Abstract
LAMMER kinases (also know as Cdc-2-like or CLKs) are a family of dual specificity serine/threonine protein kinases that are found in all sequenced eukaryotic genomes. In the fission yeast, S. pombe, the LAMMER kinase gene, Lkh1, positively regulates the expression of the antioxidant defense genes, superoxide dismutase1 (sod1+, CuZn-SOD) and catalase (ctt1+, CAT). We have shown that mutations in the Drosophila LAMMER kinase gene, Darkener of apricot (Doa), protect against the decrease in life span caused by the reactive oxygen species (ROS) generator paraquat, and at the same time show an increase in cytoplasmic (CuZn-Sod or SOD1) and mitochondrial superoxide dismutase (Mn-Sod or SOD2) protein levels and activity. The siRNA mediated knock down of the human LAMMER kinase gene, CLK-1, in HeLa and MCF-7 human cell lines leads to an increase in both SOD1 activity and mRNA transcript levels. These data suggest that SOD1 is negatively regulated by LAMMER kinases in Drosophila and human cell lines and that this regulation may be conserved during evolution.
Keywords: LAMMER kinase, Superoxide dismutase, CDC-Like-kinase, CLK, TG003, Drosophila
INTRODUCTION
The antioxidant defense enzyme, superoxide dismutase (SOD), plays a critical role in the prevention of damage by reactive oxygen species (ROS) and the in vitro activation of SOD can increase differentiation, decrease cell proliferation [1] and in Drosophila melanogaster loss of SOD1 causes early mortality [2]. Two SOD genes are found in all organisms, including bacteria [3]. The copper/zinc-containing SOD (SOD1) encoded by the SOD1 gene, is normally found in the cytoplasm, nucleus and lysosomes [3], and mutations in the human SOD1 gene have been linked to familial and sporadic forms of amyotrophic lateral sclerosis [4]. Human SOD1 is upregulated in response to phorbol ester treatment, mediated by SP-1, EGR-1 and the EGR-related protein, WT-1 [5]. In rat liver, the cis-acting C/EBP element appears to play a major role in regulating the SOD1 gene [6]. The promoter of SOD1 also has a metal response element (MRE) that upregulates the gene in response to heavy metals [7]. The mitochondrial, or iron/manganese-containing form of SOD, (SOD2), which is encoded by the nuclear gene SOD2, is highly inducible by cytokines and oxidants [3, 8]. Protein kinase C stimulating agents also induce expression of the human SOD2 gene [9], and increased levels of the transcription factor AP2 decrease its expression[10]. Single amino acid polymorphisms of SOD2 are associated with diabetic nephrology [11], and increased levels of SOD2 have been reported in mesothelioma [12] and colorectal carcinomas [13]. Overexpression of SOD2 in MCF-7 cells abolished tumor necrosis factor-mediated activation of NFκB and suppressed apoptosis [14]. In many eukaryotes there is also an extracellular SOD (EC-SOD, SOD3) which, in mammals, is secreted by a limited number of cell types and has been detected in blood plasma, lymph and cerebrospinal fluid [3, 15].
In S. pombe, the expression of SOD1 is positively regulated by the LAMMER kinase gene lkh1+, and null alleles of lkh1 are more sensitive to killing by H2O2 then wild type strains [16, 17]. LAMMER kinases are members of a family of dual-specificity protein kinases that regulate various aspects of cell growth and differentiation by an as yet unknown mechanism [18]. All LAMMER kinases assayed have been found to phosphorylate serine/arginine-rich splicing factors (SR) and regulate pre-mRNA splicing [19–22]. Human CLK-1 directly and specifically influences the activity of SR factors and may play a role in governing splice site selection [23].
The single LAMMER kinase gene in Drosophila is encoded by the Darkener of apricot (Doa) locus. Like other LAMMER kinase family members, the Doa gene encodes a dual specificity serine/threonine kinase capable of phosphorylating SR proteins and posses the highly conserved EHLAMMERILG motif [19, 24, 25]. Mutations in Doa were initially identified as dominant second site suppressors of mutations induced by copia retrotransposon insertions [24, 25]. Most Doa alleles are recessive lethal; one exception is the hypomorphic allele DoaDem [17, 25]. The amino terminal non-catalytic domain of LAMMER kinases are not well conserved across species, but the catalytic kinase domain of LAMMER kinases is well conserved across orthologues, and of the six Doa splice forms identified in Drosophila all share a common catalytic domain [26]. Based on the catalytic domains, DOA is most similar to that of the human LAMMER kinase CLK-2 with 75% identity, but is also shares a significant similarity to catalytic domain of human CLK-1 with 62% identity [17].
We observed in Drosophila melanogaster that mutations in Doa, protected flies against acute lethality caused by feeding the ROS generating agent paraquat. We focused our studies on the regulation of SOD since the over-expression of SOD1 in both S. cereviseae and Drosophila has been reported to reduce oxidative damage and extend life span [27]. We found that flies with mutant Doa have increased expression of SOD1 and smaller increases in SOD2. We extended the studies to human cancer cells that have 4 LAMMER kinase genes and found that inhibition of CLK-1 alone increased expression of SOD1 but not SOD2. Thus, an increase in SOD1 by inhibition of CLK-1 appears to be a conserved mechanism that provides a possible explanation for the increased paraquat resistance caused by Doa mutation in Drosophila. In addition, we suggest that this mechanism may provide protection against ROS-induced carcinogenesis in human cancer cells.
