Abstract
Neuronal nitric oxide (NO) synthase (nNOS) is dynamically regulated in response to a variety of physiologic and pathologic stimuli. Although the dynamic regulation of nNOS is well established, the molecular mechanisms by which such diverse stimuli regulate nNOS expression have not yet been identified. We describe experiments demonstrating that Ca2+ entry through voltage-sensitive Ca2+ channels regulates nNOS expression through alternate promoter usage in cortical neurons and that nNOS exon 2 contains the regulatory sequences that respond to Ca2+. Deletion and mutational analysis of the nNOS exon 2 promoter reveals two critical cAMP/Ca2+ response elements (CREs) that are immediately upstream of the transcription start site. CREB binds to the CREs within the nNOS gene. Mutation of the nNOS CREs as well as blockade of CREB function results in a dramatic loss of nNOS transcription. These findings suggest that nNOS is a Ca2+-regulated gene through the interactions of CREB on the CREs within the nNOS exon 2 promoter and that these interactions are likely to be centrally involved in the regulation of nNOS in response to neuronal injury and activity-dependent plasticity.
Nitric oxide (NO) is an important biological messenger that plays a prominent role in the physiology of the central nervous system. Three isoforms account for NO production and include neuronal NO synthase (nNOS; type I), inducible NO synthase (iNOS; type II), and endothelial NO synthase (eNOS; type III). In the nervous system, nNOS accounts for the majority of the physiologic actions of NO (1, 2). As a diffusible messenger molecule, NO is ideally suited to modulate and regulate synaptic function by acting as a spatial signal (3). Many investigations have shown that nNOS expression is dynamically regulated by both physiological and pathophysiological stimuli; however, the molecular mechanisms controlling the expression of nNOS in response to these stimuli are not known (1, 4–7).
The structure of the nNOS gene is extremely complicated. Its genomic structure in humans spans more than 240 kilobases, and its expression is potentially regulated by more than nine separate alternative first exons, which splice to a common exon 2 that contains a large 5′ untranslated region (UTR) before the start methionine (8). nNOS expression may be regulated at multiple levels, which could be relevant to a variety of physiologic functions of NO, ranging from a modulator of neuronal plasticity and behavior to a mediator of neuronal cell death (4, 9). To begin to understand how diverse stimuli regulate nNOS expression, we sought to identify the signaling pathways that mediate nNOS expression in neurons. In this study, using primary embryonic cortical neurons, we show that neuronal activity controls nNOS expression through influx of Ca2+ into neurons through L-type voltage-sensitive Ca2+ channels (VSCCs). Furthermore, we find that Ca2+ influx through L-type VSCCs stimulates transcription from the nNOS promoter contained within exon 2 by means of a CREB family transcription factor-dependent mechanism.
Methods
For methodological details, see supplemental materials at www.pnas.org.
Cell Culture, Transfection, and NOS Assays.
Cortical neurons were harvested from either rat or mouse embryos at the stage of embryonic day 16 (E16) and cultured by using standard procedures (10). After 5 days in vitro (DIV), cells were transfected by using a calcium phosphate precipitate method as described (11) with minor modifications. β-Galactosidase (β-gal) (CLONTECH) and luciferase (Promega) activity was measured in whole cell lysates by using chemiluminescence-based detection. NOS catalytic activity was assayed by monitoring the conversion of [3H]arginine to [3H]citrulline as described (12). Statistical significance was determined by ANOVA and the Student t test.
Immunoblotting, Northern Blotting, Reverse Transcription (RT)-PCR, and S1 Nuclease- and RNase-Protection Assays.
nNOS protein was detected with a monoclonal antibody that recognizes nNOS (Transduction Laboratories, Lexington, KY) and were performed using standard procedures (10). Total cellular RNA was isolated by using guanidinium isothiocyanate/phenol/chloroform (13). Ten micrograms of RNA from each treatment was subjected to Northern blot analysis following a standard protocol (14), using a 1.2-kb nNOS exon 2 probe (15). RT-PCR was performed as described (11), using nNOS exon 1a, 1b, 1c, and 2 5′-selective probes and a common 3′ exon 2 probe. Amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used as control. Calibration curves were prepared to obtain quantitative data from the RT-PCR assays. Using a cDNA probe specific for nNOS and nNOS exon 2 β-gal reporter mRNA, we carried out S1 nuclease assays according to protocols and reagents (S1 nuclease kit) from Ambion (Austin, TX). For RNase-protection assays, an exon 22-specific nNOS probe and a GAPDH probe were used that protected 127-bp and 316-bp fragments, respectively. RNase-protection assays were carried out according to protocols and reagents from Ambion (RPA II kit).
Electrophoretic Mobility-Shift Assays (EMSAs).
Cortical neurons were stimulated with 50 mM KCl in MEM at 37°C for 12 h at 5 DIV. EMSAs were carried out as described previously (16) by using a probe corresponding to the nNOS promoter exon 2 sequence (the probe is indicated in Fig. 4). Polyclonal anti-CREB was used as described (17) and anti-ATF-1 (SC-243), anti-ATF-2 (SC-242), and anti-c-Jun (SC822) were purchased from Santa Cruz Biotechnology and used in the supershift assays at a dilution of 1:20.
