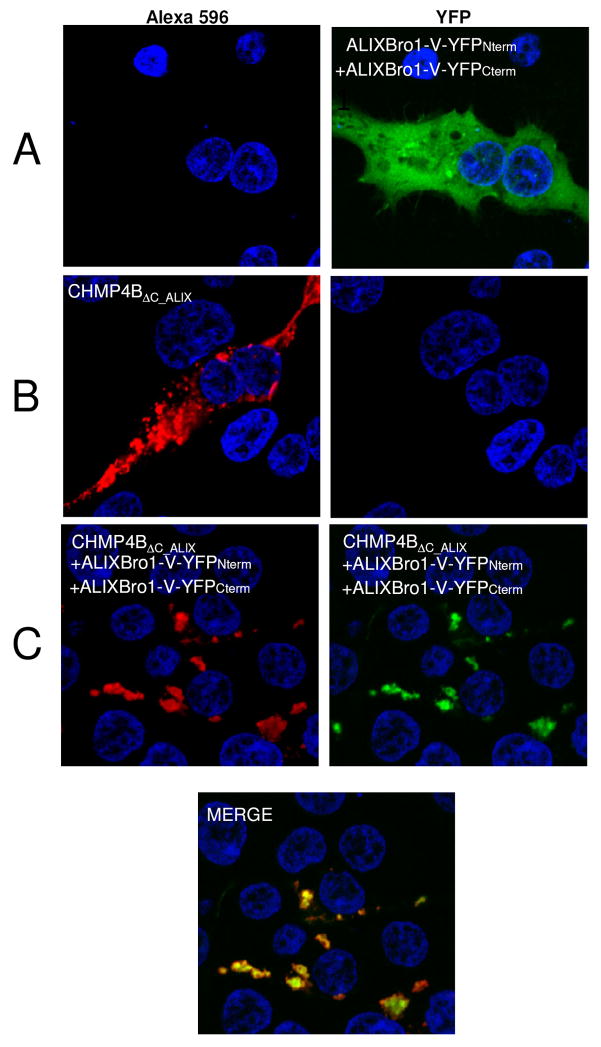
We used the split YFP complementation assay to test ALIX dimerization in cells (Michnick et al., 2007). Both N-terminal YFP and C-terminal YFP halves were fused to the C-terminus of ALIXBro1-V and expressed in HEK293 cells. Fluorescence analyses by confocal microscopy revealed detection of YFP fused to ALIXBro1-V only when both fusion proteins were co-expressed (Figure 6A, right panel). This thus indicates that ALIXBro1-V forms dimers when expressed in mammalian cells, which mainly localize to the cytoplasm.
Figure 6. ALIXBro1-V dimerizes in vivo.
ALIXBro1-V was expressed using a split YFP expression system. (A) Co-expression of both ALIXBro1-V YFP fusion protein halves in HEK293 cells resulted in complementation (right panel). ALIXBro1-V expression thus induces ALIXBro1-V dimerization in vivo.
(B) Expression of CHMP4BΔC-ALIX revealed inclusions partly localized along the plasma membrane (left panel).
(C) Co-expression of ALIXBro1-V YFP fusion constructs and CHMP4BΔC-ALIX; red fluorescence CHMP4BΔC-ALIX (left panel), green fluorescence dimeric ALIX (middle panel) and the lower panel show the co-localization: CHMP4BΔC-ALIX recruits ALIXBro1-V dimers in vivo into large inclusions. Control staining with DAPI is shown in all panels.
Both ALIXV monomers and dimers interact with a late domain peptide
In order to determine whether dimeric ALIX might affect enveloped virus budding, isothermal calorimetry was performed to determine the interaction of ALIXV with a peptide derived from EIAV Gag p9. This revealed equilibrium dissociation constants (KD) of 5 +/− 0.3 μM for monomeric ALIXV, consistent with the reported interactions of ALIX monomers and retroviral late domains (Fisher et al., 2007; Lee et al., 2007; Zhai et al., 2008) and of 8 +/− 1.2 μM for dimeric ALIXV. Thus dimeric ALIX constitutes a target for retroviral Gag.
ALIX dimerization is important for HIV-1 budding
We tested the effect of ALIX dimerization on HIV-1 budding since RFP-ALIXV-PRD expression has been shown to exert a dominant negative effect on HIV-1 particle release (Strack et al., 2003). Expression of RFP-ALIXVmut2-PRD (still able to form dimers in solution) in HIV-1 infected cells had no consequence on the dominant negative effect imposed by RFP-ALIXV-PRD. Particle release was similarly reduced as in case of wild type RFP-ALIXV-PRD expression (Figure 7B, left panel, lanes 2 and 3). In contrast, expression of RFP-ALIXVmut1-PRD reversed the dominant negative effect of the wild type because no effect on particle release was observed (Figure 7A, left panel lane 3). Cellular expression levels of RFP-ALIXV-PRD constructs were comparable (Figure 7C) as well as the presence of intracellular Gag products. This thus indicates that the potential to dimerize is required for this ALIX construct in order to exert a dominant negative effect, which confirms a physiological role for ALIX dimerization.
