Abstract
As drug delivery, therapy, and medical imaging are becoming increasingly cell-specific, there is a critical need for high fidelity and high-throughput screening methods for cell surface interactions. Cell membrane-mimicking surfaces, i.e., supported lipid bilayers (SLBs), are currently not sufficiently robust to meet this need. Here we describe a method of forming fluidic and air-stable SLBs through tethered and dispersed cholesterol groups incorporated into the bottom leaflet. Achieving air stability allows us to easily fabricate SLB microarrays from direct robotic spotting of vesicle solutions. We demonstrate their application as cell membrane-mimicking microarrays by reconstituting peripheral as well as integral membrane components that can be recognized by their respective targets. These demonstrations establish the viability of the fluidic and air-stable SLB platform for generating content microarrays in high throughput studies, e.g., the screening of drugs and nanomedicine targeting cell surface receptors.
Introduction
The majority of drugs under development target cell surfaces. Examples include molecules that bind G-protein coupled receptors1 and ion channels,2 the bivalent monoclonal antibody (mAb)3 or engineered multivalent mAb fragments4 that recognize cell surface antigens, and other small molecules5 or nanoparticles6 that attack disease cells based on multivalent recognition. In view of this cell-centered drug development, there is a growing need of high throughput analytical techniques incorporating the fluidic cell membrane environment. Fluidic supported lipid bilayers (SLBs) constitute a benchmark in studying cell surface processes, providing an analytical platform which mimicks the cell membrane environment.7–10 However, due to its instability under a variety of sample processing or handling conditions, particularly exposure to air, developing the SLB into a practical and high-throughput technique has been extremely difficult. In this paper, we report a novel method of fabricating air-stable and fluidic SLBs based on the unique properties of cholesterol. Cholesterol, a crucial and naturally occurring component of all mammalian plasma membranes, is recognized for its biochemical and biophysical importance. The presence of cholesterol increases the stability and rigidity of lipsomes by increasing their area-expansion modulus and bending energies.11 The rigid and flat cholesterol molecule imposes conformational ordering locally and increases the packing density of lipid molecules in the immediate surrounding.12 In nature, cholesterol molecules in the cell membrane are known to phase separate into cholesterol-rich and cholesterol-deficient domains.12 To fully take advantage of the stabilizing effect, we disperse and immobilize cholesteryl by covalent linking to a hydrophilic polymer [poly(ethyleneglycol) (PEG)] brush. This allows a uniform interaction of cholesteryl groups with the entire bottom leaflet of an SLB, resulting in the air stability of the SLB while maintaining the fluidity of the lipid membrane environment. The fluidic and air-stable SLB is not only a robust model for biophysical studies but also an efficient cell-mimicking platform for high-throughput analysis.
Experimental Methods
Materials
Egg phophatidylcholine (EggPC), 1,2-dioleoyl-3-trimethyl ammonium propane (DOTAP), and ganglioside-GM1 were from Avanti Polar Lipids (Alabaster, AL). Texas Red tagged dihexadecanoyl-phosphatidylethanolamine (TR-DHPE) was purchased from Invitrogen (Carlsbad, CA). The negatively charged PEG brush coated glass coverslips were from MicroSurfaces, Inc. (Minneapolis, MN). Primary antibodies against PLN (L-15) and pS16-PLN (Ser16) were from Santa Cruz Biotechnology (Santa Cruz, CA). FTIC tagged secondary antibodies [AffiniPure Donkey Anti-Goat IgG(H+L)] were from Jackson ImmunoResearch Laboratories (West Grove, PA). All other chemical reagents were from Sigma-Aldrich (St. Louis, MO). Phospholamban (PLN) monomer (C36A, C41F, C46A construct) and pentamer (wild-type) were expressed and purified as previously described. 13 Phosphorylation at S16 (pS16-PLN) was carried out on monomeric PLN as described previously using the catalytic subunit of protein kinase A (Sigma-Aldrich, St. Louis, MO).14 Unlabeled cholera toxin B (CTB) and FTIC dye-tagged CTB were both purchased from Sigma-Aldrich (St. Louis, MO).