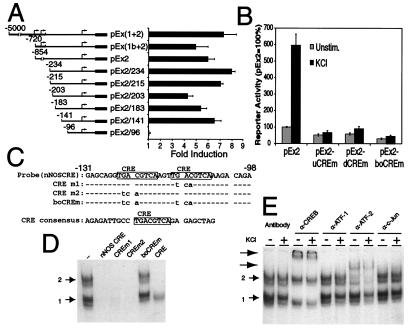
Figure 4.
Effect of nNOS promoter mutations on β-gal reporter gene expression. (A) Analysis of nNOS promoter activation by the β-gal assay. A series of reporter genes containing variant 5′-lengths of nNOS were constructed and fused to β-gal and were transiently transfected into cultured cortical neurons. In the bar graph, each value is the average of at least three to five independent determinations, and the error bars indicate the SEM. Arrows indicate the transcriptional initiation sites. The numbers indicate the position of the promoters 5′-end relative to the translation initiation ATG. The black boxes indicate the β-gal gene. (B) Disruption of the nNOS CRE sites abolishes nNOS promoter activation in cultured cortical neurons. Cultured cortical neurons were transfected with indicated nNOS-β-gal constructs and plasmid RSV-Luc. Two days after transfection, cells were left unstimulated or were stimulated with 50 mM KCl for 12 h. Each value is the average of at least three to five independent determinations, and the error bars indicate the SEM. (C) The nucleotide sequences of the nNOS CRE probe (nNOS-CRE) and the mutant probes (CREm1, CREm2, and boCREm) and a consensus CRE probe. (D) CREB and ATF-2 bind to the nNOS CREs. Binding of nuclear proteins to radiolabeled nNOS-CRE in an electrophoretic mobility-shift assay. Nuclear extracts were obtained from cultured cortical neurons that were incubated with the radiolabeled probe in the presence of competitors. Numbers 1 and 2 indicate complexes 1 and 2, respectively. This experiment was replicated two times with similar results. (E) CREB and ATF-2 bind to nNOS CRE. Nuclear extracts obtained from cultured cortical neurons that were unstimulated (−) or stimulated with 50 mM KCl for 12 h (+) were incubated with the radiolabeled probe in the presence of the indicated antibodies (α). Arrows indicate supershifted complexes. This experiment was replicated two times with similar results.
Adenoviral Vector-Mediated Gene Transfer of Cultured Neurons.
Recombinant inducible adenovirus expressing A-CREB was generated by using a Cre-recombinase/LoxP system (18, 19). The shuttle plasmid pAdEGI-A-CREB was made by insertion of the A-CREB cDNA into pAdEGI [containing the ecdysone promoter, green fluorescent protein (GFP), IRES, and a multiple cloning site]. Two-day-old immature neurons in mixed rat primary cortical cultures were coinfected with either AdEGI control/AdVgRXR or AdEGI-A-CREB/AdVgRXR by exposure to 1 × 109 plaque-forming units of adenovirus vectors per ml in 0.3 ml of serum-free medium for 1 h at 37°C. Virus-containing solution was removed from the cells and complete medium containing 1 mM muristerone was returned for 24-h incubation to allow transgene expression. GFP expression was used to monitor infection, and nNOS expression was detected by using immunohistochemistry as described (10).
Results
Membrane Depolarization Induces nNOS Gene Expression.
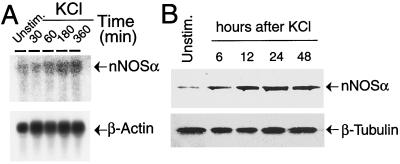
To characterize the mechanisms that link neuronal activity with nNOS gene expression, we determined whether membrane depolarization regulates nNOS expression. Accordingly, we depolarized E15 cortical cultures at 5 days in vitro (E15 + 5 DIV) with 50 mM KCl to activate VSCCs. A clear increase in nNOS message is detected within 30 min of membrane depolarization (Fig. 1A). nNOS message peaks at approximately 180 min of the initial stimulus and remains elevated for at least 6 h after membrane depolarization (Fig. 1A). Depolarization of cortical neurons also dramatically induces the expression of nNOS protein as determined by Western blot analysis (Fig. 1B). Accompanying the increase in message and protein is an increase in Ca2+-dependent NOS catalytic activity (Fig. 6 in supplemental material at www.pnas.org).
Figure 1.
Membrane depolarization induces nNOS expression. (A) Northern blot analysis of nNOS. E15–16 cortical cultures were depolarized with 50 mM KCl. A probe to β-actin was used indicate that an equivalent amount of RNA was loaded in each lane. This experiment was replicated three times with similar results. (B) Western blot analysis of nNOS protein. nNOS isoform α (nNOSα) was detected with an antibody to C-terminal nNOS. β-Tubulin was used to indicate that an equivalent amount of protein was loaded in each lane. This experiment was replicated three times with similar results.
nNOS Exon 2 Is the Major Inducible Transcript in Cortical Neurons.