Figure 7. ALIX dimerization is important for HIV-1 budding.
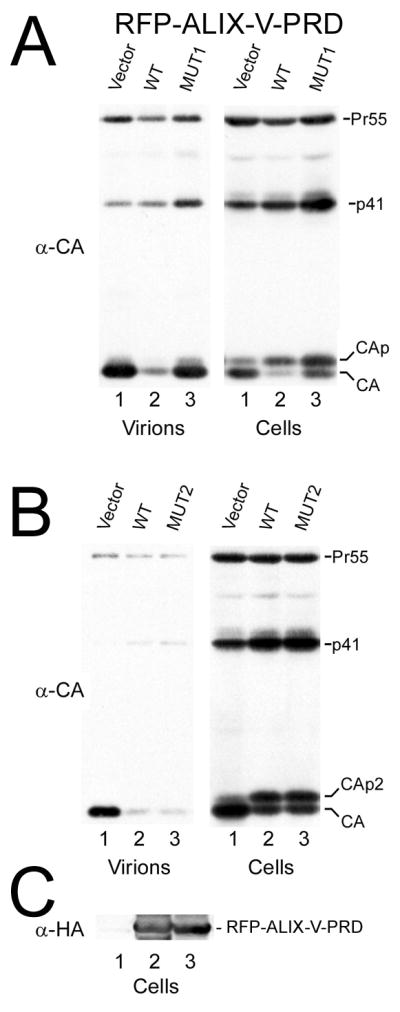
HIV-1 particle release detected upon expression of RFP-ALIXV-PRD wild type and the dimerization mutants.
(A) Expression of RFP-ALIXVmut1-PRD reverses the dominant negative effect observed for expression of wild type RFP-ALIXV-PRD. Lane 1, vector control; lane 2, wild type RFP-ALIXV-PRD; lane 3, RFP-ALIXVmut1-PRD. Detection of extracellular Gag levels (virions, left panel) and intracellular Gag levels (right panels) are shown.
(B) Expression of RFP-ALIXVmut2-PRD does not influence the dominant negative effect observed for expression of wild type RFP-ALIXV-PRD. Lane 1, vector control; lane 2, wild type RFP-ALIXV-PRD; lane 3, RFP-ALIXVmut2-PRD.
(C) Expression levels of RFP-ALIXV-PRD are shown for wild type (lane 2) and RFP-ALIXVmut1-PRD (lane 3) expression (lane 1, RFP vector expression control).
ALIX binds to CHMP4 polymers
We next analyzed whether ALIXBro1-V interacts with CHMP4 polymers. Both ALIXBro1-V conformers are found in the upper fractions of a sucrose gradient consistent with their monodispersity in solution (Figure 8A and B). In contrast, C-terminally deleted CHMP4B fused to MBP (MBP-CHMP4BΔC) forms polymers found in the bottom fractions of a sucrose gradient (Figure 8C). Since MBP-CHMP4BΔC lacks the ALIX binding site, we fused the ALIX binding site motif (McCullough et al., 2008) via a linker to MBP-CHMP4BΔC (MBP-CHMP4BΔC_ALIX). MBP-CHMP4BΔC_ALIX localizes to the upper and to the bottom fractions of a sucrose gradient (Figure 8D) similar to MBP-CHMP4BΔC. Removal of MBP by TEV cleavage renders all of CHMP4BΔC_ALIX polymeric and is thus present in the bottom of a sucrose gradient (Figure 8E). Incubation of MBP-CHMP4BΔC_ALIX with either monomeric or dimeric ALIXBro1-V redistributes ALIXBro1-V from the upper fractions to the bottom fractions upon sucrose gradient centrifugation (Figures 8F and G). Since both MBP-CHMP4BΔC_ALIX and ALIXBro1-V migrate at the same positions on SDS-PAGE their presence is confirmed by western blot analyses (Figure 8H). Similar complex formation is observed when CHMP4BΔC_ALIX (MBP removed by TEV cleavage) is incubated with monomeric or dimeric ALIXBro1-V (Figure 8I and J).
Figure 8. ALIXBro1-V interacts with CHMP4 polymers in vitro.