MATERIALS AND METHODS
Drosophila stocks and genetics
Stocks of Drosophila melanogaster were maintained on standard cornmeal-molasses medium and all crosses were carried out at 25° C unless otherwise noted. Stocks of wa; Doaγ3B/TM6B, eTb, ywa; Pr DoaHD/TM6B, e Tb and wa; DoaDem/TM6B, eTb were a generous gift from L. Rabinow (Univ. Paris). All other stocks were obtained from the Bloomington Drosophila Stock Center.
Paraquat resistance test
To examine the effects of genetic background on resistance to the O2.- generator paraquat (1,1′-dimethyl-4,4′-bipyridylium dichloride) (Sigma Chemical, St. Louis, MO) a modification of the method of Seong et al [28] was used. Animals eclosing within a 2-day period were pooled, starved on 1.5% agar for 6 h and transferred (20 flies/vial) to vials containing GF/A filter disks (Whatman) soaked in 10 mM paraquat in 5% sucrose. Control flies received 5% sucrose without paraquat. At least 100 flies were used for each genetic background, and approximately equal numbers of males and females were tested. Dead flies were counted twice daily for 5 days, and the time of 50% mortality was determined from a second order polynomial regression curve fitted to the data using Microsoft Excel.
Cell culture
HeLa and MCF-7 cells were maintained in Dulbecco’s modified eagles medium supplemented with 10% fetal bovine serum (Hyclone, South Logan, UT). The LAMMER kinase inhibitor TG003 (EMD, San Diego, CA) [29], was dissolved in DMSO as a 20 mM stock solution and added to log-phase cultures at a final concentration of 10 μM. Control cells received DMSO without inhibitor. Following incubation at 37° C for the times indicated cells were harvested for SOD assays, and Western blot analyses, as described below. To assess the effect of TG003 on cell growth, AlamarBlue was added to a final concentration of 10%, and its reduction was monitored at 570 nm and 600 nm [30].
SOD assays
To measure the effect of Doa alleles on SOD activity, female flies heterozygous for the desired allele were crossed with Ore R males, and male progeny were collected 1 day after eclosion. Groups of 10 males were homogenized on ice in pre-chilled homogenization buffer (20 mM HEPES, pH 7.2, 1 mM EGTA, 210 mM mannitol, and 70 mM sucrose) at a ratio of 1 fly per 40 μl, using a PowerGen 125 homogenizer (Fisher Scientific, Pittsburgh, PA). Tumor cells were rinsed briefly in PBS, and homogenized in homogenization buffer (20 mM Hepes, pH 7.2, 1mM EGTA, 210 mM mannitol, and 70 mM sucrose). Homogenates were centrifuged at 3000 × g for 10 min at 4°C, and the supernatants were serially diluted with 50 mM Tris-HCl, pH 8.0, to dilutions ranging from 1:40 to 1:640. Quadruplicate 10 μl aliquots of each dilution were assayed for SOD activity using an SOD Assay kit (Cayman Chemical Co, Ann Arbor, MI). SOD2 activity was determined in the presence of 5 mM NaCN. SOD1 activity was estimated by subtracting the SOD2 activity from the total activity. Protein concentrations in the homogenates were measured with a Coomassie Plus Protein Assay Reagent (Pierce Chemical, Rockford, IL). Data are presented as means ± SEM, and the significant variation was determined using a 2-tailed t-test.
siRNA treatment, mRNA Measurement and RT-PCR
Total RNA was isolated from MCF-7 and HeLa cells using the PARIS kit (Ambion, Applied Biosystems) according to the manufacturer’s protocol. TaqMan quantitative reverse transcription-PCR was done on the ABI 7300 system using the TaqMan One-Step reverse transcription-PCR Master Mix kit and predesigned primer/probe pairs for LAMMER kinases and β2-microglobulin (Applied Biosystems). Normalization and analyses were carried out with β2-microglobulin as the internal reference by the -CT method [31] using the Applied Biosystems GeneAmp 5700 SDS software. HeLa and MCF-7 cells were transfected with siRNA in 6-well plates with Dharmafect 2 (Dharmacon, Chicago, IL) according to the manufacture’s protocol. SMARTpool siCLK1, siCLK2, siCLK3, siCLK4 and non-targeting siSCR RNAs were obtained from Dharmacon (Lafayette, CO). One microgram of total RNA was reverse-transcribed and amplified with the following primers: SOD1 Exon 1 forward CCTCGGAACCAGGACCTC, SOD1 Exon 3 reverse GTGAGGACCTGCACTGGTA, SOD1 Exon 3 forward GGTGGGCCAAAGGATGAAGAG, and SOD1 Exon 5 reverse TTCACAGGCTTGAATGACAAAC.
Electrophoresis and Western blots
Adult flies, 1–3 days post-eclosion, were homogenized in lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.5% Triton-X100, 1% deoxycholate, 2 mM EDTA and protease inhibitor cocktail (Sigma). Human tumor cells grown in 6-well plates were lysed in 250 μl of the same lysis buffer and briefly sonicated. Homogenates were centrifuged at 16,000 ×g for 5 min. Supernatants (25 μg/lane) were fractionated on 10% NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred to PVDF membranes. Blots were probed with rabbit anti-human SOD1 antiserum (Upstate) diluted 1:300 (Drosophila samples) or 1:1,000 (human samples) or with rabbit anti-human SOD2 antiserum (Upstate) diluted 1:1000 (Drosophila samples) or 1:5,000 (human samples), followed by anti-rabbit IgG-horseradish peroxidase conjugate diluted 1:2500 (Invitrogen). Bands were visualized with Western Lightning Plus chemiluminescent reagent (Perkin Elmer). Subsequently, blots were stripped in 0.2 N NaOH for 5 min at room temperature, then reprobed with mouse anti-α-actin diluted 1:20,000 (Chemicon) and anti-mouse IgG conjugated to horseradish peroxidase diluted 1:20,000 (Invitrogen).