SUV
Preparation of SUV solution was carried out via the extrusion method of Avanti Polar Lipids. Briefly, mixed lipids were evaporated under an argon flow until dry and the lipid mixtures were reconstituted in Tris buffer (50 mM Tris-hydroxymethyl-aminomethane in 100 mM NaCl, pH 7.5) to give a total lipid concentration of 1 mM. A suspension of the lipid mixtures after prefiltration through 0.45 μm pores was forced through a polycarbonate filter with 50 nm pores more than 11 times. This SUV solution was stored at 4 °C until use. To incorporate GM1, we added 2% GM1 (in methanol) to the mixed lipid solution before the drying step. The reconstitution of proteins, monomeric and pentameric phospholamban (PLN), into SUVs has been described elsewhere.15,16 Briefly, PLN was first dissolved into a 10% SDS solution to give a final protein concentration of ~1 mg/ml. The protein/detergent mixture was then added to the SUV solution (~7 mg/ml) to give a final protein concentration of ~0.2 mg/mL, a lipid/protein (monomer) molar ratio of ~100:1, and a total lipid concentration of ~3.5 mg/mL. The mixture was then subjected to 2–3 days of dialysis using a 10 kDa molecular weight cutoff membrane, until all SDS was removed.
Cholesteryl-PEG Surface
To covalently attach cholesteryl groups, we incubated the PEG brush surface with 30 mM cholesteryl chloroformate in DMF/CH2Cl2 (1:1) in the presence of a catalytic amount of pyridine at room temperature. The reaction time was varied between 0.5 and 5 h to give a different surface cholesteryl density, which was quantitatively determined for each surface by X-ray photoelectron spectroscopy (Phi 540) based on the C1s peak areas. The estimated error in the surface cholesteryl density is ±20%. Details on the XPS analysis are available in the Supporting Information.
SLBs
A sufficient amount of the SUV solution was placed on each functionalized surface of interest and incubated for 1 h at room temperature. Excess vesicles were removed from the surface by flushing with copious amounts of Tris buffer. To test air stability, we removed the SLB-covered surface from the buffer solution and left it to dry in air for ~2 h. We rehydrated each surface by placing it back in the buffer solution. Note that all SLB covered surfaces were handled horizontally to avoid excessive shear force from liquid fronts running across the surface. Most washing/rinsing steps were carried out with the SLB covered surfaces facing up at the bottom of the Petri dishes and with buffer solutions added and withdrawn at localizations away from the sample surfaces.
Arraying and Immunostaining
SUV solutions were deposited on cholesteryl-PEG surfaces with a robotic arrayer (Molecular Dynamics). To avoid rapid drying of the nanoliter droplet, we added 10% glycerol to each SUV solution. We incubated the arrayed coverslips in a high humidity environment for 30 min to 1 h and then rinsed each sample with Tris buffer to remove excess lipids on the surface. For CTB incubation, the sample was incubated with a diluted CTB solution (1 nM) for 3 h. For immunostaining, we incubated the coverslips with a primary antibody (1:200 dilution in TRIS buffer) for 2 h, followed by sufficient washing and then incubation with a secondary antibody solution (1:200 dilution in TRIS buffer) for 1 h. We washed each sample with copious amounts of buffer before imaging.
FRAP
We carried out all FRAP experiments using a Nikon fluorescence microscope. Bleaching was done with a 60× objective and full lamp power for 1 min; the diameter of the bleached spot was 40 μm. Images for recovery were taken at a reduced excitation power with a 20× objective.
