Abstract
The p53 tumor suppressor is mutated in the majority of human tumors. MDM2, a well-known inhibitor of p53, is overexpressed in a large number of tumors suggesting that increased levels of MDM2 also contribute to tumorigenesis. A novel p53 inhibitor, MDM4, was more recently identified. The role of MDM4 in cancer development is not well understood. We set out to examine the levels of MDM4 by immunohistochemistry in Head and Neck Squamous Carcinomas (HNSC) to ask whether high MDM4 levels could contribute to its development and progression. In addition, MDM2 and p53 levels were examined to identify overlapping expression patterns. MDM4 is present at high levels in 50% of HNSC. Additionally, overexpression of MDM2 was detected in 80% of tumors, many of which were also positive for MDM4. A subset of tumors displayed high levels of all three proteins. Sequencing of the p53 gene revealed that tumors with positive immunoreactivity for MDM2 or MDM4, some of which also had high levels of p53, did not carry mutations in this gene. Thus, the detection of p53 by immunohistochemistry was not synonymous with the presence of p53 mutations. Expression of both MDM2 and MDM4 in tumors without p53 mutations strongly suggests that MDM2 and MDM4 inhibit the activity of this tumor suppressor in HNSC.
Keywords: MDM2, p53 mutations, immunohistochemistry
1. Introduction
The p53 tumor suppressor is the most frequently altered gene in human cancer, being mutated in approximately half of all human tumors [1]. Importantly, more than 80% of these alterations are single nucleotide substitutions that lead to missense mutations in p53, the majority of which occur within exons 5-9 and disrupt the DNA-binding domain [2]. This event results in the failure of p53 to function as a transcriptional activator of genes involved in cell cycle arrest, apoptosis, and senescence [3-4]. Previous studies in head and neck squamous carcinomas (HNSC) have revealed that p53 is mutated in 40-50% of these tumors [5].
p53 is negatively regulated by numerous factors. The first identified p53 inhibitor, MDM2, is an E3 ubiquitin ligase that binds and ubiquitinates p53 leading to its degradation by the 26S proteasome [6-8]. The significance of MDM2 as an inhibitor of p53 has been established in mouse models. Mice lacking MDM2 die early in embryogenesis by initiating a p53-dependent apoptotic response [9]. Importantly, this lethal phenotype is completely rescued by loss of p53 [10-11]. Notably, both amplification of the MDM2 gene and overexpression of MDM2 by unknown mechanisms have been observed in a subset of human tumors, some of which retain wild-type p53 [12-14]. These data suggest that increased levels of MDM2 may substitute for mutations in p53, which represents an alternative mechanism by which tumor cells escape from the tumor suppressive activities of p53. In HNSC, overexpression of MDM2 has been observed [15-16], but a clear correlation between the levels of MDM2 protein and p53 status has not been established [17].
Mouse models have more recently verified the importance of another p53 inhibitor, MDM4 (also known as MDMX). MDM4 has significant structural homology to MDM2 in the p53-binding and RING-finger domains that are located at the amino and carboxyl-terminal regions of the proteins, respectively [18-19]. Despite this structural similarity, MDM4 does not share all functional properties of MDM2. For example, although MDM4 binds and inhibits the transcriptional activation domain of p53 [18], it does not target p53 for degradation [20-22]. However, loss of MDM4 in the mouse leads to defects in cell proliferation and death during embryogenesis, a phenotype that is completely rescued by concomitant deletion of p53 [23-25]. Loss of MDM4 in other cell types such as cardiomyocytes and neural epithelium also leads to defects completely rescued by loss of p53 [26-28]. These data indicate that MDM4 is another critical inhibitor of p53 in vivo.
Previous observations indicate that MDM4 plays an important role in the development of malignant gliomas as amplification and overexpression of MDM4 was found in a subset of these tumors [29]. Absence of p53 mutations in gliomas with increased MDM4 expression suggested that high levels of MDM4 could substitute for p53 mutations. Limited studies using human cancer cell lines and various tumors such as breast, lung, and colon also revealed overexpression of MDM4 in cells lacking p53 alterations [30-31]. However, the expression of MDM4 has yet not been investigated in HNSC. Given that HNSC is the sixth most common cancer worldwide and that the overall survival rate of the disease has not changed with time [5], we sought to investigate the role of MDM4 in this tumor type. In this study, we generated a MDM4 antibody and show that high levels of MDM4 are present in HNSC. Moreover, the majority of tumors with positive immunoreactivity for MDM4 also showed high levels of MDM2 suggesting a relationship between the two proteins. All tumors examined that expressed MDM2 and MDM4, had wild type p53 suggesting that increased levels of MDM4, like MDM2, can substitute for p53 mutations in HNSC.