RESULTS
Doa regulates resistance to oxidative stress in Drosophila
In order to determine whether Drosophila heterozygous for mutations in Doa showed sensitivity to oxidative stress, we fed Doa mutants the O2.− generator paraquat [32]. To reduce genetic background and balancer chromosome effects, virgin wild type (Ore R) females were mated with males that were either Doaγ3B or DoaHD, and the newly-eclosed F1 progeny of the genotypes Doaγ3B/+ and DoaHD/+ were fed 10mM paraquat. The DoaHD allele is due to a transposable element insertion and the Doa3B allele is caused by a chromosomal breakpoint [17]. Approximately equal numbers of males and females were assayed, and age-matched Ore R animals were used as controls. The results are shown in (Figure 1). During the first 24 hours few animals of any genotype died. By 74 hours exposure, approximately half the Ore R animals had died and by 112 hours all were dead. In contrast, the Doaγ3B/+ and the DoaHD/+ animals survived considerably longer than the controls. The estimated times of 50% mortality (TM50) were 74.3 hours for Ore R, 109.2 hours for Doaγ 3B/+ and 116.2 hours for DoaHD/+. Less than 1% of the control animals maintained in the absence of paraquat died during the experimental period (data not shown). The results suggest that decreased levels of Doa in adult flies significantly increases their resistance to oxidative stress.
Figure 1.
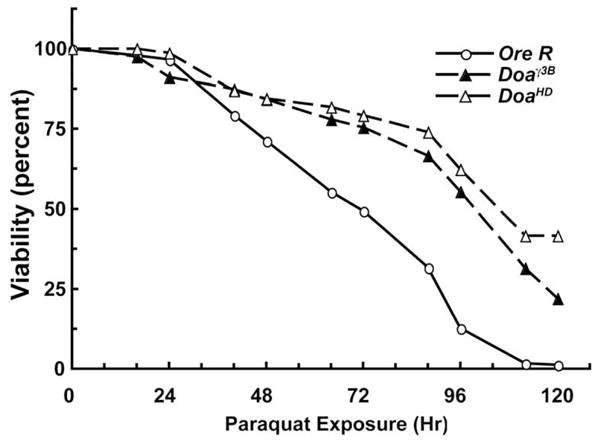
Mutations in Doa cause resistance to paraquat. Ore R, Doaγ3B/+ and DoaHD/+ animals 1–3 days post eclosion were starved for 6 hr, and then fed continuously with 10 mM paraquat dissolved in 5% sucrose. The number of dead animals was counted twice each day for 5 days and the data was expressed as percent survival. The estimated times of 50% mortality were 74.3 h for Ore R, 109.2 h for Doaγ3B/+ and 116.2 h for DoaHD/+. Less than 1% of the sibling control animals for each genotype that were fed 5% sucrose without paraquat died during the 5-day counting period. n ≥ 100 for each genotype.
Doa alleles affect SOD expression in Drosophila
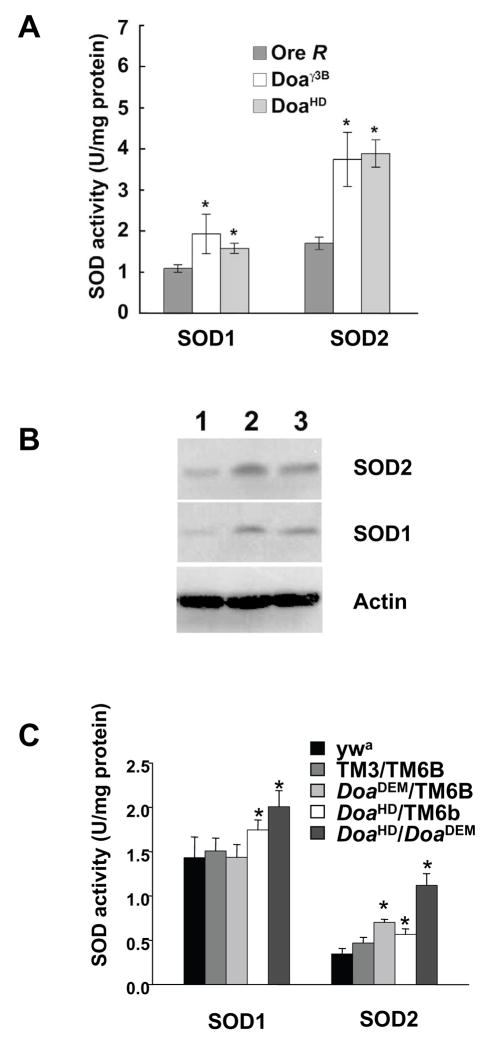
To determine whether the increased resistance of the Doa mutant flies to paraquat was due, at least in part, to increased SOD activity in the Doa mutants, homogenates of young adult Ore R, Doaγ3B/+ and DoaHD/+ flies were assayed for total SOD activity and SOD2 activity. Because the only SOD genes in the Drosophila genome are Sod1 and Sod2 [33], the SOD1 activity could then be estimated by subtracting SOD2 activity from total SOD activity. The Doaγ3B/+ and DoaHD/+ animals exhibited approximately 2-fold increases in both SOD1 and SOD2 activity over those measured in wild type (Ore R) controls (Figure 2A). Western blots revealed that SOD1 and SOD2 protein levels corresponded with activity (Figure 2B). Animals heterozygous for the apparent null allele, Doaγ 3B, appeared to express more SOD2 than either the wild-type controls or those carrying the hypomorphic allele, DoaHD. These results suggest that Doa regulates the levels of SOD2 and SOD1 expression in Drosophila.