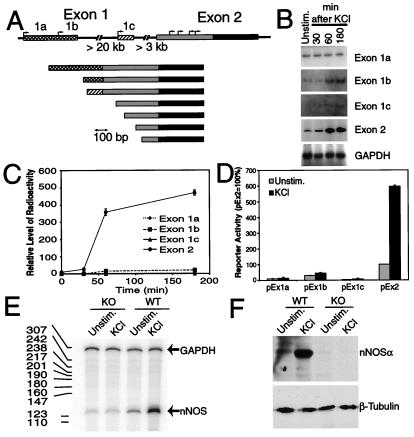
Given the finding that membrane depolarization induces nNOS message and protein, experiments were carried out to determine which of the nNOS exons is capable of mediating a depolarization-induced response. We sought to characterize the transcript(s) that are regulated by membrane depolarization by first identifying neuronally enriched transcripts by using the method of rapid amplification of 5′-cDNA ends (5′-RACE) from mRNA isolated from adult mouse cortex and cerebellum and embryonic (E17) cortex. Primers were located in the exon 2 coding region, thus allowing the isolation of mouse nNOS cDNA sequences upstream of exon 2. A total of 12 5′-RACE clones were analyzed (Fig. 2A). 5′-RACE reveals the presence of at least three alternative first exons, which we have designated exon 1a, 1b, and 1c, which splice to a common exon 2 containing a large 5′-UTR region (GenBank accession nos. AF135825–AF135827) (Fig. 2A). Mouse exon 1a and 1b correspond to human exons 1f and 1g, respectively, with the exception that human exons are separated by a short intron (8). These exons correspond to the human exons that are enriched in the neuronal cluster (8). Mouse exon 1c is unique. To investigate whether sequences contained within the 5′-UTR of rodent nNOS exons 1a, 1b, 1c, and 2 are responsive to membrane depolarization, a quantitative RT-PCR assay was developed to measure the level of each nNOS mRNA. The GAPDH gene, which is unresponsive to membrane depolarization, was used as a control for variations in sample handling (11). The exon 2 transcript is most responsive to membrane depolarization, with a time course of responsiveness similar to the nNOS message (Fig. 2 B and C). Exon 1b is modestly responsive to membrane depolarization (Fig. 2B), whereas exons 1a and 1c fail to respond to a significant degree (Fig. 2 B and C).
Figure 2.
Expression of alternative nNOS transcripts. (A) Schematic representation of the nNOS gene and mature transcripts. 5′-RACE clones isolated from adult mouse cortex and cerebellum and embryonic (E17) cortex. The 5′-UTRs encoded by the 5′ exons are indicated by cross-hatched, hatched, and shaded boxes. The putative initiation start sites determined by 5′-RACE are indicated with arrows. (B) RT-PCR analysis of nNOS transcripts after membrane depolarization. PCR products were visualized by PhosphorImager analysis. (C) Quantification of each exon-specific product. The levels of radioactivity were obtained from the image and normalized against the GAPDH level of the same sample. Each value is the average from two independent determinations. (D) Analysis of nNOS exon activation by reporter analysis. A series of reporter plasmids containing exon 1a, exon 1b, and exon 1c (pEx1a, pEx1b, and pEx1c) as well as exon 2 alone (pEx2) fused to β-gal were transfected into cortical cultures, and 2 days later the cultures were stimulated with 50 mM KCl for 12 h. Plasmid RSV-Luc was cotransfected to monitor the transfection efficiency. Each value is the average of three to five independent determinations, and the error bars indicate the SEM. (E) The response of nNOS mRNA in wild-type mice (WT) versus nNOS−/− mice (knock-out, KO), which lack exon 2, was examined by RNase-protection assay using an exon 22 probe, which is common to all known nNOS transcripts. A probe to GAPDH was used to control for RNA input. The sizes (bp) of the RNA ladder are indicated by the numbers in the left margin. This experiment was replicated three times with similar results. (F) The response of nNOS protein in wild-type mice (WT) versus nNOS−/− mice (KO) measured with an antibody to C-terminal nNOS. The blot was stripped and probed with an antibody to β-tubulin to indicate that an equivalent amount of protein was loaded in each lane. This experiment was replicated three times with similar results.
To confirm that depolarization induces the expression of nNOS through sequences contained within the 5′-UTR of exon 2, we constructed a series of reporter plasmids containing exon 1a (pEx1a), exon 1b (pEx1b), and exon 1c (pEx1c) without the downstream exon 2 sequences, as well as exon 2 (pEx2) alone fused to the β-gal reporter gene. Each reporter construct was transfected into embryonic cortical cultures by using a modified calcium phosphate transfection procedure (20). Two days later the cultures were depolarized and the neurons were harvested to assess reporter activity. Consistent with the RT-PCR analysis, exons 1a and 1c have minimal reporter activity in response to membrane depolarization (Fig. 2D). Exon 1b is modestly stimulated by membrane depolarization and exon 2 is stimulated almost 6-fold in response to membrane depolarization. To verify that the sequences contained within exon 2 regulate nNOS mRNA in response to membrane depolarization, we examined the response of both nNOS mRNA and protein in wild-type mice compared with nNOS−/− mice, which lack exon 2 (15) (Fig. 2 E and F). For purposes of mRNA detection we used an RNase-protection assay with an exon 22 probe that is common to all known nNOS transcripts, and for protein detection we used an antibody to the C terminus that recognizes all known nNOS isoforms in both wild-type and nNOS−/− mice. Using the exon 22 nNOS probe, we detected robust elevations in nNOS mRNA in response to membrane depolarization in wild-type cultures, but failed to detect increases in nNOS message in cultures from nNOS−/− mice (Fig. 2E). In a similar manner, we observed a robust increase in nNOS protein in wild-type cultures, but failed to detect an increase in nNOS−/− cultures (Fig. 2F).
nNOS mRNA Is Regulated by Ca2+ Influx Through L-Type VSCCs Through a cAMP/Ca2+ Response Element (CRE)-Like Sequence in the nNOS Exon 2 Promoter.