Results and Discussions
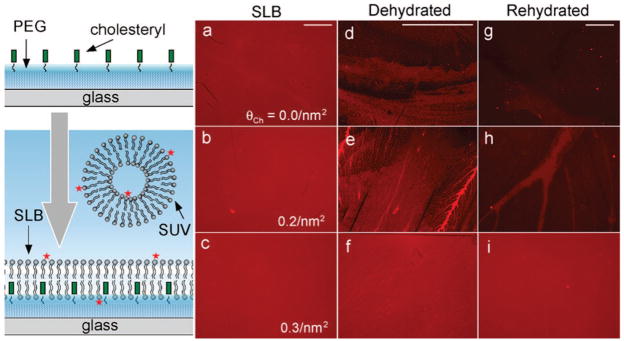
To form SLBs on the cholesteryl-PEG/glass surfaces, we use the vesicle fusion method7–10 (illustration in Figure 1). The fluorescence microscopy images in Figure 1 are from SLBs formed on surfaces with cholesteryl densities of θCh = 0.0, 0.2, and 0.3/nm2, respectively. While SLBs with spatially uniform fluorescence intensity distributions are formed on all surfaces in buffer solutions (a–c), the influence of cholesteryl becomes apparent when the SLB is exposed to air and dehydrated. On surfaces with low cholesteryl densities (θCh < 0.3/nm2), moving through the water–air interface and drying in air results in desorption and extensive restructuring of surface adsorbed lipid molecules, as evidenced by the lower and nonuniform fluorescence intensity of images (d, e). Upon rehydration of these dried surfaces in buffer solution, most lipid molecules are washed off the surfaces (g, h). In contrast, when the surface cholesteryl density reaches a critical value of θCh = 0.3/nm2, fluorescence images of the SLBs remain spatially uniform upon drying (f) and rehydration (i), thus establishing air stability. Results for θCh = 0.5/nm2 (data not shown) are similar to those of θCh = 0.3/nm2.
Figure 1.
Fluorescence microscopy images of supported lipid bilayers formed on a PEG brush surface with the indicated densities of surface tethered cholesteryl groups: θCh = 0.0 (a, d, g), 0.2 (b, e, h), and 0.3/nm2 (c, f, i). The scale bar (0.4 mm) for each column is shown at the top. The SLBs are formed from the fusion of SUVs containing 0.5% Texas-Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE), 50% egg phophatidylcholine (EggPC), and 50% 1,2-dioleoyl-3-trimethyl ammonium propane (DOTAP). Images for the as-formed SLBs in buffer solution (a, b, c) and those after dehydration (g, h, i) are taken with a 4× objective, while those in the dehydrated state (d, e, f) are taken with 10× objective to reveal more details.
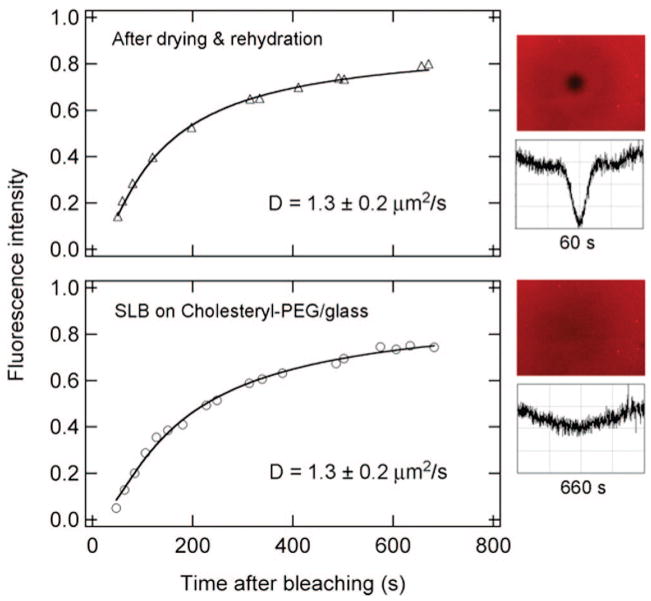
Within the air-stable SLB, lipids remain mobile even after the membrane has gone through dehydration and rehydration, as shown by fluorescence recovery after photobleaching (FRAP) in Figure 2. Sample fluorescence microscopy images (along with cross-sectional profiles) for the rehydrated SLB are shown on the right side. Similar images are obtained for the as-prepared SLB in the buffer solution before drying (not shown). Fitting the FRAP data in Figure 2 gives lipid diffusion constants of D = 1.3 ± 0.2 μm2/s and a total recovery percentage > 90% at long times, both before and after the dehydration–rehydration cycle. This diffusion constant is typical for tethered lipid bilayers.17 Within the range of cholesterol density (0.1–0.5/nm2) investigated, we find no significant change in lipid diffusion constants.