Figure 2.
SOD activity is elevated in adult flies carrying mutations in Doa. (A) Total SOD activity was measured in homogenates of age-matched Doaγ3B/+ and DoaHD/+ males. Ore R males were assayed as wild type controls. SOD2 activity was measured in parallel aliquots containing 5 mM NaCN. SOD1 activity was then estimated by subtracting the SOD2 activity from the total activity. (B) Representative Western blot of homogenates of adult males 1–3 days post eclosion. Lane 1, wild type (Ore R); lane 2, Doaγ3B/+; lane 3, DoaHD/+. Staining for actin is included on all Western blots as a control for loading. (C) SOD activity in trans-heterozygous DoaDem/DoaHD flies along with heterozygous DoaDem/TM6B and DoaHD/TM6B siblings. Because the DoaDem/TM6B and DoaHDTM6B animals were in yw and wa backgrounds, respectively, stocks of yw and TM6B/TM3 animals were used as controls. Error bars indicate standard error. * SOD activities significantly different from control values (p < 0.05).
Because both the Doaγ3B and the DoaHD alleles are homozygous lethal, the SOD studies described above were carried out using animals heterozygous for Doa. In crosses of DoaHD and the hypomorphic allele DoaDem a small percentage of the progeny that eclose are of the trans-heterozygous genotype, DoaDem/DoaHD. To examine SOD activity in the presence of Doa levels lower than those found in either Doaγ3B/+ or DoaHD/+ flies, we collected F1 male progeny trans-heterozygous ywa/Y; DoaDem/Pr DoaHD, along with their ywa/Y;Pr DoaHD/TM6B, and ywa/Y; DoaDem/TM6B siblings, and determined their levels of SOD (Figure 2C). Age-matched ywa/Y males were assayed as wild-type controls. In these experiments, the SOD2 activity in DoaDem/DoaHD trans-heterozygotes was 3.2-times that in wild-type controls. The activity in DoaDem/TM6B and DoaHD/TM6B animals was increased by 2.0-fold and 1.6-fold, respectively. In contrast, SOD1 activity in DoaDem/DoaHD animals was increased only 1.4-fold compared to controls, and the SOD1 activity in DoaDem/TM6B and DoaHD/TM6B remained close to control levels. The activities of both SOD1 and SOD2 in TM3/TM6B males were comparable to those in wild-type controls. Taken together, these data suggest that, in Drosophila, the LAMMER kinase gene Doa regulates the expression of SOD2 and SOD1.
CLK-1 negatively regulates SOD activity in mammalian cells
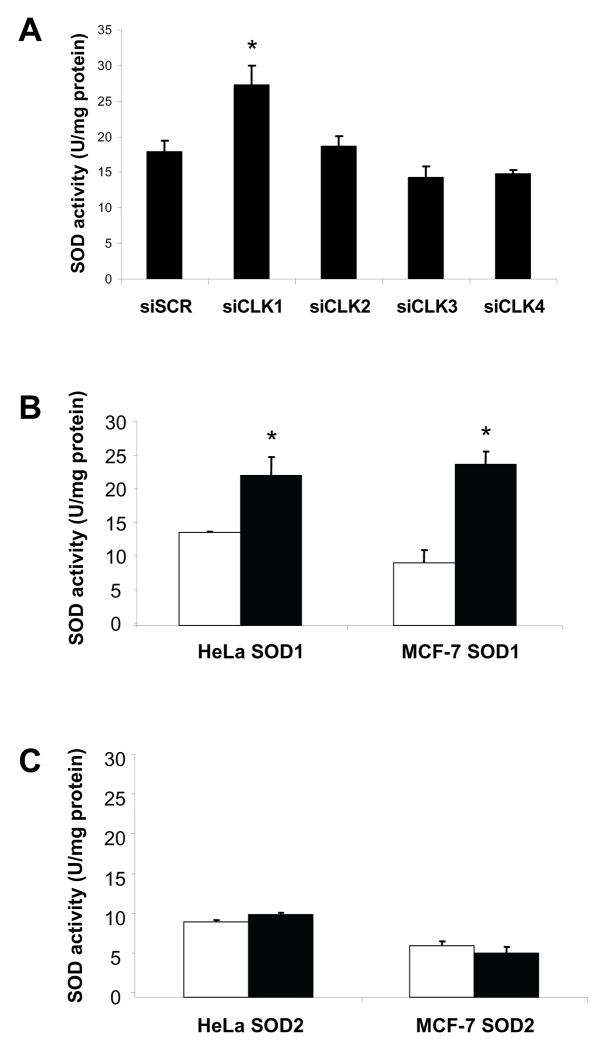
In order to test whether SOD is regulated by LAMMER kinases in human cells, we treated human adenocarcinoma (HeLa) cells with siRNA against the human LAMMER kinase genes CLK-1, CLK-2, CLK-3 or CLK-4 and assayed total SOD activity 72 hours later. We found that knockdown of CLK-1 led to a 1.5 fold increase in total SOD activity (P< .05), while individual knockdown of CLK-2, CLK-3 and CLK-4 had no significant effect on total SOD activity (Figure 3A).
Figure 3.
Knockdown of CLK-1 increases SOD activity in HeLa and MCF-7 cells. (A) Total SOD activity in HeLa cells treated with siRNA against CLK-1, CLK-2, CLK-3 and CLK-4. (B) SOD1 activity in HeLa and MCF-7 cells treated with CLK-1 siRNA. (C) SOD2 activity in HeLa and MCF-7 cells treated with CLK-1 siRNA.. White bars indicate non-targeting siSCR controls; black bars indicate siCLK-1. In each assay RT-PCR confirmed that the target transcript level was reduced at least 80%. Error bars indicate standard error. *SOD activities significantly different from control values (p < 0.05).