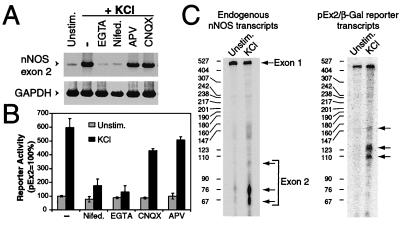
Having obtained evidence that sequences contained within exon 2 control the induction of nNOS mRNA, we sought to determine the potential mechanism by which the expression of nNOS mRNA is controlled by membrane depolarization. PCR analysis of exon 2 mRNA in response to membrane depolarization revealed that nNOS mRNA expression is regulated by Ca2+ influx through L-type VSCCs, as the extracellular chelator of Ca2+, EGTA (2 mM), and the Ca2+ channel blocker nifedipine (100 μM) markedly reduce depolarization-induced nNOS mRNA expression (Fig. 3A). Secondary activation of N-methyl-d-aspartate (NMDA)-glutamate receptor or α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-glutamate receptor was excluded by the failure of the selective NMDA receptor blocker 2-amino-5-phosphonovaleric acid (APV; 25 μM) and the selective AMPA antagonist 6-cyano-7-nitroquinoxaline-2,3-dione(CNQX; 25 μM) to attenuate the induction of nNOS mRNA (Fig. 3A). Cycloheximide fails to superinduce or block depolarization-induced nNOS mRNA expression, thus nNOS transcription does not require new protein synthesis (data not shown). To determine whether the induction of the nNOS exon 2-driven β-gal gene expression by membrane depolarization requires Ca2+ influx through L-type VSCCs, we tested the reporter's response when Ca2+ influx was blocked. Similar to the blockade of nNOS mRNA by nifedipine and EGTA, nNOS exon 2 reporter activity is also attenuated (Fig. 3B). APV and CNQX also fail to influence nNOS exon 2-driven β-gal activity in response to membrane depolarization (Fig. 3B). To verify that the functional depolarization/Ca2+-responsive element resides within the exon 2 promoter, we used an S1 nuclease-protection assay to assess whether transcription is initiated within exon 2 in response to membrane depolarization. For the S1 nuclease-protection assay, two separate antisense single-stranded DNA probes were synthesized that are complementary to the exon 2 mRNA and the β-gal reporter mRNA and are capable of recognizing transcripts synthesized from either the endogenous nNOS gene or the transfected nNOS-β-gal reporter gene, respectively (Fig. 3C). The antisense exon 2 DNA probe protects two major fragments of approximately 60–76 nucleotides (located at −59 to −75 bp 5′ of the ATG translation start site of nNOSα) that are clearly induced within 6 h after membrane depolarization, and it also protects several larger transcripts, between 100 and 300 nucleotides, that are also modestly induced by membrane depolarization (Fig. 3C). A large fragment of approximately 500 nucleotides is also protected in both unstimulated and membrane-depolarized cultures (Fig. 3C). This protected fragment most likely represents exon 1 spliced to exon 2. Thus, exon 1 fails to respond to membrane depolarization, consistent with the exon 1a, 1b, and 1c reporter assays (Fig. 3C and see Fig. 2D). On the basis of the design of the antisense single-stranded DNA probes, the probe that recognizes the β-gal reporter mRNA should protect a fragment that is 50 nucleotides longer then the probe complementary to the exon 2 mRNA. When the exon 2 nNOS-β-gal reporter gene is transfected into cortical neurons, membrane depolarization of neurons leads to a robust increase in correctly initiated nNOS-β-gal transcripts of approximately 110 and 126 nucleotides as well as a minor fragment of 150 nucleotides (Fig. 3C). A larger fragment of approximately 425 nucleotides that fails to respond to membrane depolarization is also protected; it corresponds to the large 5′-UTR of exon 2 up to the exon splice acceptor site that is contained within the exon 2 nNOS-β-gal reporter construct. Taken together, these results indicate that the essential regulatory elements that are critical for nNOS transcription in response to membrane depolarization are contained within the exon 2 DNA sequence elements just 5′ of the initiation sites of nNOS mRNA synthesis.