2. Materials and methods
2.1. Tumor Samples
Paraffin-embedded tumor samples were obtained from the Department of Pathology at The University of Texas MD Anderson Cancer Center. Samples were fixed in 10% buffered formalin, embedded in paraffin, and sectioned.
2.2. Generation of anti-MDM4 antibody
The DNA sequence encompassing amino acids 109-198 of the human MDM4 cDNA was cloned into the pGEX-2T GST Fusion vector (Promega). After purification, the GST-Mdm4 fusion protein was used to immunize rabbits. The MDM4 antibody from crude serum was purified by the GST Orientation Kit (Pierce).
2.3. Cell Transfections and Western Blot Analyses
MCF-7 or HeLa cell lines were transfected with 2 μg of pcDNA3.1 vector containing the MDM4 cDNA, a pCMV plasmid expressing MDM2, or empty vector using FuGENE6 reagent (Roche). Twenty-four hours later, cells were harvested and whole cell lysates were prepared. Proteins were loaded on a 8% SDS-PAGE gel and transferred to nitrocellulose membrane. Membranes were incubated with antisera against MDM4 at a 1:1000 dilution. Signal was detected using the ECL kit (Amersham Pharmacia Biotech). Membranes were stripped and blotted with SMP14 monoclonal antibody (1:1000 dilution; Santa Cruz Biotechnology) for MDM2, and subsequently stripped and blotted again with antisera against β-actin (1:2000 dilution; Sigma). Immunohistochemistry on cells grown on cover slips was performed using the Vectastain ABC Elite kit (Vector Laboratories). Twenty-four hours after transfection, cells were washed with Phosphate Buffered Saline (PBS) and fixed in 50% methanol/50% acetone for 3 min, then rinsed again in PBS. Fixed cells were incubated with 50 mM glycine/PBS for 10 min, washed with PBS, and subsequently treated with 0.2% Triton /PBS for 10 min followed by three washes in PBS. Cells were then treated with 0.3% H2O2 for 3 min to inhibit endogenous peroxidase activity and rinsed with PBS. Cells were blocked for 10 min at room temperature in 2% horse serum/PBS and incubated for 1 hr at room temperature with anti-MDM2 (SMP14, 1:200, Santa Cruz Biotechnology) or with anti-MDM4 polyclonal antibody (Ab112, 1:200). Secondary antibody and ABC reagents were added as described in the Vectastain protocol. Diaminobenzidine substrate (Vector Laboratories) was used according to the manufacturer's instructions. Nuclear Fast Red was used to counterstain cells.
2.4. Immunohistochemistry in paraffin embedded tissues
The expression of MDM2, MDM4 and p53 in paraffin embedded cells was analyzed by performing immunohistochemistry as described [14]. Briefly, sections were deparaffinized in a xylene-to-alcohol gradient. The slides were steamed for 30 min in citric buffer (pH 6.0). Endogenous peroxidases were inhibited by incubating the samples in 3% H2O2 for 15 min. Samples were blocked in 2% horse serum in PBS-Tween 20 for 30 min. After blocking, samples were incubated with primary antibodies for MDM2 (SMP14, 1:200, Santa Cruz Biotechnology), MDM4 (AB112, 1:150) and p53 (Bp5312, 1:200, Santa Cruz Biotechnology) for 2 hours at 37°C Proteins were detected using the Vectastain ABC Elite kit. The Diaminobenzidine substrate (Vector Laboratories) was used as indicated by the company. Nuclear Fast Red was used to counterstain. Histological evaluation was based on cellular localization (nuclear, cytoplasmic, both); intensity of staining [weak (1+), moderated (2+) and strong (3+)]; and percentage of positive cells in the sample. Cases were considered positive if 10% or more of the dysplastic or tumor cells were stained. Since normal tissue does not stain with any of these antibodies, a 10% threshold eliminates false positives. As a positive control for MDM4 staining, we used paraffin embedded brain sections of a mouse model that expresses MDM4. The Ab112 antibody was made to a conserved epitope between mouse and human MDM4. Surrounding normal tissue served as a negative control.