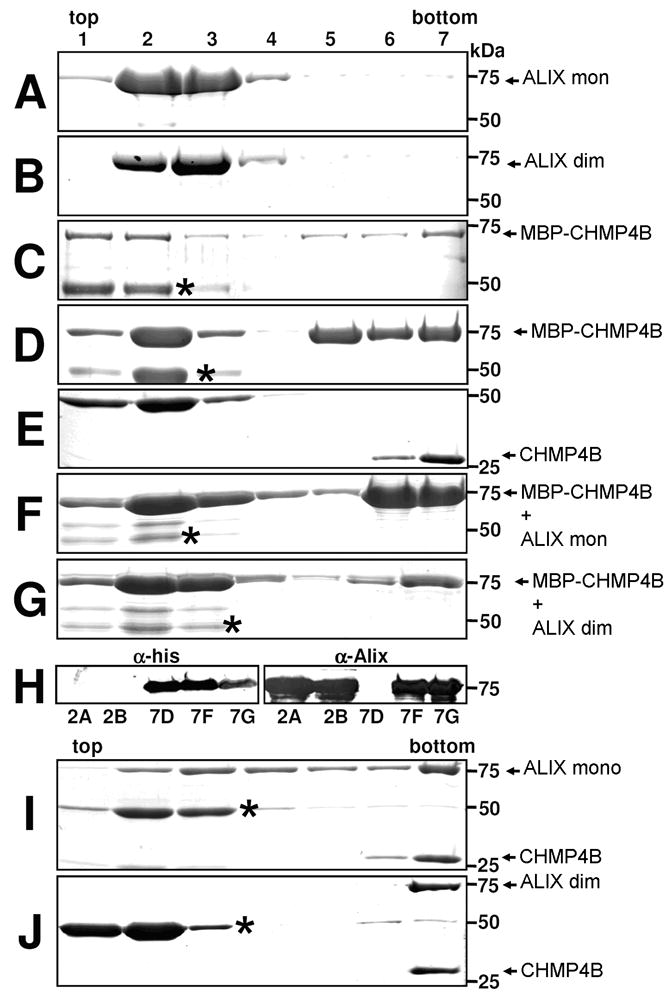
Sucrose gradient centrifugation analysis of monomeric (A) and dimeric (B) ALIXBro1-V, (C) MBP-CHMP4BΔC, (D) MBP-CHMP4BΔC-ALIX, (E) CHMP4BΔC-ALIX, (F) MBP-CHMP4BΔC-ALIX and monomeric ALIXBro1-V, (G) MBP-CHMP4BΔC-ALIX and dimeric ALIXBro1-V, (H) Western blot detection of both MBP-CHMP4BΔC-ALIX and ALIXBro1-V present in the bottom fraction of gradients F and G; (I) CHMP4BΔC-ALIX and monomeric ALIXBro1-V, (J) CHMP4BΔC-ALIX and dimeric ALIXBro1-V. (Protein bands migrating below the 50kDa marker protein correspond to MBP* (*; C, D, E, F, G, I, J)).
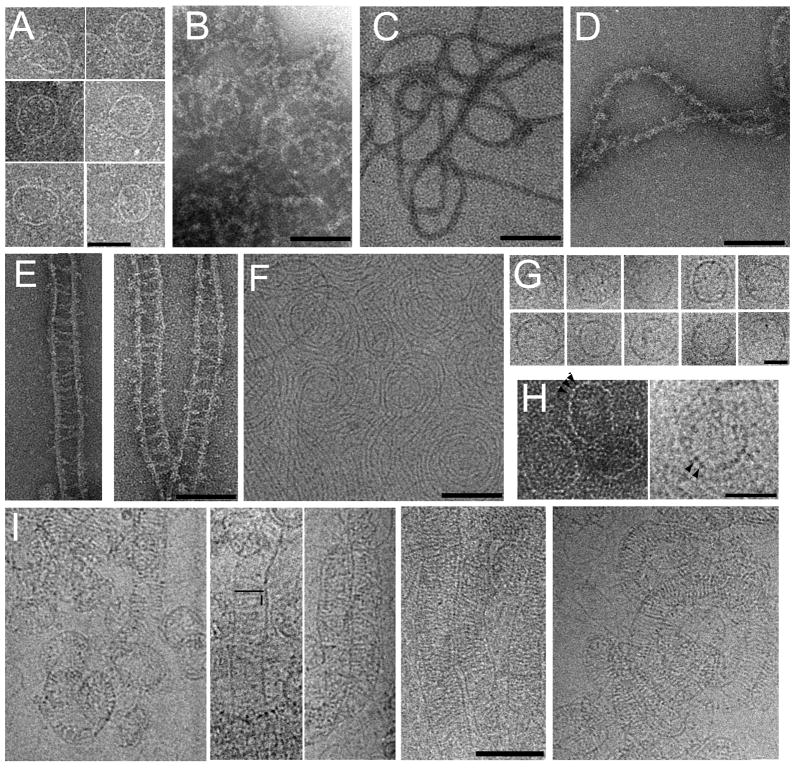
The bottom fractions of the single proteins and the complexes were analyzed by negative staining electron microscopy. MBP-CHMP4BΔC assembles into ring-like structures of varying diameters (~ 40 to ~ 70 nm) (Figure 9A). The width of the rings measures ~25Å and the length of the potential repeating units is ~40 Å (Figure 9H, left panel). Thicker appearing (~100 Å) more filamentous structures are formed by MBP-tagged CHMP4BΔC_ALIX (Figure 9B). Removal of MBP from CHMP4BΔC_ALIX reveals more defined filaments, which have diameters of ~100 Å (Figure 9C). These filaments can be decorated with monomeric ALIXBro1-V, which render the surface to appear fuzzy (Figure 9D). Dimeric ALIXBro1-V bridges two CHMP4BΔC_ALIX filaments and generates ladders made up of “legs” of CHMP4B and “rungs” of ALIXBro1-V (Figure 9E). The rungs are ~400Å wide and are ~100 Å longer than dimeric ALIXBro1-V in solution since SAXSanalysis revealed a Dmax of ~300Å (Figure 2B). The presence of these structures was further confirmed by cryo electron microscopy imaging. The cryo images reveal ~30 Å thick circular arrays (Figure 9F and S2) and ring-like structures for CHMP4BΔC_ALIX (Figure 9G). The length of a potential repeating unit in the rings measures ~50Å, similar to the negatively stained images (Figure 9H, left and right panel). This thus suggests that the filaments or rings are most likely constructed by a head to tail interaction of single CHMP4B molecules. When these structures are decorated with dimeric ALIXBro1-V linear and spiral structures are observed with dimeric ALIXBro1-V spanning a distance of ~ 450Å, with a spacing of ~50 Å between rungs (Figure 9I). The cryo images also show mostly ladders as observed by negative staining although it cannot be excluded that some cross linking between ladders