When CLK-1 was knocked down there was a 1.6 fold increase in SOD1 activity in HeLa cells, and a 2.5 fold increase in SOD1 activity in breast carcinoma (MCF-7) cells (Figure 3B). However, in neither cell line did knockdown of CLK-1 cause a significant change in SOD2 activity (Figure 3C). This suggests that CLK-1 is sufficient for regulation of SOD1 in human cells. However, CLK-1 either does not regulate SOD2, or CLK regulation of SOD2 is at least partially redundant, in human cells. This effect on SOD1 activity in human cells lacking CLK-1 is similar to what is seen in Drosophila Doa mutants (Figure 2).
SOD1 protein and transcript are regulated by CLK-1 in mammalian cells
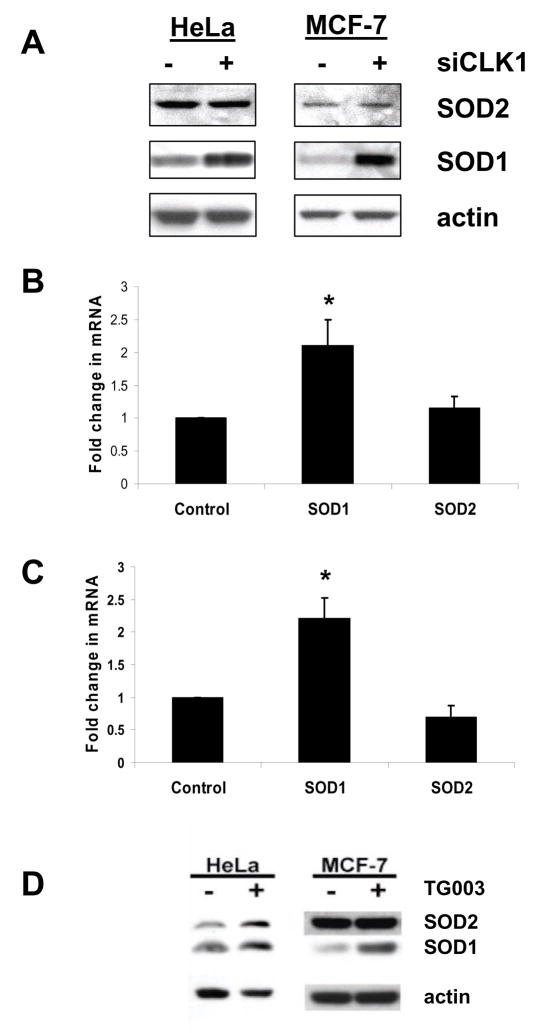
LAMMER kinases are known to regulate pre-mRNA splicing [19–22]. We therefore asked if knockdown of CLK-1 would affect SOD protein and transcript levels in HeLa and MCF7 cells. We used a 72-hour timepoint after treatment with siRNA against CLK-1. We found that HeLa and MCF-7 cells showed significantly increased SOD1 protein and transcript after treatment. SOD1 protein increased approximately 2.5 fold in HeLa cells and 1.6 fold in MCF-7 cells. SOD2 protein did not change in HeLa or MCF-7 cells (Figure 4A). RT-PCR analysis showed that the SOD1 transcript increases 2.1 fold in MCF-7 and (Figure 4B). 2.2 fold in HeLa cells (Figure 4C). There was no significant difference in the SOD2 transcript.
Figure 4.
Knockdown of CLK-1 increases SOD transcript and protein. (A) Western blots of lysates from HeLa cells and MCF-7 cells treated with CLK-1 siRNA. (B) SOD1 and SOD2 transcript levels in MCF-7 cells treated with CLK-1 siRNA were measured using Taqman real-time PCR. (C) SOD1 and SOD2 transcript levels in HeLa cells treated with CLK-1 siRNA were measured using Taqman real-time PCR. (D) Western blots of lysates from HeLa cells and MCF-7 cells grown for 48 hr in 10 μM TG003. In each assay RT-PCR confirmed that the target transcript level was reduced at least 80%. Error bars indicate standard error. *SOD- transcripts significantly different from control values (p < 0.05). Staining for β-actin was done to control for sample loading.
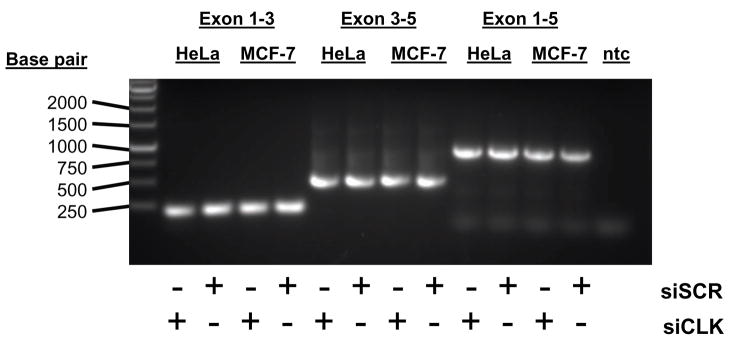
To determine if the change seen in SOD1 transcript levels were due to changes in splicing we performed RT-PCR on total RNA from HeLa and MCF-7 cells treated with siRNA against CLK-1. RT-PCR amplification of SOD1 did not show any evidence of alternate splicing of the SOD1 transcript (Figure 5).
Figure 5.
RT-PCR analysis of SOD transcripts in HeLa and MCF-7 cells treated with siRNA against CLK-1. (A) Results of amplification of SOD1 cDNA using primers spanning exon 1 through 3, exon 3 through 5 and and exon 1 through 5 (ntc: no template control).