2.4. p53 Mutation Analysis
Genomic DNA from 25 microdissected tumors was extracted by incubating samples in 100 μL TE9 buffer (500 mM Tris, 20 mM EDTA, 10 mM NaCl, pH 9.0) and 10 μL of 10X PK solution (5mg/mL Proteinsase K, 10% SDS) at 42°C overnight. To extract the DNA, an equal volume of 1:1 phenol/chloroform solution was added to the samples followed by vortex and spin for 1 min at 14,000rpm. DNA was precipitated by using NH4 Acetate/ethanol. DNA was screened for p53 mutations by performing sequencing analysis using primer sets that expand exons 5-9 of the gene [32].
3. Results
3.1. The Anti-MDM4 Antibody Specifically Recognizes MDM4
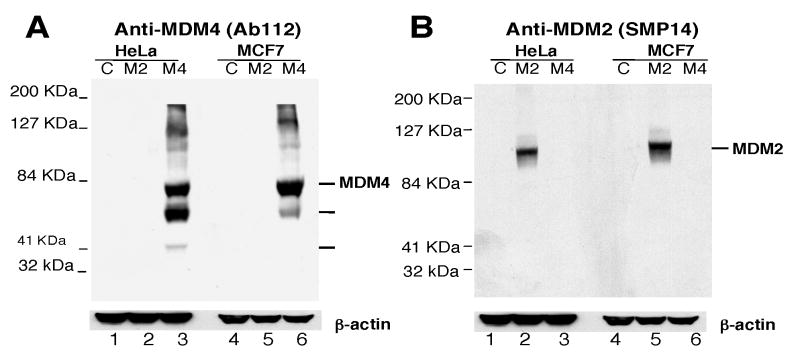
In order to study the role of MDM4 in tumorigenesis, we generated an antibody against human MDM4 and examined a number of tumor samples for increased expression of MDM4. Since MDM4 shares sequence homology with MDM2, the antibody was generated against the unique central region of MDM4 encompassing amino acids 109-198 (Fig. 1). To verify that the MDM4 antibody (Ab112) specifically recognizes MDM4 and not MDM2 or other proteins, we transiently transfected MCF-7 and HeLa cell lines with plasmids containing full length MDM4 or MDM2, and performed Western blot analysis. Several bands were only visible in cells transfected with MDM4-containing plasmids suggesting they were MDM4 products (Fig. 2-A). The highest molecular weight band migrated at the expected size for full length MDM4 (∼80 KDa). The smaller bands (∼55 and 40 KDa) might be degradation products or other MDM4 variants since these products were not observed in cells transfected with MDM2 or vector control (Figure 2-A). The same membrane was stripped and blotted with the MDM2 antibody in order to verify the efficiency of MDM2 transfection. MDM2 was detected in both MCF-7 and HeLa cells after transfection (Fig. 2-B). The levels of β-actin expression demonstrated equal protein loading in the gel. Thus, the Ab112 antibody showed high specificity for MDM4 in Western blot assays.
Fig. 1.

Generation of an MDM4 polyclonal antibody (Ab112). A, Schematic representation of the MDM2 and MDM4 structural domains. B. Alignment between MDM2 and MDM4 proteins. Ab112 antibody was raised against human MDM4 using amino acids 109-198 as epitope. Underlined sequence represents the epitope used to generate the antibody.
Fig. 2.
Anti-MDM4 (Ab112) antibody specifically recognizes MDM4 and not MDM2 by Western Blot analysis. A, MCF-7 and HeLa cell lines were transiently transfected with MDM2 (M2), MDM4 (M4) or empty vector (C). Cell lysates were used in Western blot assays to detect MDM4 using Ab112 antibody. B, The same blot was stripped and blotted with SMP14 monoclonal antibody to detect MDM2. Human β-actin was used as loading (lower panels A and B).