occurs (Figure S3). The difference in spacing observed by negative staining and cryo EM might be either due to the fact that (i) binding of ALIXBro1-V is sensitive to the heavy atom solution used for negative staining or due to the fact that (ii) ALIXBro1-V, which is in contact with the carbon is embedded with heavy atom and visualized, a phenomenon observed previously (Gaillard et al., 2008). Each CHMP4BΔC_ALIX monomer extends the ALIX binding site from the predicted core CHMP4 structure by ~30 amino acids that could span up to ~100 Å in an extended conformation. The extension of the ALIX binding site peptide from the CHMP4B polymer and the positions of the CHMP4B binding sites in the ALIXBro1-V dimer ( ~230 Å apart) cannot explain the observed spacing between two CHMP4B filaments induced by dimeric ALIXBro1-V interaction. The flexible linkage of a ~300Å long ALIXBro1-V dimer would not produce the rather evenly spaced CHMP4 filaments. It is thus conceivable that further conformational changes induced by binding to CHMP4B filaments have to occur that produce longer ALIXBro1-V dimers as indicated by the electron microscopy images. Such changes might include the long extended conformation spanning the whole concave side of ALIXBro1 and connecting ALIXBro1 helix 10 and ALIXV helix 11. Notably part of this region becomes more accessible to D2O exchange in the dimer as compared to the monomer (Figure 3B). The preferred detection of ladders might be explained by conformational constraints imposed by the cross linking of two CHMP4 filaments by ALIXBro1-V, since the ALIX binding site could be positioned towards the interior of the CHMP4B ladder.
Figure 9. Electron microscopy analyses of CHMP4BΔC-ALIX and ALIXBro1-V complexes.
Negative staining EM images of (A) MBP-CHMP4BΔC, (B) MBP-CHMP4BΔC-ALIX, (C) CHMP4BΔC-ALIX, (D) CHMP4BΔC-ALIX - ALIXBro1-V (monomer) complexes, (E) CHMP4BΔC-ALIX- ALIXBro1-V (dimer) complexes. (F) Cryo-electron microscopy image of CHMP4BΔC-ALIX. (G) Cryo EM images of selected ring structures of CHMP4BΔC-ALIX; (H) Ring-like structures revealing the potential repeating unit, marked by arrows (left panel, negative staining, right panel cryo image). (I) 5 panels of cryo EM images showing CHMP4BΔC-ALIX - ALIXBro1-V (dimer) complexes. The length of one rung and the distance between rungs are indicated schematically in panel 2. The scale bars are 50 nm (A), 100 nm (B-F, I) and 30 nm (G, H).