The LAMMER kinase inhibitor TG003 increases SOD expression in mammalian cells
To determine whether pharmacological inhibition of LAMMER kinase would increase SOD expression in human cells, we used the LAMMER kinase inhibitor TG003. TG003 acts as a competitive inhibitor of ATP binding to the kinase domain. It has a Ki of 10 nM for CLK-1/Sty in HeLa cells, and exhibits IC50s of 20 nM, 200 nM, >10 μM and 10 nM for mouse CLK-1, CLK-2, CLK-3 and CLK-4, respectively [29].
As reported previously for HeLa cells [29], treatment of cells with TG003 had no significant effect on the growth of HeLa or MCF-7 cells (data not shown). Western blot analysis of lysates from HeLa and MCF-7 cells grown for 48 hours in 10 μM TG003 showed an approximate 2 fold increase in SOD1 protein (Figure 4D). The response of SOD1 protein levels to TG003 was similar to what we observed in CLK-1 siRNA treated cells (Figure 4A) suggesting that this response is due to CLK-1 inhibition by TG003. SOD2 protein levels did not change in MCF-7 cells, but did increase approximately 3 fold in HeLa cells (Figure 4D).
The effect of treating MCF-7 cells with TG003 on SOD2 activity (Figure 4D) is different from what is seen when CLK-1 alone is knocked down (Figure 4A), but is similar to what is seen in Drosophila Doa mutants (Figure 2C). One explanation for the difference between the siRNA and TG003 findings is that in MCF-7 cells SOD2 regulation requires simultaneous inhibition of CLK-1, CLK-2 and CLK-4 activity which is only seen in the case of TG003 treated cells.
DISCUSSION
Our studies have shown that mutations in Drosophila Doa, cause increased levels of both SOD1 and SOD2 protein and activity, providing a possible explanation for the antioxidant mechanism behind the increase in Drosophila resistance to ROS caused by loss of DOA function. We also found that when human CLK-1 is knocked down by treatment with siRNA in HeLa and MCF-7 human tumor cells, there is an increase in SOD1 protein, activity and transcript. While the sequence of the conserved catalytic domain of CLK-2 suggest that it is the closest human ortholog to Doa (75% identity), there is still considerable conservation between the catalytic domains of Doa and CLK-1 (62% identity). From these data we can not discount the possibility that CLK-2 or one of the other two LAMMER kinase genes contribute to SOD regulation. In fact our results using the pan LAMMER kinase inhibitor TG003 support the possibility that more then one of the human LAMMER kinases is involved in SOD regulation. Our findings in Drosophila and human cell lines contrast with reports in S. pombe where the LAMMER kinase homolog lkh1+ appears to positively regulate SOD, and mutations in lkh1+ decrease resistance to ROS and decrease sod1+ mRNA [16]. Although the direction of regulation we observed in Drosophila and HeLa and MCF-7 cells was opposite to that reported in yeast, taken together these data suggest that the ability of LAMMER kinases to regulate SOD is evolutionarily conserved between humans and Drosophila.
When newly eclosed adult Drosophila were exposed to oxidative stress by feeding them the O2.− generator paraquat [28], animals carrying mutations in the LAMMER kinase homolog Doa survived significantly longer than wild-type controls. One possible explanation for this resistance to paraquat is the coincident increase in SOD protein and activity seen in Doa mutants. This is in concert with previous reports that Drosophila SOD loss of function mutants are more sensitive to paraquat [34]. While overexpression of bovine SOD in Drosophila is sufficient to increase resistance to paraquat [35, 36] overexpression of transgenic Drosophila SOD1 was insufficient to increase resistance to paraquat in two different studies [34, 37]. The difference in paraquat resistance between Doa mutants and the Drosophila SOD1 transgenic lines may be due to differences in SOD activity. For example, while we saw a greater than two fold increase in SOD1 activity in Doaγ3Band DoaHD mutants, the transgenic lines in the previous studies increased SOD activity by at most 70% [34]. Alternatively, paraquat resistance in Doa mutants might due to the simultaneous increase in both SOD1 and SOD2 activity or in the regulation of other stress resistance genes by Doa.
We found that SOD2 is negatively regulated by the Doa gene in Drosophila, and that SOD2 protein levels are increased in HeLa cells treated with the pan LAMMER kinase inhbitorTG003, but not in TG003 treated MCF-7 cells, nor in HeLa or MCF-7 cells treated with siRNA against the CLK-1. In S. pombe, the regulation of the two SOD genes is different; SOD1 transcript is increased by Atf1 in response to H2O2, while SOD2 is Atf1 independent [38, 39], and there is no report of SOD2 being regulated by lkh1+. While there is only one LAMMER kinase gene in Drosophila [17], encoded by Doa, there are four reported LAMMER kinase genes in humans. At 10 μM TG003, as used in our study, there would most likely be inhibition of ATP binding to CLK-1, CLK-4 (and possibly CLK-2) [29]. Therefore, we may not have detected regulatory activities that CLK-3 or CLK-2 may exert on SOD. This suggests that in MCF-7 and HeLa cells, SOD2 is regulated by a LAMMER kinase other then CLK-1, or a combination of LAMMER kinases.
The mechanism for the increase in SOD1 by CLK-1 remains unclear. CLK-1 phosphorylates SR proteins to regulate pre-mRNA splicing, specifically influencing the activity of SR protein splicing factors [23]. It was possible that CLK-1 regulates SOD1 through alternate splicing, at least five alternatively spliced forms of human SOD1 have been reported [40, 41], but we found no evidence of that in our studies. Examination of Drosophila genomic sequence and EST data suggest that the Drosophila SOD1 and SOD2 genes are not alternatively spliced. There may be indirect effects of CLK-1 on SOD1 expression since the promoter of human SOD1 is regulated by Egr-1 and two splicing variants of the Egr-related protein WT1 exist [5].