Figure 3.
nNOS mRNA is regulated by Ca2+ influx through L-type VSCCs. (A) Blocking Ca2+ influx inhibits nNOS exon 2 mRNA expression. RT-PCR analysis of nNOS exon 2 mRNA after membrane depolarization was carried out in the presence or absence of EGTA (2 mM), nifedipine (100 μM), 2-amino-5-phosphonovaleric acid (APV; 25 μM), or 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; 25 μM). GAPDH was used as a control for RNA input and reverse transcription efficiency. PCR products were visualized by PhosphorImager analysis. This experiment was replicated two times with similar results. (B) Preventing Ca2+ influx blocks nNOS exon 2 reporter activity. Cortical neurons were transfected with the nNOS exon 2 β-gal reporter construct (pEx2). Before membrane depolarization, cells were left untreated (−) or treated with same drugs as in A. Each value is the average of three to five independent determinations, and the error bars indicate the SEM. (C) Mapping of transcription initiation sites. Cortical neurons were either transfected with the pEx2 construct or left untreated. Two days later the cells were treated with 50 mM KCl (6 h) or were unstimulated, and total RNA was isolated. S1 nuclease-protection assays were performed with probes selective for nNOS exon 2 and pEx2-β-Gal. Transcription start sites are indicated by arrows. The sizes (bp) of the RNA ladder are indicated by the numbers in the left margin. This experiment was replicated three times with similar results.
Because Ca2+ influx triggers the induction of nNOS mRNA through the 5′-UTR contained in exon 2, we sought to identify the cis-acting DNA sequences that direct Ca2+-dependent nNOS transcription. Toward this end, a series of reporter plasmids containing various lengths of nNOS exon 2 5′-flanking region were fused to the β-gal reporter gene (see Fig. 4A). Because the S1 nuclease assays indicated that nNOS transcripts containing exon 2 also contain exon 1, we examined the effects of exon 1 on exon 2's response to Ca2+ by fusing exon 1 at the appropriate splice-acceptor site to exon 2. After 2 days, transfected cortical neurons were membrane depolarized by exposure to KCl. Exon 2 (pEx2) containing the entire 5′-UTR and exon 2 fused with exon 1 [pEx(1+2) and pEx(1b+2)] have a significant Ca2+ response, which is reproducibly induced 5- to 7-fold in response to membrane depolarization (Fig. 4A). Deletion of exon 2 sequences between −854 and −234 5′ of the nNOS exon 2 start ATG and deletion of sequences between −234 to −141 have minimal effects on the Ca2+ inducibility of the β-gal reporter gene. However, deletion of −141 to −96 completely eliminates the Ca2+ inducibility of the β-gal reporter gene, indicating that this region contains the key Ca2+-responsive regulatory element.
Since the deletion analysis suggested that the key regulatory sequence(s) for Ca2+ inducibility of nNOS were between −141 and −96, we compared this DNA sequence with other promoter sequences that are known to contain Ca2+-response elements. Analysis of the mouse nNOS genomic sequence reveals the presence of two consensus CREs, 5′-TGACGTCA-3′ (21) located at −122 to −115 and −111 to −104 bp 5′ of the ATG translation start site (5′-−122TGACGTCAAGTTGACGTCA−104-3′, with the consensus sites in boldface; Fig. 4C), which are upstream of the major initiation starts sites determined by S1-nuclease mapping. Both CRE sites are completely conserved in human, rat, and mouse nNOS exon 2. CRE-binding sites regulate both Ca2+ and cAMP responsiveness of a variety of genes through the CREB transcription factor family (21, 22).
To determine the importance of the CRE sites within the −122 to −104 region of the nNOS exon 2 5′-UTR, in context substitution mutants of the entire exon 2 promoter were generated that were expected to disrupt transactivating factor binding to the nNOS promoter (Fig. 4B). Mutation of the first, second, or both CRE sites completely eliminated the Ca2+ inducibility of the β-gal reporter gene (Fig. 4B). These same in-context mutations were made within the promoter construct containing both exon 1 and 2 [pEx(1+2)/boCREm] and the Ca2+ inducibility of the β-gal reporter gene is also eliminated (data not shown).
A CREB/Family Member Interacts with nNOS CRE Sites and Mediates nNOS Induction.
We next investigated the nature of factors present in nuclear extracts that interact with the downstream CREs in nNOS exon 2. To identify the factors present in nuclear extracts obtained from cortical neurons, electrophoretic mobility-shift assays were performed with a 34-bp fragment of 32P-labeled DNA corresponding to the nNOS promoter sequence that includes the two CRE sites (Fig. 4C). When the DNA probe was incubated with nuclear extracts prepared from unstimulated neurons, two major and two minor complexes were identified (Fig. 4D). The binding of proteins to the two CRE sites is specific, inasmuch as the formation of the major complexes was completely inhibited by the addition of excess unlabeled sequence, but not by an excess of unlabeled sequence in which both CRE elements were mutated (Fig. 4D). Addition of excess of unlabeled sequence containing a single mutation of either CRE site inhibited the formation of the major protein/DNA complexes. A consensus CRE site oligonucleotide disrupted both major complexes and one minor complex was retained, suggesting that within these protein/DNA complexes a protein unrelated to the CREB transcription factor family may bind to these regulatory sequences.