Figure 2.
FRAP results for an SLB formed on a PEG/glass surface with a tethered cholesteryl density of 0.3/nm2. The lower panel is for the as-formed SLB, and the upper panel, for the SLB after air-drying and rehydration. The diffusion constants are obtained from fits (solid lines) to the FRAP data, and average values from repeated measurements are shown. Right: Two fluorescence images (240 μm × 400 μm) and cross-sectional profiles for the rehydrated SLB taken at 60 and 660 s after photobleaching.
We have also carried out FRAP measurements on the SLB in the dehydrated state and find no lipid mobility (data not shown). Note that we use the term “air-stable SLB” to describe the supported lipid bilayer which remains intact after dehydration and rehydration. While fluorescence microscopy shows no difference between the hydrated and the dehydrated SLB, it is possible that, in the dehydrated state, reorganization of lipids may occur on scales smaller than what is resolvable optically. Even after rehydration, defects with sizes smaller than optical resolution may also form in the SLB after repeated cycles of dehydration and rehydration. However, the insensitivity of the FRAP data to dehydration/rehydration cycles suggests that an accumulation of defects, if present, is not significant.
A supported lipid bilayer is known to be unstable upon exposure to air.18 This is because dehydration of the hydrophilic head groups makes the organized two-dimensional lamellar configuration energetically unstable. Extensive reorganization of lipid molecules into three-dimensional structures occurs upon air exposure, resulting in the destruction of the SLB and desorption of lipids from the surface.10 The key to our success lies in the dispersed (statistically from the attachment reaction) and immobile nature of tethered cholesteryl groups. The critical surface cholesteryl density of 0.3/nm2 corresponds to a cholesteryl-to-lipid ratio of ~1:6 in the bottom leaflet. At this critical density, the local ordering and stabilization by cholesteryl are distributed throughout the entire bottom leaflet. This effect is combined with covalent tethering of the cholesteryl groups to provide a sufficient energy barrier which prevents the restructuring of the lipid bilayer when the top surface is dehydrated, thus, leading to air stability. Supporting our conclusion, we note that air stability was not achieved in previous studies of supported lipid bilayers on tethered cholesteryl groups in binary thiol self-assembled monolayers (SAMs) on gold surfaces,19 where extensive phase separation resulted in domains of cholesteryl thiol separated from those of short hydrophilic thiols.20 Vesicle deposition on such a phase-separated surface led to lipid monolayers on the cholesteryl domains while lipid bilayers formed on the hydrophilic thiol domains, without achieving air stability.
There have been previous attempts in making stable supported membranes. Plant and co-worker demonstrated the approach of a hybrid bilayer21,22 where a stable lipid monolayer was assembled onto an alkanethiol self-assembled monolayer. The hybrid bilayer is not suitable for the incorporation of transmembrane proteins. Saavedra and co-workers demonstrated a stable supported lipid membrane by polymerizing diacetylene–lipid conjugates.23 The lack of lipid mobility in such a polymerized membrane limits it application. Cremer and coworkers achieved air stability by covering the SLB surface with proteins and polymer brushes.24,25 However, the protection functionalities on the surface of the SLB may inhibit or prevent proper interaction of membrane proteins or surface glycans with their targets, thus limiting their applications. This group also demonstrated that an SLB becomes air-stable if the solution in contact with the SLB before drying contains a relatively high concentration of trehalose,26 an approach adapted from anhydrobiotic organisms.27 This approach requires the introduction of trehalose every time the SLB is exposed to air. In contrast, the air-stable SLB demonstrated here can be continuously dehydrated and rehydrated without the need of adding special components to the solutions. As shown below, it also allows the reconstitution of peripheral as well as transmembrane components.