Cellular localization of activated CHMP4B and ALIXBro1-V
To determine whether activated CHMP4BΔC_ALIX localizes to membranes and/or induces membrane remodeling (Hanson et al., 2008) in the absence of other ESCRT factors, CHMP4BΔC_ALIX was expressed with a C-terminal flag-tag in HEK293 cells. Immunostaining revealed that CHMP4BΔC_ALIX concentrates in inclusions that appear to be close to the plasma membrane (Figure 6B) similar to a previous report (Lin et al., 2005); upon co-expression with dimeric ALIXBro1-V fused to the two YFP Venus halves most of ALIXBro1-V is also recruited into CHMP4BΔC_ALIX positive inclusions (Figure 6C), thus indicating that dimeric ALIX targets ESCRT-III CHMP4 polymers in vivo.
Discussion
ALIX is a cellular adaptor protein that engages in a number of seemingly unrelated processes ranging from endosomal trafficking (Saksena et al., 2007), apoptosis (Vito et al., 1999)(Missotten et al., 1999), cytoskeleton reorganization (Cabezas et al., 2005) (Pan et al., 2006), retrovirus budding (Usami et al. 2009) and cytokinesis (Carlton and Martin-Serrano, 2007)(Morita et al., 2007). Thus ALIX underlies tight regulation that requires the C-terminal proline-rich region (PRD) harboring numerous protein-protein interacting motifs important for function and regulation of Alix (Odorizzi, 2006). PRD keeps ALIX in an auto-inhibited conformation (Zhou et al., 2008b)(Zhou et al., 2008a)(Lazert et al., 2008),which suggests that ALIX has to be activated. Our data indicate that activation of ALIX induces dimerization. Oligomerization and dimerization of ALIX was observed previously(Munshi et al., 2007)(Fisher et al., 2007), and we confirm that the V-domain is sufficient for dimerization. It is, however, yet unclear which signal induces dimerization in vitro and in vivo. We show that both ALIXBro1-V and ALIXV monomers and dimers are stable entities and do not exchange in vitro (Fisher et al., 2007).
ALIXBro1-V contains a Bro1 domain (Kim et al., 2005) and a central V-shaped domain (Fisher et al., 2007)(Lee et al., 2007). Bro1 interacts with ESCRT-III CHMP4 and the V-domain harbors the retroviral late domain binding site (Kim et al., 2005)(Zhai et al., 2008)(McCullough et al., 2008)(Lee et al., 2007). SAXS analyses of dimeric ALIXBro1-V revealed an elongated ALIXBro1-V structure that measures ~ twice the length of monomeric ALIXBro1-V. Interestingly, the shape of dimeric ALIXBro1-V resembles that of BAR domains such as amphipysin (Peter et al., 2004) and endophilin (Weissenhorn, 2005). Although we observe membrane interaction of ALIXBro1-V in vitro (data not shown) no specific membrane remodeling effect could be assigned. Thus, the architecture might be preferentially used to bridge two effector molecules.
Since no concentration dependent equilibrium between monomers and dimers could be detected (Fisher et al., 2007), dimer formation must include specific conformational changes. Hydrogen/deuterium exchange experiments coupled to mass spectrometry peptide mapping revealed several sites of conformational flexibility, notably not only within the V-dimerization domain but also within the Bro1 domain. Such conformational flexibility is consistent with the structure of ALIXBro1-V, which shows that Bro1 and V are connected via a short linker as well as some degree of flexibility between the two arms of the V domain (Fisher et al., 2007). The mutagenesis studies suggest that the two arms of ALIXV are connected via a hinge region, which is stabilized in the crystal structure by a salt bridge between K640 and E650, that also hydrogen bonds to N400 (Lee et al., 2007)(Fisher et al., 2007). A set of mutations within this region including K640 destabilized the monomer and in addition prevented dimer formation. Since the hydrogen/deuterium exchange experiments suggest that this region is more protected in dimeric ALIXBro1-V, changes within this region might prevent important dimer contacts. Alternatively, the mutations could artificially stabilize an open monomer conformation that itself prevents dimerization. The latter possibility is supported by the SAXS model of ALIXVmut1 that indicates an elongated form of ALIXV; the crystal structure of ALIXV would fit into the SAXS envelope when one arm of the V-domain is rotated by ~ 140°. Based on these data we suggest the following dimerization model: Displacement of PRD serves as a first signal to release auto-inhibition (Zhou et al., 2008b)(Zhou et al., 2008a), a second signal then leads to an opening of the V-domain and subsequent anti-parallel dimerization via the V-domain arms (Figure 5E).