Since the CRE site is a common recognition site of the CREB/ATF transcription factor family, supershift assays were carried out to determine which member(s) of the family is a component of the protein complex that binds to the CREs. In this assay, specific polyclonal antibodies to CREB and monoclonal antibodies to ATF-1, ATF-2, and c-Jun were added to the protein/DNA binding reaction mixture, and the effect on complex formation from nuclear extracts prepared from KCl-stimulated or unstimulated neurons was assessed (Fig. 4E). We found that in the presence of the anti-CREB antibody the protein/DNA complex 1 was disrupted and that a supershifted band was formed. The DNA protein complex 2 was disrupted in the presence of anti-ATF-2 antibody, producing a supershifted band. In contrast, anti-ATF-1 and anti-c-Jun antibodies had no effect on the protein/DNA association. Identical results were obtained when the consensus CRE oligonucleotide was used as a probe (data not shown). Thus, the two CRE sites and flanking sequences contained within the nNOS exon 2 probe do not confer additional protein/DNA interactions. There was also no reproducible change in the protein/DNA complexes when extracts obtained from unstimulated and membrane-depolarized cells were compared (Fig. 4E). Thus, the protein/DNA complexes are already present in unstimulated neurons, and membrane depolarization does not change their DNA-binding activity. In addition, forebrain nuclear extracts from CREB−/− and ATF-2−/− mutant mice lack DNA protein complex 1 and 2, respectively (S.A. and D.D.G., unpublished observations). Taken together, these results suggest that CREB and ATF-2 are components of CRE/protein complex 1 and 2, respectively, and that CREB/ATF-2 or a related protein may be a critical mediator of the membrane depolarization-induced transcription of nNOS promoter exon 2.
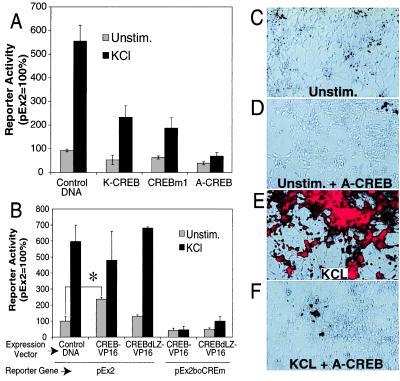
To further investigate the importance of a CREB family member as a mediator of Ca2+-induced nNOS transcription, several CREB mutants were introduced into cortical neurons, and their effects on nNOS exon 2-driven β-gal expression were assessed. Expression of three functionally different CREB dominant-negatives, K-CREB (23), CREBm1 (24), and A-CREB (16), reduce membrane depolarization-induced nNOS exon 2 β-gal reporter gene activity (Fig. 5A). It is important to mention that DNA-binding activity of CREB and its closely related family members is potently and selectively inhibited by A-CREB, whereas the DNA-binding activity of other B-ZIP transcription factors, including ATF-2, are not altered, even at very high concentrations of A-CREB (16). Thus, ATF-2 is not sufficient to mediate nNOS promoter activation in the absence of CREB. A constitutively active CREB (CREB-VP16) (11) induces nNOS-β-gal reporter gene activity even in the absence of membrane depolarization (Fig. 5B). Deletion of the DNA-binding domain of CREB (CREB-dLZ-VP16) abolishes the constitutive activation of the reporter gene, but has minimal effects on nNOS exon 2 β-gal reporter gene activity induced by membrane depolarization. Thus, the induction of nNOS exon 2 β-gal reporter gene activity by CREB-VP16 is due to the specific interaction between CREB and the CRE. An intact CRE is required for nNOS-promoter induction, since an nNOS exon 2 β-gal reporter gene containing mutations of both CRE sites (pEx2-boCREm) is not responsive to CREB-VP16 (Fig. 5B). The finding that a constitutively active CREB-VP16 triggers Ca2+-independent nNOS exon 2 reporter-dependent transcription as well as the findings that both CRE mutations and dominant inhibitory forms of CREB block membrane depolarization induced nNOS exon 2 reporter dependent transcription strongly suggests that CREB or a closely related family member binds to the nNOS CRE elements and mediates Ca2+-dependent nNOS transcription.
Figure 5.
A CREB family member mediates nNOS transcription after membrane depolarization. (A) Overexpression of dominant-negative K-CREB, CREBm1, or A-CREB inhibits promoter activation. K-CREB, CREBm1, or A-CREB expression and control vectors were cotransfected with pEx2 into cultured cortical neurons. Two days after transfection, cells were left unstimulated or stimulated with 50 mM KCl for 12 h. Cell extracts were then assayed for β-gal and luciferase activities. Each value is the average of at least three to five independent determinations, and the error bars indicate the SEM. (B) A constitutively active CREB triggers Ca2+-independent promoter activation. Cultured cortical neurons were transfected with the indicated constructs, and β-gal and luciferase activities were assayed. CREB-VP16 contains full-length CREB and the transcriptional activation domain of VP16, a potent herpes virus transcriptional activator. CREBdLZ-VP16 is similar to CREB-VP16 except that the DNA-binding domain and dimerization domain of CREB are deleted. Each value is the average of at least three to five independent determinations, and the error bars indicate the SEM. ∗, P ≤ 0.002. (C–F) Dominant-negative A-CREB attenuates KCl-induced expression of the endogenous nNOS. Cultured cortical neurons were coinfected with AdVgRXR and AdEGI-control or AdEGI-A-CREB viruses and treated with muristerone to induce transgene expression. Twenty-four hours later cortical neurons were left unstimulated or were stimulated with 50 mM KCl. nNOS expression was monitored by immunofluorescence with merged Hoffman modulation/immunofluorescence photomicrographs. Unstimulated neurons infected with control viruses (AdVgRXR and AdEGI-control) show a modest amount of nNOS immunostaining in the absence of stimulation (C), but in response to membrane depolarization show striking nNOS immunoreactivity (D). In contrast, neurons infected with viruses containing A-CREB and VgRXR show minimal nNOS immunoreactivity in unstimulated cultures (E) and minimal response to KCl stimulation (F). These experiments were repeated in four different cultures in replicates of three wells per condition. Infection efficiency as monitored by GFP immunofluorescence varied between 50% and 95%. Shown are representative fields in which expression of GFP was >90%. (20× objective.)