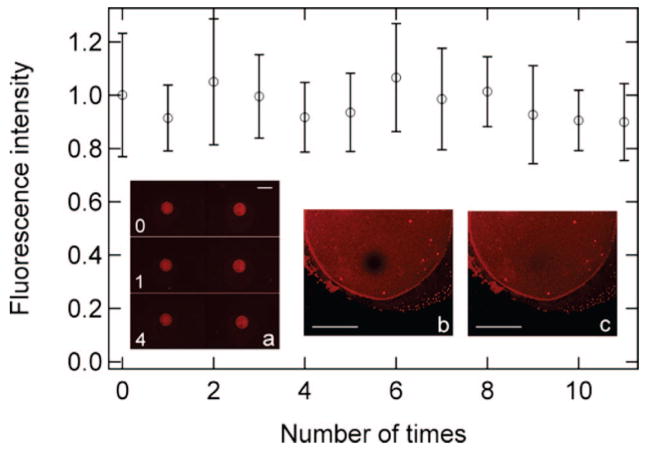
The success in achieving air stability has enabled us to directly form SLB microarrays simply from robotic spotting. Drying of the nanoliter droplets after arraying is not a problem because of the air stability of the SLB and because excess lipids are easily washed off. Figure 3a shows fluorescence images of a 2 × 1 array of SLB formed from robotic spotting of the SUV solution on the cholesteryl-PEG surface and the same array after it has been withdrawn from the air–water interface once and four times, respectively. Insets (b) and (c) show zoomed-in images on one spot taken at 47s and 600s, respectively, after photobleaching. Lipid molecules in the SLB spot are clearly in the fluidic state; fitting the FRAP data (not shown) gives similar diffusion constants as that in Figure 2. The data points in Figure 3 show that the spot intensity remains constant after the array has been withdrawn from the air–water interface for as many as 11 times (without complete drying), indicating the remarkable robustness of the SLB spots.
Figure 3.
(a) Fluorescence microscope images of a 2 × 1 array of SLBs on the cholestery-PEG/glass surface θCh = 0.3/nm2). The as-formed array is shown at the top, and those after air-exposure once and four times are in the middle and bottom, respectively. The data points are fluorescence intensity as a function of the number of times of air exposure. The zoomed-in images are for a single spot (transferred through the air–water interface once after arraying and washing) taken at (b) 47 s and (c) 600 s after photobleaching. All fluorescence measurements were carried out with the sample in buffer solution. The scale bar is 400 μm in (a) and 100 μm in (b) and (c).
The formation of such an air-stable SLB microarray does not require surface prepatterning to form corrals28,29 but can be achieved from simple robotic spotting. Note that Fang et al. previously reported air-stable G-protein coupled receptor arrays formed from the direct deposition of membrane solution on γ-aminopropylsilane (GAPS) coated glass surface.30 However, control experiments reported by McBee and Saavedra showed that SUVs deposited on the GAPS surface did not form SLBs and did not possess sufficient stability upon withdrawing the sample from the air–water interface.31 Our ability to form fluidic and air-stable SLB microarrays from simple robotic spotting has opened the door to large scale screening of cell surface interactions. We demonstrate this using two model systems.
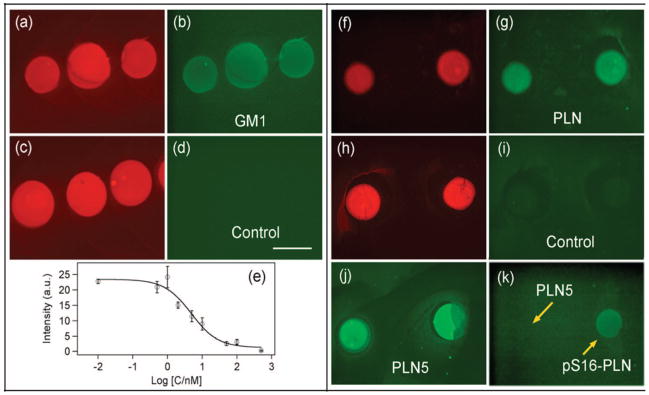
In the first system, we incorporate into the SLB 2% ganglioside GM1 a glycolipid containing the oligosaccharide unit for specific binding by cholera toxin B (CTB) subunits. We choose this model system because the pentameric CTB is known to bind to five GM1’s, and this multivalent interaction is expected to require fluidity in cluster formation.32 The fluorescence microscopy images in the left panel in Figure 4 show SLB arrays with (a and b) or without (c and d) GM1. As expected, the green dye-tagged CTB binds specifically to GM1 containing SLB arrays. We carry out FRAP measurements on SLB spots after CTB binding and find that the TR-DHPE lipids (red channel) remain mobile while the CTB-GM1 complexes (green channel) are not, in agreement with previous studies.33 We also carry out a quantitative analysis of binding on the SLB microarray using a competition assay. Panel (e) in Figure 4 shows the amount of surface bound FITC-CTB (green fluorescence intensity) as a function of solution phase CTB (without dye label) in the presence of 1 nM FITC-CTB. The data give an IC50 value of ~6 nM, in agreement with previous measurements on CTB binding to ganglioside GM1 in lipid membranes supported on microspheres.34
Figure 4.