Because our goal was to study the expression pattern of MDM4 in human tumors, we next determined if Ab112 antibody could recognize MDM4 by immunohistochemistry. For this purpose, we transfected HeLa cells with plasmids expressing MDM4, MDM2, or vector alone and performed immunohistochemistry. We detected MDM4 only in cells transfected with MDM4 and not in cells with MDM2 or cells with empty vector (Fig. 3A, upper panels and data not shown). Expression of MDM4 was localized either in the nucleus (filled arrow) in some cells or in the nucleus and cytoplasm in others (open arrow). MDM2 transfected HeLa cells also showed high MDM2 levels (Fig. 3A, lower panels). MDM2 was generally localized to the nucleus, although occasional cytoplasmic localization was also noted. Additionally, we analyzed paraffin-embedded H1299 human cells transfected with MDM4 by immunohistochemistry. We detected positive staining in cells transfected with MDM4, but not in untransfected control cells (Fig. 3B). These data indicated that Ab112 recognized MDM4 protein by immunohistochemical assays. More importantly, even though MDM2 and MDM4 are highly homologous, Ab112 only detected MDM4 by Western blot and immunohistochemical assays.
Fig. 3.
MDM4 is specifically recognized by Ab112 antibody in immunohistochemical assays. A, HeLa cells were transfected with plasmids that contain MDM2 (left panels) or MDM4 (right panels) genes and analyzed by immunohistochemistry using Ab112 to detect MDM4 or SMP14 to recognize MDM2 as control for the transfection efficiency. Cellular localization of MDM4 includes nuclear (filled arrow) and cytoplasmic (open arrow) compartments. Duplicates of transfected cells that were incubated in SMP14 antibody (lower panel) showed detection of MDM2 only in cells overexpressing MDM2; original magnification X200. B, H1299 human cell lines transfected with MDM4 (right) or non-transfected (NT; left). Cell pellets were formalin fixed, paraffin embedded, sectioned and analyzed by immunohistochemistry. Arrow points to positive staining for MDM4; original magnification X600.
3.2. MDM4 Is Overexpressed In HNSC
Human HNSC tissue sections were evaluated for MDM4 expression by immunohistochemistry. In addition, we analyzed the expression pattern of p53 and MDM2 in these tumors in order to determine the relationship between these proteins. A total of 56 HNSC samples were analyzed (Supplementary Table 1). Cases were considered positive if 10% or more of the dysplastic or tumor cells were stained. Of these, twenty-eight (50%) had high expression of MDM4 (Table 1; Fig. 4, and Fig. 5). Sixteen (57%) of the tumors expressing MDM4 showed simultaneous expression of the protein in both cytoplasm and nucleus (Fig.4B, E; Fig.5B, E, and H) and twelve (43%) showed nuclear expression exclusively (Fig. 5K). Twenty-three (82%) samples also had high MDM4 levels in dysplastic epithelium (Fig. 5B). Importantly, normal squamous epithelium was negative for MDM4 expression.
TABLE 1.
Differential levels of MDM2 and MDM4 with p53 in HNSC
| MDM2 + | MDM2 − | MDM4 + | MDM4 − | |
|---|---|---|---|---|
| p53+ | 29 | 6 | 19 | 16 |
| p53− | 16 | 5 | 9 | 12 |
| Total | 45 | 11 | 28 | 28 |
Fig. 4.
HNSC exhibit different expression patterns of MDM2, MDM4 and p53 by immunohistochemistry. From left to right, HNSC samples where stained for MDM2 (A, D, G and J), MDM4 (B, E, H, and K), and p53 (C, F, I and L). Panels A, B, and C show a sample that was positive for MDM4 and p53. Panels D, E, and F represent a HNSC tumor with high levels of MDM2 and MDM4, but not p53. Panels G, H, and I show a tumor with elevated levels of MDM2 and p53, but not MDM4. Panels J, K, and L are characteristic of samples with high levels of MDM2 alone; original magnification X100.
Fig 5.
Concomitant overexpression of MDM2, MDM4, and p53 in HNSC. From left to right, HNSC samples where stained for MDM2 (A, D, G, and J), MDM4 (B, E, H, and K), and p53 (C, F, I, and L). Panels A-C represent a tumor (*) with MDM2, MDM4 and p53 staining in dysplastic epithelium (**); original magnification X100. Panels D-F show the same sample as in A-C showing co-expression of the three proteins in the tumor area; original magnification X200. Panels G-I illustrate co-localization of the three proteins analyzed in the same area of the tumor; original magnification X100. Panels J-L show the only sample with exclusive nuclear localization of all three proteins; original magnification X200.