To ascertain whether CREB is a critical regulator of the response of the endogenous nNOS gene to membrane depolarization, we constructed an ecdysone-inducible adenoviral vector containing A-CREB. Cortical neurons were infected with the inducible A-CREB adenovirus, and expression of A-CREB was induced by the ecdysone analog muristerone. The effect of A-CREB expression on endogenous nNOS transcription was then determined by immunohistochemistry (Fig. 5 C–F). A virus expressing GFP was used to control for nonspecific effects due to viral infection. In neurons infected with the GFP adenovirus, membrane depolarization elicited a robust induction of nNOS protein in the majority of neurons that was comparable to that in noninfected neurons (Fig. 5C). In neurons infected with A-CREB, unstimulated neurons expressed less nNOS protein, and membrane depolarization-induced nNOS expression was significantly reduced in neurons that contained A-CREB (Fig. 5 E and F).
Discussion
One of the major findings of this study is our observation that nNOSα is a Ca2+-regulated gene. In response to membrane depolarization, nNOS message and protein are induced severalfold by means of the activation of L-type VSCCs. We have explored the molecular mechanisms by which Ca2+ influx via VSCCs regulates the expression of nNOS in cortical neurons. We have focused on the analysis of the promoter contained within exon 2, as this is the major activity-regulated exon of the nNOS gene. Two of the elements identified in this study are the two CRE sites located approximately 38 nucleotides 5′ of the first initiation site of nNOS exon 2 transcription. Our discovery that nNOS expression is regulated by membrane depolarization through VSCCs and the subsequent interaction of CREB and/or CREB-related family members with the CRE sites of the nNOS exon 2 promoter sheds considerable light on the mechanisms underlying the inducibility of nNOS and how nNOS might contribute to the regulation of diverse pathologic and physiologic processes.
Originally, nNOS was thought to be a constitutively expressed enzyme (25). However, numerous investigations suggest that nNOS is inducible in response to a variety of physiologic and pathologic stimuli (1, 4–6). The importance of understanding the molecular mechanisms that regulate nNOS expression is illustrated by the diversity of processes, that are thought to be regulated by NO, including neuronal injury, plasticity, and gene regulation (1, 4–6). Many of these processes are Ca2+-dependent, and because nNOS catalytic activity is also Ca2+-dependent, there is a unique potential for both simultaneous transcriptional and catalytic activity-mediated regulation of nNOS/NO. This may be important in both pathological and physiological events. For instance, NO is thought to play a major role in Ca2+-dependent excitotoxic neuronal injury (26), and nNOS message and protein are dramatically induced in these same neuronal injury paradigms (1, 4–6). In a similar manner, several Ca2+-dependent neuronal plasticity models are thought to be regulated in part by NO, and nNOS expression is dynamically regulated in many of these models (1, 4–6). Given the evidence that nNOS-derived NO may be involved in synaptic strengthening as well as its importance in neuronal injury and development, it will be useful to characterize further the mechanisms by which Ca2+ regulates CREB and/or CREB family members activate nNOS transcription. Because CRE/CREB signaling is also regulated by cAMP, it is likely that there maybe a convergence of cAMP and Ca2+ in the regulation of nNOS expression.
Regulation of nNOS transcription may be controlled in a complex fashion by multiple promoters (27). At least nine alternative first exons have been identified in the human gene, and the alternative splicing of exon 1 to a common exon 2 yields an identical protein (8). Other alternative transcripts also exist because of alternative splicing downstream of exon 2 (6, 9, 28, 29). In both rats and mice, several alternative first exons and splice variants have been identified (9, 29, 30), although the complexity identified in human nNOS has not been revealed in rodent nNOS. This diversity of alternative promoters and different nNOS transcripts probably helps to control the diversity of nNOS expression in many different organs and also regulates its expression during activity-dependent synaptic plasticity and development and in response to injury (6, 9, 27). The alternative promoter usage probably also accounts for the high expression of nNOS in the embryonic (E15–E19) cortical plate neurons and dorsal root ganglia cells and nNOS's restricted distribution to approximately 1–2% of neurons in the adult brain (31, 32). We show here that the sequences between −141 and −96 within the 5′-UTR of mouse nNOS exon 2 contains binding elements that regulate the response of nNOS to membrane depolarization. The importance of exon 2 in regulating nNOS levels is supported by observations in mice in which exon 2 was deleted by homologous recombination (15). Deletion of exon 2 leads to a >95% reduction in nNOS catalytic activity (15) and the loss of inducibility in response to membrane depolarization (see Fig. 3 B and C). The unusual location of a promoter within the 5′-UTR of nNOS exon 2 and alternative promoters provides a unique complexity for regulation of nNOS expression, which probably involves both transcriptional and translation control (8).