Expression of truncated ALIX conformers such ALIXV-PRD and ALIXV have been shown to induce dominant negative effects on retroviral budding (Strack et al., 2003)(Munshi et al., 2007). We show that such an effect is inhibited in the presence of the dimerization mutations since RFP-ALIXVmut1-PRD expression is no longer a dominant negative inhibitor of HIV-1 budding. It is thus conceivable that truncated forms of ALIX which retain the V dimerization domain form heterodimers with endogenous ALIX. The function of such mixed dimers might be compromised by the lack of adaptor protein binding which might position the complex within the correct network of interactions required to trigger down stream effects. Thus ALIXBro1-Vmut1 that no longer dimerizes has no effect on the budding process. The dominant negative effect may further depend on the interaction with Gag, since dimeric ALIXV interacts with a EIAV late domain peptide with a similar Kd as reported for monomeric ALIXV (Munshi et al., 2007)(Fisher et al., 2007).
Several lines of evidence suggest that ALIX functions directly on membranes. Firstly, ALIX localizes to the plasma and endosomal membranes (Welsch et al., 2006). Secondly, it plays a role in the formation of internal endosomal membranes in conjunction with lysobiphosphatidic acid (LBPA) (Matsuo et al., 2004). Thirdly, expression of ALIXV-PRD was shown to induce cytoplasmic vacuolization into tubulo-vesicular structures that co-localize with ALIX (Chatellard-Causse et al., 2002); this effect might be also due to the formation of mutant-wild type ALIX heterodimers that lack important binding sites. Fourthly, it is recruited by interaction with ESCRT-III CHMP4, which acts at membranes (Katoh et al., 2003)(von Schwedler et al., 2003)(Strack et al., 2003). ESCRT-III proteins CHMP2A and CHMP3 have been shown to assemble into heteromeric helical polymers that may catalyze late budding steps such as abscission (Lata et al., 2008b). Similarly, ESCRT-III CHMP4 expression in mammalian cells was shown to form circular filaments (Hanson et al., 2008) consistent with ring-like and filamentous structures of CHMP4 observed in vitro (Ghazi-Tabatabai et al., 2008). Here we show that activated CHMP4, which most likely requires detachment of the C-terminal MIM domain from the N-terminal core (Shim et al., 2007)(Lata et al., 2008a), forms linear filaments as well as spiral and ring-like structures. The ring and spiral CHMP4B structures (Figure 9A, F, G) are most likely formed by head to tail interactions of single CHMP4B molecules. Thus CHMP4B polymerization differs from CHMP2A-CHMP3 polymerization since it does not require homo- or heterodimerization. The latter is consistent with the excess of CHMP4 over its potential binding partner CHMP6 present in the yeast ESCRT-III complex (Saksena et al., 2009). The spiral CHMP4B structures observed here might thus reflect the polymer structures observed in vivo (Hanson et al., 2008) that drive early steps in HIV-1 assembly and budding (Carlson et al., 2008).
Both ALIX and CHMP4B are present at the midbody (Carlton and Martin-Serrano, 2007)(Morita et al., 2007) and CHMP4-ALIX-CEP55 interactions are essential for midbody formation (Carlton et al., 2008). Since dimeric ALIXBro1-V bridges preferentially two CHMP4B filaments or spirals in vitro, ALIX could support CHMP4 polymer positioning on membranes during viral budding as well as at the midbody supported by CEP55 (Carlton et al., 2008)(Morita et al., 2007). ALIX polymerizes itself (Carlton et al., 2008) whereas dimeric ALIX might constitute the the building block for CHMP4 induced polymerization at budding sites. Because ALIXBro1-V lacks the multimerization site (Carlton et al., 2008), we do not observe ALIX polymers in vitro. Polymerization of ALIX is further supported by the stochiometry of ESCRT-III. ESCRT-III contains multiple copies of CHMP4 (Teis et al., 2008)(Saksena et al., 2009) supporting CHMP4 polymerization which in turn provides multiple binding sites for ALIX polymerization.
The formation and physiological role of ALIX dimers was further proven by using a YFP complementation assay (Michnick et al., 2007). Dimeric ALIXBro1-V-YFP is mostly found in the cytosol, indicating that dimerization is not sufficient for membrane targeting. When it is expressed together with CHMP4BΔC-ALIX both structures co-localize in large inclusions, demonstrating that polymeric CHMP4B recruits dimeric ALIX in vivo. Thus our data suggest that recruitment of activated ESCRT-III CHMP4B entails the function of dimeric ALIXBro1-V. This might help to position ESCRT-III at sites of membrane remodeling by providing access to the cytoskeleton (Cabezas et al., 2005)(Pan et al., 2006).