Increased formation of ROS plays an important role in various human pathologies [3] Mutations in SOD1 are associated with 20% of familial amyotrophic lateral sclerosis cases [42, 43]. In human tumor cells, overexpressing SOD2 inhibits cell proliferation, increases differentiation, and can reverse a malignant phenotype to a nonmalignant phenotype[44–46]. Given the role of SOD in reducing the damaging effects of ROS, increasing SOD activity through inhibition of CLK-1 may provide an effective way for treating some of these diseases.
Acknowledgments
This work was supported by Grant CA077204 from the National Cancer Institute to G.P. and CA096812 to E.M. We thank Leonard Rabinow for fly stocks and sharing unpublished data about Doa, and Marc Brabant for helpful discussions of the data. A portion of this work was carried out in the Model Organism Shared Service at the Arizona Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.North S, Moenner M, Bikfalvi A. Recent developments in the regulation of the angiogenic switch by cellular stress factors in tumors. Cancer Lett. 2005;218(1):1–14. doi: 10.1016/j.canlet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci U S A. 1989;86(8):2761–5. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337–49. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 4.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279(15):15499–504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 5.Minc E, de Coppet P, Masson P, Thiery L, Dutertre S, Amor-Gueret M, Jaulin C. The human copper-zinc superoxide dismutase gene (SOD1) proximal promoter is regulated by Sp1, Egr-1, and WT1 via non-canonical binding sites. J Biol Chem. 1999;274(1):503–9. doi: 10.1074/jbc.274.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Kim YJ, Rhee SK. H+-pumping ATPase has little stimulatory effect on in vitro translocation of a model protein into Vibrio alginolyticus inside-out membrane vesicles. Mol Cells. 1997;7(4):473–7. [PubMed] [Google Scholar]
- 7.Yoo HY, Chang MS, Rho HM. Heavy metal-mediated activation of the rat Cu/Zn superoxide dismutase gene via a metal-responsive element. Mol Gen Genet. 1999;262(2):310–3. doi: 10.1007/s004380051088. [DOI] [PubMed] [Google Scholar]
- 8.Soini Y, Kaarteenaho-Wiik R, Paakko P, Kinnula V. Expression of antioxidant enzymes in bronchial metaplastic and dysplastic epithelium. Lung Cancer. 2003;39(1):15–22. doi: 10.1016/s0169-5002(02)00392-6. [DOI] [PubMed] [Google Scholar]
- 9.Kim HP, Roe JH, Chock PB, Yim MB. Transcriptional activation of the human manganese superoxide dismutase gene mediated by tetradecanoylphorbol acetate. J Biol Chem. 1999;274(52):37455–60. doi: 10.1074/jbc.274.52.37455. [DOI] [PubMed] [Google Scholar]
- 10.Zhu CH, Huang Y, Oberley LW, Domann FE. A family of AP-2 proteins down-regulate manganese superoxide dismutase expression. J Biol Chem. 2001;276(17):14407–13. doi: 10.1074/jbc.M009708200. [DOI] [PubMed] [Google Scholar]
- 11.Nomiyama T, Tanaka Y, Piao L, Nagasaka K, Sakai K, Ogihara T, Nakajima K, Watada H, Kawamori R. The polymorphism of manganese superoxide dismutase is associated with diabetic nephropathy in Japanese type 2 diabetic patients. J Hum Genet. 2003;48(3):138–41. doi: 10.1007/s100380300021. [DOI] [PubMed] [Google Scholar]
- 12.Kahlos K, Anttila S, Asikainen T, Kinnula K, Raivio KO, Mattson K, Linnainmaa K, Kinnula VL. Manganese superoxide dismutase in healthy human pleural mesothelium and in malignant pleural mesothelioma. Am J Respir Cell Mol Biol. 1998;18(4):570–80. doi: 10.1165/ajrcmb.18.4.2943. [DOI] [PubMed] [Google Scholar]
- 13.Janssen AM, Bosman CB, Sier CF, Griffioen G, Kubben FJ, Lamers CB, van Krieken JH, van de Velde CJ, Verspaget HW. Superoxide dismutases in relation to the overall survival of colorectal cancer patients. Br J Cancer. 1998;78(8):1051–7. doi: 10.1038/bjc.1998.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappaB and activated protein-1. J Biol Chem. 1998;273(21):13245–54. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 15.Karlsson K, Marklund SL. Heparin-induced release of extracellular superoxide dismutase to human blood plasma. Biochem J. 1987;242(1):55–9. doi: 10.1042/bj2420055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YD, Kang WH, Yang WS, Shin KS, Sook Bae K, Park HM. LAMMER kinase homolog, Lkh1, is involved in oxidative-stress response of fission yeast. Biochem Biophys Res Commun. 2003;311(4):1078–83. doi: 10.1016/j.bbrc.2003.10.110. [DOI] [PubMed] [Google Scholar]
- 17.Yun B, Farkas R, Lee K, Rabinow L. The Doa locus encodes a member of a new protein kinase family and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 1994;8(10):1160–73. doi: 10.1101/gad.8.10.1160. [DOI] [PubMed] [Google Scholar]
- 18.Tang Z, Mandel LL, Yean SL, Lin CX, Chen T, Yanagida M, Lin RJ. The kic1 kinase of schizosaccharomyces pombe is a CLK/STY orthologue that regulates cell-cell separation. Exp Cell Res. 2003;283(1):101–15. doi: 10.1016/s0014-4827(02)00022-8. [DOI] [PubMed] [Google Scholar]
- 19.Du C, McGuffin ME, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 1998;2(6):741–50. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- 20.Nikolakaki E, Du C, Lai J, Giannakouros T, Cantley L, Rabinow L. Phosphorylation by LAMMER protein kinases: determination of a consensus site, identification of in vitro substrates, and implications for substrate preferences. Biochemistry. 2002;41(6):2055–66. doi: 10.1021/bi011521h. [DOI] [PubMed] [Google Scholar]