Supplementary Material
Acknowledgments
We are grateful to Drs. M. E. Greenberg (CREB-VP16 and CREBdLZ-VP16) and P. L. Huang (pBam9) for plasmids and Dr. David Johns for adenoviral constructs used in this study and for Hoke Morita's assistance with construction of the adenoviral vectors. This research was supported by grants from the National Institutes of Health, the American Heart Association, the Muscular Dystrophy Association, and the Boehringer Ingelheim Fonds (Stuttgart, Germany).
Abbreviations
- nNOS
neuronal NO synthase
- UTR
untranslated region
- VSCCs
voltage-sensitive Ca2+ channels
- En
embryonic day n
- DIV
days in vitro
- β-gal
β-galactosidase
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- 5′-RACE
rapid amplification of 5′-cDNA ends
- CRE
cAMP/Ca2+ response element
Footnotes
References
- 1.Garthwaite J, Boulton C L. Annu Rev Physiol. 1995;57:683–706. doi: 10.1146/annurev.ph.57.030195.003343. [DOI] [PubMed] [Google Scholar]
- 2.Yun H Y, Dawson V L, Dawson T M. Crit Rev Neurobiol. 1996;10:291–316. doi: 10.1615/critrevneurobiol.v10.i3-4.20. [DOI] [PubMed] [Google Scholar]
- 3.Schuman E M. Semin Cell Biol. 1994;5:251–261. doi: 10.1006/scel.1994.1031. [DOI] [PubMed] [Google Scholar]
- 4.Forstermann U, Boissel J P, Kleinert H. FASEB J. 1998;12:773–790. [PubMed] [Google Scholar]
- 5.Dawson T M, Gonzalez-Zulueta M, Kusel J, Dawson V L. Neuroscientist. 1998;4:96–12. [Google Scholar]
- 6.Boissel J P, Schwarz P M, Forstermann U. Nitric Oxide. 1998;2:337–349. doi: 10.1006/niox.1998.0189. [DOI] [PubMed] [Google Scholar]
- 7.Dawson T M, Snyder S H. J Neurosci. 1994;14:5147–5159. doi: 10.1523/JNEUROSCI.14-09-05147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Newton D C, Robb G B, Kau C L, Miller T L, Cheung A H, Hall A V, VanDamme S, Wilcox J N, Marsden P A. Proc Natl Acad Sci USA. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson T M, Sasaki M, Gonzalez-Zulueta M, Dawson V L. Prog Brain Res. 1998;118:3–11. doi: 10.1016/s0079-6123(08)63196-9. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Zulueta M, Ensz L M, Mukhina G, Lebovitz R M, Zwacka R M, Engelhardt J F, Oberley L W, Dawson V L, Dawson T M. J Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao X, Finkbeiner S, Arnold D B, Shaywitz A J, Greenberg M E. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 12.Bredt D S, Snyder S H. Proc Natl Acad Sci USA. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Huang P L, Dawson T M, Bredt D S, Snyder S H, Fishman M C. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- 16.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty D D, Vinson C. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 18.Johns D C, Marx R, Mains R E, O'Rourke B, Marban E. J Neurosci. 1999;19:1691–1697. doi: 10.1523/JNEUROSCI.19-05-01691.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Z, Dudek H, Miranti C K, Greenberg M E. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montminy M R, Sevarino K A, Wagner J A, Mandel G, Goodman R H. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 23.Walton K M, Rehfuss R P, Chrivia J C, Lochner J E, Goodman R H. Mol Endocrinol. 1992;6:647–655. doi: 10.1210/mend.6.4.1350057. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 25.Bredt D S, Snyder S H. Neuron. 1992;8:3–11. doi: 10.1016/0896-6273(92)90104-l. [DOI] [PubMed] [Google Scholar]
- 26.Samdani A F, Dawson T M, Dawson V L. Stroke. 1997;28:1283–1288. doi: 10.1161/01.str.28.6.1283. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Newton D C, Marsden P A. Crit Rev Neurobiol. 1999;13:21–43. doi: 10.1615/critrevneurobiol.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- 28.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, et al. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 29.Brenman J E, Xia H, Chao D S, Black S M, Bredt D S. Dev Neurosci. 1997;19:224–231. doi: 10.1159/000111211. [DOI] [PubMed] [Google Scholar]
- 30.Lee M A, Cai L, Hubner N, Lee Y A, Lindpaintner K. J Clin Invest. 1997;100:1507–1512. doi: 10.1172/JCI119673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bredt D S, Glatt C E, Hwang P M, Fotuhi M, Dawson T M, Snyder S H. Neuron. 1991;7:615–624. doi: 10.1016/0896-6273(91)90374-9. [DOI] [PubMed] [Google Scholar]
- 32.Bredt D S, Snyder S H. Neuron. 1994;13:301–313. doi: 10.1016/0896-6273(94)90348-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.