The number of HNSC with MDM2 expression was greater than the number of tumors that showed high MDM4 levels. Forty-five of fifty-six (80%) tumors were positive for MDM2 (Table 1; Fig. 4; and Fig. 5). Similar to MDM4, thirty-seven (82%) samples expressing high MDM2 levels showed staining in the dysplastic mucosa (Fig. 5A). The expression of MDM2 in the basal and immediate suprabasal cells of the epithelium adjacent to the tumor cells has been previously reported in HNSC [33-34]. High MDM2 levels were present in both nuclear and cytoplasmic compartments in the majority of the tumors (96%) (Fig. 4D, G, J; Fig. 5D, G), whereas exclusive nuclear staining was detected in only two sample (4%) (Fig.5J). Interestingly, these two tumors also showed exclusive nuclear staining for MDM4 and p53 (Fig. 5J-L).
In accordance with studies performed by others [35], p53 was also found overexpressed in about half of HNSC studied (35/56 or 63% of tumors). However, the expression of p53 in dysplastic epithelium was less common than the expression of MDM2 and MDM4. Twenty (57%) samples expressed p53 in dysplastic epithelium (Fig. 5C). Of interest, overexpression of p53 in the displastic mucosa was frequently accompanied by overexpression of MDM2, MDM4 or both in the same region (Fig. 5C).
Since both MDM2 and MDM4 are important regulators of p53, we compared expression of all three proteins in HNSC. High MDM4 levels were rarely observed in the absence of high MDM2 levels. Only six of twenty-eight (21%) tumors were positive for MDM4 staining and negative for MDM2 staining (Table 2, and Fig. 4A-C). p53 was also overexpressed in four of these tumors (Fig. 4A-C). Of 45 samples that stained positive for MDM2, 23 were negative for MDM4, and these were evenly split into samples that did (14 samples) or did not (9 samples) stain for p53, which was always exclusively nuclear (Table 2; Fig. 4G-I and J-L, respectively). Additionally, expression of all three proteins occurred in fifteen (27%) tumors. Only 11 of 56 HNSC did not stain for MDM2. Two of these were positive for MDM4, two were positive for p53, and four were positive for MDM4 and p53. Three samples were negative for all three proteins studied (Table 2). When stained with more than one antibody, the majority of HNSC samples display overlapping expression patterns (Fig 5).
TABLE 2.
Relationship of p53 immunoreactivity to the expression of MDM2 and MDM4 in HNSC
| p53+ | p53− | |
|---|---|---|
| MDM2+ MDM4+ | 15 | 7 |
| MDM2+ MDM4− | 14 | 9 |
| MDM2− MDM4+ | 4 | 2 |
| MDM2− MDM4− | 2 | 3 |
As we detected high levels of MDM2, MDM4, or both proteins combined in tumors that also expressed high levels of p53 (33/56 or 59%), we investigated the status of the p53 gene in 24 HNSC tumors that had high levels of MDM2, MDM4 or both. Of these, thirteen had high levels of p53. In addition, we examined one tumor that had no detectable expression of any protein analyzed. None of the 25 samples analyzed carried mutations in exons 5-9 of p53 (Supplementary Table 1). This region encompasses greater than 90% of all reported p53 mutations found in human tumors. Sequence analysis successfully identified polymorphisms in adjacent intronic sequences.
Lastly, we compared the relationship between expression of p53, Mdm2 and Mdm4, and tumor grade. Even though we were not able to find a clear statistical significance between protein expression and degree of malignancy, we observed two interesting features. First, of six HNSC samples that were positive for MDM4 staining and negative for MDM2 staining, three were classified as high-grade (poorly differentiated) tumors, and the other three samples were classified as moderate (intermediate) grade tumors with an advanced stage of progression (Supplementary Table 1). Second, of twelve HNSC samples graded as dysplastic or well-differentiated tumors, all of them showed high levels of MDM2. Of these, four expressed only MDM2, two expressed MDM2 and p53 together, and one expressed MDM2 and MDM4 together. Five of these samples show high levels of all three proteins.