- 21.Nayler O, Stamm S, Ullrich A. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem J. 1997;326(Pt 3):693–700. doi: 10.1042/bj3260693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. Embo J. 1996;15(2):265–75. [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19(10):6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabinow L, Birchler JA. A dosage-sensitive modifier of etrotransposoninduced alleles of the Drosophila white locus. Embo J. 1989;8(3):879–89. doi: 10.1002/j.1460-2075.1989.tb03449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabinow L, Chiang SL, Birchler JA. Mutations at the Darkener of apricot locus modulate transcript levels of copia and copia-induced mutations in Drosophila melanogaster. Genetics. 1993;134(4):1175–85. doi: 10.1093/genetics/134.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kpebe A, Rabinow L. Alternative promoter usage generates multiple evolutionarily conserved isoforms of Drosophila DOA kinase. Genesis. 2008;46(3):132–43. doi: 10.1002/dvg.20374. [DOI] [PubMed] [Google Scholar]
- 27.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126(3):365–79. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Seong KH, Ogashiwa T, Matsuo T, Fuyama Y, Aigaki T. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology. 2001;2(3):209–17. doi: 10.1023/a:1011517325711. [DOI] [PubMed] [Google Scholar]
- 29.Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, Furuichi K, Shibuya H, Krainer AR, Suzuki M, Hagiwara M. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279(23):24246–54. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed SA, Gogal RM, Jr, Walsh JE. A new rapid and simple nonradioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170(2):211–24. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Castello PR, Drechsel DA, Patel M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J Biol Chem. 2007;282(19):14186–93. doi: 10.1074/jbc.M700827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 34.Seto NO, Hayashi S, Tener GM. Overexpression of Cu-Zn superoxide dismutase in Drosophila does not affect life-span. Proc Natl Acad Sci U S A. 1990;87(11):4270–4. doi: 10.1073/pnas.87.11.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reveillaud I, Kongpachith A, Park R, Fleming JE. Stress resistance of Drosophila transgenic for bovine CuZn superoxide dismutase. Free Radic Res Commun. 1992;17(1):73–85. doi: 10.3109/10715769209061090. [DOI] [PubMed] [Google Scholar]
- 36.Reveillaud I, Niedzwiecki A, Bensch KG, Fleming JE. Expression of bovine superoxide dismutase in Drosophila melanogaster augments resistance of oxidative stress. Mol Cell Biol. 1991;11(2):632–40. doi: 10.1128/mcb.11.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orr WC, Sohal RS. Effects of Cu-Zn superoxide dismutase overexpression of life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1993;301(1):34–40. doi: 10.1006/abbi.1993.1111. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Kwon ES, Kim DW, Cha J, Roe JH. Regulation and the role of Cu,Zn-containing superoxide dismutase in cell cycle progression of Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2002;297(4):854–62. doi: 10.1016/s0006-291x(02)02290-8. [DOI] [PubMed] [Google Scholar]
- 39.Jeong JH, Kwon ES, Roe JH. Characterization of the manganesecontaining superoxide dismutase and its gene regulation in stress response of Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2001;283(4):908–14. doi: 10.1006/bbrc.2001.4853. [DOI] [PubMed] [Google Scholar]
- 40.Hirano M, Hung WY, Cole N, Azim AC, Deng HX, Siddique T. Multiple transcripts of the human Cu,Zn superoxide dismutase gene. Biochem Biophys Res Commun. 2000;276(1):52–6. doi: 10.1006/bbrc.2000.3427. [DOI] [PubMed] [Google Scholar]
- 41.Kawata A, Kato S, Shimizu T, Hayashi H, Hirai S, Misawa H, Takahashi R. Aberrant splicing of human Cu/Zn superoxide dismutase (SOD1) RNA transcripts. Neuroreport. 2000;11(12):2649–53. doi: 10.1097/00001756-200008210-00009. [DOI] [PubMed] [Google Scholar]
- 42.Rachakonda V, Pan TH, Le WD. Biomarkers of neurodegenerative disorders: how good are they? Cell Res. 2004;14(5):347–58. doi: 10.1038/sj.cr.7290235. [DOI] [PubMed] [Google Scholar]
- 43.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–93. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 44.Chuang TC, Liu JY, Lin CT, Tang YT, Yeh MH, Chang SC, Li JW, Kao MC. Human manganese superoxide dismutase suppresses HER2/neumediated breast cancer malignancy. FEBS Lett. 2007;581(23):4443–9. doi: 10.1016/j.febslet.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 45.Kinnula VL, Crapo JD. Superoxide dismutases in malignant cells and human tumors. Free Radic Biol Med. 2004;36(6):718–44. doi: 10.1016/j.freeradbiomed.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y, Chaiswing L, Oberley TD, Batinic-Haberle I, St Clair W, Epstein CJ, St Clair D. A mechanism-based antioxidant approach for the reduction of skin carcinogenesis. Cancer Res. 2005;65(4):1401–5. doi: 10.1158/0008-5472.CAN-04-3334. [DOI] [PubMed] [Google Scholar]