Experimental procedures
Protein expression and purification
Cloning, expression, purification and SEC characterization including MALLS of ALIX and CHMP4B constructs is described in the supporting online material (SOM).
Small Angle Scattering Data Collection and Analysis
The synchrotron radiation X-ray scattering data were collected on the X33 SAXS beam line (DESY and EMBL, Hamburg) (Koch and Bordas, 1983; Roessle et al., 2007) and at the ESRF beam line ID02 (Grenoble). A MAR345 image plate with online readout (MarResearch, Germany) was used at a sample - detector distance of 2.4 m covering the range of momentum transfer 0.1 < s < 4.5 nm−1 (s = 4π sin(θ)/λ, where θ is the scattering angle and λ = 0.15 nm is the X-ray wavelength). The s-axis was calibrated by the scattering pattern of Silver-behenate salt (d-spacing 5.84 nm). The scattering patterns from ALIXBro1-V in the monomeric and dimeric phase were measured at protein concentrations of 10.0, 7.4, 5.0 and 2 mg/ml and 2.9, 1.4 and 1.2 mg/ml, respectively. ALIXVmut1 was measured at a concentration of 7mg/ml at the ESRF (Grenoble) beam line ID02. Further details on SAXS data analysis and model calculation are described in SOM.
Mammalian expression constructs and HIV-1 release assay
The mammalian expression vector pDSRed2-C1/AIP1ΔBRO, encoding ALIXV-PRD, with the N-terminal Bro1-domain replaced by RFP (Strack et al., 2003), was mutated to produce both V-domain dimerization mutants (RFP-ALIXVmut1-PRD and RFP-ALIXVmut2-PRD) using standard procedures. See SOM for further details.
For indirect immunofluorescence detection of CHMP4BΔC_ALIX its cDNA was cloned with a C-terminal Flag-epitope into the mammalian expression vector pCAGGS by PCR using pBADM41-CHMP4BΔC_ALIX as a template. The plasmid was transfected alone or together with ALIXBro1-V fused to the N-terminal halve of YFP and the C-terminal halve of YFP in HEK293 cells using TransIT (Mirus) and OptiMem (Invitrogen) according to the manufacturer’s instructions. For further details see SOM.
Hydrogen/deuterium (H/D) exchange mass spectrometry (MS)
Deuterium labeling of monomeric and dimeric ALIXBro1-V was carried out by 20-fold dilution of the protein samples in deuterated buffer (5mM Hepes, 100mM NaCl in D2O, pD 7.6) followed by incubation at 4°C for different time intervals (from 10 s to 30 min). The reaction was quenched by lowering the pD to 2.1 with 0.1 M HCl solution. Pepsin (Sigma-Aldrich) digestion was performed at 4°C and pH 2.5 for 2 min in 0.1 M HCl and a protein:protease ratio of 1:1 (w/w). Further details are reported in SOM.
Electron microscopy
CHMP4BΔC-Alix and ALIXBro1-V were purified by sucrose gradient centrifugation (Lata et al., 2008b) and samples present in the bottom fractions were analyzed by electron microscopy using negative staining with uranyl acetate or sodium silicotungstate (pH 7.0) (Lata et al., 2008b). For cryo electron microscopy CHMP4BΔC-Alix alone or in complex with ALIXBro1-V the sucrose was removed and 4 μl sample were loaded onto a Quantifoil R2/1 holey grid (Quantifoil Micro Tools GmbH, Germany). The grids were treated and observed using a JEOL 2010 FEG electron microscope (Schoehn et al., 2008).