Discussion
Mouse models have demonstrated the importance of MDM2 and MDM4 in negative regulation of p53. While numerous studies have evaluated the role of MDM2 in human cancers [12-14], few have analyzed the role of MDM4. In this study, we analyzed the role of MDM4 in tumorigenesis and showed that MDM4 was present at high levels in 50% of HNSC suggesting its involvement in the etiology of this disease. Moreover, high levels of MDM2 and MDM4 were detected in dysplastic mucosa suggesting that inhibition of the p53 tumor suppressive activity occurs in early stages of this disease. Additionally, since all HNSC samples that were graded as dysplastic or well-differentiated tumors in our study show high levels of MDM2, it appears likely that MDM2 is a major player in the initiation of this disease. All tumors with high levels of MDM4 and without alterations in the levels of MDM2 showed an advanced tumor grade. Therefore, it may be possible that those HNSC tumors that did not express high levels of MDM2 in early stages of the disease may require overexpression of MDM4 in late stages to inhibit p53 activity and allow tumor progression.
MDM2 is another p53 inhibitor present in high levels in HNSC [15-16]. In this study, we observed that many tumors expressed high levels of both, MDM2 and MDM4. The interaction between MDM2 and MDM4 inhibits autoubiquitination and degradation of MDM2 [20-22]. Thus, MDM4 overexpression in HNSC may stabilize MDM2, enhancing p53 inhibition. The mechanism(s) by which MDM4 expression is increased is unknown. MDM4 maps to chromosome 1q32, a region that is amplified in some human cancers [36]. A small fraction of gliomas and breast tumors have amplification of the MDM4 gene [29, 31]. Thus, gene amplification may be one mechanism by which the levels of MDM4 increase in human HNSC.
Interestingly, many HNSC samples that expressed high levels of MDM2 also showed high levels of p53. Sequencing analysis revealed absence of p53 mutations in samples expressing high levels of MDM2, MDM4 or both. MDM2 binds and targets wild type p53 for degradation and, consequently, samples with high MDM2 are expected to have undetectable levels of p53 protein. A possible explanation for this phenomenon is that MDM2 is not able to degrade p53 in these tumors, although it is still capable of binding p53 and inhibiting its transactivation domain.
Some HNSC had high levels of all three proteins. Our data suggest that MDM2 and MDM4 cooperate to inhibit p53 regardless of its stability. In support of this, overexpression of MDM4 in cancer cell lines inhibits p53 activity although it blocks MDM2-mediated p53 degradation [20-22]. Other tumors had high MDM2 and MDM4 yet did not have stable p53. Perhaps in this case, MDM4 is modified so that it cannot bind MDM2, which allows MDM2 to degrade p53. In our study, a small percentage (5%) of HNSC samples did not show expression of any of the proteins analyzed. The absence of any alterations in these three proteins suggests that either the p53 pathway is intact or disrupted by unknown mechanisms. Finally, the localization of MDM2 and MDM4 in the cytoplasm of tumor cells suggests that these oncogenes may also have p53-independent functions in these malignant cells.
Our data indicate that disruption of the p53 pathway by MDM2 and MDM4 overexpression is a common event in the head and neck squamous tumorigenesis. Moreover, high levels of MDM2 and/or MDM4 in HNSC that retain wild type p53 suggest that these inhibitors may substitute for mutations in p53 and, therefore, contribute to the severity and progression of the disease. Together, our data strongly suggest that overexpression of MDM2 or MDM4 and p53 mutations in HNSC are mutually exclusive events. The presence of high levels of MDM2 and MDM4 in many HNSC suggests that these data should be considered in the treatment of HNSC. Drugs such as Nutlin and RITA that disrupt the MDM2/p53 interaction (and presumably MDM4/p53 interaction since both inhibitors bind the same domain of p53 with similar affinities) may be useful in the treatment of this disease [37-38].
Supplementary Material
Acknowledgments
This study was supported by a H&N SPORE project to Guillermina Lozano and Adel K. El-Naggar (CA97007). Yasmine A. Valentin-Vega was supported by the Schissler Fellowship Foundation and with Juan A. Barbosa by the Cancer Genetics Training Grant (CA009299).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Rice K, Greenblatt MS, et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–5. [PMC free article] [PubMed] [Google Scholar]
- 3.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 4.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–26. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Nylander K, Dabelsteen E, Hall PA. The p53 molecule and its prognostic role in squamous cell carcinomas of the head and neck. J Oral Pathol Med. 2000;29(9):413–25. doi: 10.1034/j.1600-0714.2000.290901.x. [DOI] [PubMed] [Google Scholar]
- 6.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 7.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 8.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420(1):25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 9.Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63(24):8664–9. [PubMed] [Google Scholar]
- 10.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 11.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 12.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–3. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–9. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20(30):4041–9. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- 15.Matsumura T, Yoshihama Y, Kimura T, Shintani S, Alcalde RE. p53 and MDM2 expression in oral squamous cell carcinoma. Oncology. 1996;53(4):308–12. doi: 10.1159/000227578. [DOI] [PubMed] [Google Scholar]
- 16.Schoelch ML, Le QT, Silverman S, Jr, et al. Apoptosis-associated proteins and the development of oral squamous cell carcinoma. Oral Oncol. 1999;35(1):77–85. doi: 10.1016/s1368-8375(98)00065-7. [DOI] [PubMed] [Google Scholar]
- 17.Partridge M, Kiguwa S, Emilion G, Pateromichelakis S, A'Hern R, Langdon JD. New insights into p53 protein stabilisation in oral squamous cell carcinoma. Oral Oncol. 1999;35(1):45–55. doi: 10.1016/s1368-8375(98)00051-7. [DOI] [PubMed] [Google Scholar]
- 18.Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. Embo J. 1996;15(19):5349–57. [PMC free article] [PubMed] [Google Scholar]
- 19.Shvarts A, Bazuine M, Dekker P, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43(1):34–42. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 20.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274(53):38189–96. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 21.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20(3):1001–7. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stad R, Ramos YF, Little N, et al. Hdmx stabilizes Mdm2 and p53. J Biol Chem. 2000;275(36):28039–44. doi: 10.1074/jbc.M003496200. [DOI] [PubMed] [Google Scholar]
- 23.Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29(1):92–5. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 24.Migliorini D, Denchi EL, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22(15):5527–38. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finch RA, Donoviel DB, Potter D, et al. Mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62(11):3221–5. [PubMed] [Google Scholar]
- 26.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A. 2006;103(9):3226–31. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Fernandez-Garcia B, Lozano G. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. doi: 10.1161/CIRCULATIONAHA.107.689901. Submitted. [DOI] [PubMed] [Google Scholar]
- 28.Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103(9):3232–7. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riemenschneider MJ, Buschges R, Wolter M, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59(24):6091–6. [PubMed] [Google Scholar]
- 30.Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61(5):1839–42. [PubMed] [Google Scholar]
- 31.Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24(13):5835–43. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans SC, Mims B, McMasters KM, et al. Exclusion of a p53 germline mutation in classic Li-Fraumeni syndrome family. Hum Genet. 1998;102:681–686. doi: 10.1007/s004390050761. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal S, Mathur M, Srivastava A, Ralhan R. MDM2/p53 co-expression in oral premalignant and malignant lesions: potential prognostic implications. Oral Oncol. 1999;35(2):209–16. doi: 10.1016/s1368-8375(98)00092-x. [DOI] [PubMed] [Google Scholar]
- 34.Yanamoto S, Kawasaki G, Yoshitomi I, Mizuno A. p53, mdm2, and p21 expression in oral squamous cell carcinomas: relationship with clinicopathologic factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94(5):593–600. doi: 10.1067/moe.2002.127404. [DOI] [PubMed] [Google Scholar]
- 35.Osman I, Sherman E, Singh B, et al. Alteration of p53 pathway in squamous cell carcinoma of the head and neck: impact on treatment outcome in patients treated with larynx preservation intent. J Clin Oncol. 2002;20(13):2980–7. doi: 10.1200/JCO.2002.06.161. [DOI] [PubMed] [Google Scholar]
- 36.Pimkhaokham A, Shimada Y, Fukuda Y, et al. Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J Cancer Res. 2000;91(11):1126–33. doi: 10.1111/j.1349-7006.2000.tb00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 38.Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.