Supplementary Material
Acknowledgments
We thank Dr. A. Grichine from the Institute Albert Bonnoit (Grenoble) microscopy platform for his assistance and Dr. B. Blot and C. Chatellard-Causse for helpful discussions. This work was supported by the ANRS (WW), the DFG SPP1175 (WW), the EU FP6 Program (SAXIER, RIDS 011934) (DS, MR), the National Institute of Allergy and Infectious Diseases (R37AI029873 to HG), the ANR (“Jeune Chercheur”, G.S.), the French Ministry of Research (C.M.) and postdoctoral fellowships from the European Molecular Biology Organization (S.L. and B. H.) and the International Human Frontier Science Program Organization (S.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. The HIV-1 p24 monoclonal antibody (183-H12-5C) (provided by Drs. Bruce Chesebro and Kathy Wehrly) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci U S A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falguieres T, Luyet PP, Bissig C, Scott CC, Velluz MC, Gruenberg J. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol Biol Cell. 2008;19:4942–4955. doi: 10.1091/mbc.E08-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol Biol Cell. 2008;19:4534–4544. doi: 10.1091/mbc.E08-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature Reviews Molecular Cell Biology. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MHJ, Bordas J. X-ray diffraction and scattering on disordered systems using synchrotron radiation. Nucl Instrum Methods. 1983;208:461–469. [Google Scholar]
- Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008a;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008b;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Solomons J, Pires R, Gottlinger HG, Weissenhorn W. Structure and function of ESCRT-III. Biochem Soc Trans. 2009;37:156–160. doi: 10.1042/BST0370156. [DOI] [PubMed] [Google Scholar]
- Lazert C, Chazal N, Briant L, Gerlier D, Cortay JC. Refined study of the interaction between HIV-1 p6 late domain and ALIX. Retrovirology. 2008;5:39. doi: 10.1186/1742-4690-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7–1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- Mahul-Mellier AL, Hemming FJ, Blot B, Fraboulet S, Sadoul R. Alix, making a link between apoptosis-linked gene-2, the endosomal sorting complexes required for transport, and neuronal death in vivo. J Neurosci. 2006;26:542–549. doi: 10.1523/JNEUROSCI.3069-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul-Mellier AL, Strappazzon F, Petiot A, Chatellard-Causse C, Torch S, Blot B, Freeman K, Kuhn L, Garin J, Verna JM, et al. Alix and ALG-2 are involved in TNF-R1 induced cell death. J Biol Chem. 2008;283(50):34954–65. doi: 10.1074/jbc.M803140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz P. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313 – 1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Nat Acad Sci USA. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999;6:124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. Journal of Cell Science. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev PV, Round A, Fiedler S, Hermes S, Svergun DI. Upgrade of the Small Angle X-ray scattering Beamline X33 at the EMBL Hamburg. J Appl Crystallogr. 2007;40:190–194. [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends in Biochemical Sciences. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Chen B, Randazzo LM, Bogler O. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J Cell Sci. 2003;116:2845–2855. doi: 10.1242/jcs.00522. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, Bogler O. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol Cell Biol. 2004;24:8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoehn G, El Bakkouri M, Fabry CM, Billet O, Estrozi LF, Le L, Curiel DT, Kajava AV, Ruigrok RW, Kremer EJ. Three-dimensional structure of canine adenovirus serotype 2 capsid. J Virol. 2008;82:3192–3203. doi: 10.1128/JVI.02393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Yamada K, Mizuno T, Yorikawa C, Takahashi H, Satoh H, Kitaura Y, Maki M. The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1. J Biochem (Tokyo) 2004;135:117–128. doi: 10.1093/jb/mvh014. [DOI] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-Terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Popova E, Gottlinger H. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689 – 699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kawasaki M, Inuzuka T, Okumura M, Kakiuchi T, Shibata H, Wakatsuki S, Maki M. Structural basis for Ca2+-dependent formation of ALG-2/Alix peptide complex: Ca2+/EF3-driven arginine switch mechanism. Structure. 2008;16:1562–1573. doi: 10.1016/j.str.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Tanzi GO, Piefer AJ, Bates P. Equine Infectious Anemia Virus Utilizes Host Vesicular Protein Sorting Machinery during Particle Release. J Virol. 2003;77:8440–8447. doi: 10.1128/JVI.77.15.8440-8447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Trioulier Y, Torch S, Blot B, Cristina N, Chatellard-Causse C, Verna JM, Sadoul R. Alix, a protein regulating endosomal trafficking, is involved in neuronal death. J Biol Chem. 2004;279:2046–2052. doi: 10.1074/jbc.M309243200. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and Specific Rescue of Human Immunodeficiency Virus Type 1 Budding Defects by a Nedd4-like Ubiquitin Ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, H GG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J Mol Biol. 2005;351:653–661. doi: 10.1016/j.jmb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Welsch S, Habermann A, Jager S, Muller B, Krijnse-Locker J, Krausslich HG. Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic. 2006;7:1551–1566. doi: 10.1111/j.1600-0854.2006.00489.x. [DOI] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lee YC, Lin SH, Kuang J. The CHMP4b and Src docking sites in the Bro1 domain are autoinhibited in the native state of Alix. Biochem J. 2008a;418(2):277–84. doi: 10.1042/BJ20081388. [DOI] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lin SH, Kuang J. The HIV-1 p6/EIAV p9 docking site in Alix is autoinhibited as revealed by a conformation-sensitive anti-Alix monoclonal antibody. Biochem J. 2008b;414(2):215–20. doi: 10.1042/BJ20080642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.