Fluorescence microscopic images of SLB arrays deposited on the cholesteryl-PEG/glass surface θCh = 0.3/cm2). Left: Arrays with (a and b) or without (c and d) 2% ganglioside GM1. The red channel shows TR-DHPE (0.5%), and the green channel detects the binding of FITC-CTB. (e) Competitive assay of the binding of FITC-CTB to the GM1 containing an SLB array from a solution containing 1 nM FITC-CTB and varying concentrations (C/nm) of CTB without dye label. Right: (f and g) Spots containing PLN with a protein/lipid ratio of 1:100; (h and i) controls without protein; (j) spots containing PLN5 with a protein cluster/lipid ratio of 1:500, and (k) spots containing PLN5 (left) with a protein cluter/lipid ratio of (1:500) and pS16-PLN (right) with a ratio of (1:100). The red channel is for TR-DHPE, and the green channel, for the FITC-2nd antibody. A primary antibody against PLN is used for (g, i, j) and that specific for pS16-PLN in (k). The scale bar in (d) is 400 μm.
In the second model, we use phospholamban (PLN), a single-pass integral membrane protein which inhibits the sarcoplasmic reticulum calcium ATPase (SERCA), thus regulating heart muscle contraction and relaxation.15 We incorporate both monomeric (PLN, 6 kDa)15 and pentameric (PLN5, 30 kDa)16 forms into SLB microarrays and use an antibody (Anti-PLN) against PLN for specific detection. The right panel in Figure 4 shows the specific detection of PLN (f–j). The red channel (TR-DHPE) shows the position of the spots, and the green channel is immunostaining (FITC-2nd antibody). The control (h and i) shows spots without protein incorporation. Anti-PLN recognizes PLN5 (j) with a higher affinity than that for the monomer (g), indicating greater accessibility of antibody binding sites on PLN5. In addition, we demonstrate that our approach is sensitive to post-translational modifications. In fact, PLN is phosphorylated at S16 by protein kinase A, a process which releases SERCA inhibition and re-establishes calcium flux.14 We re-constituted phosphorylated PLN at serine 16 (pS16-PLN) in SLB arrays, and using the anti-pS16-PLN antibody we are able to localize this protein (k). While the above experiment successfully demonstrates the incorporation of a single domain transmembrane protein, potential applications in drug screening require the incorporation of more complicated systems, such as the G-protein coupled receptor complex. It may be necessary to control the distance between the bottom leaflet and the polymer cushion by varying the length of the tether linking the cholesteryl group.
Conclusions
We achieve both air stability and fluidity in supported lipid bilayers based on the unique properties of cholesterol and demonstrate the easy fabrication of SLB microarrays from direct robotic spotting. The SLB microarray can serve as a general platform for the analysis of cell surface interactions. The screening of ligands for membrane proteins, such as G-protein coupled receptors1 and ion channels,2 is an obvious example. Another exciting application is the high-throughput analysis of multivalent cell surface interactions.35 In the burgeoning field of nanomedicine, multivalent interaction is often employed in targeting nanoparticle based drugs to particular cells.6 It is now possible to implement all these cell surface interactions in a high-throughput format based on the fluidic and air-stable SLB microarray reported here.
Supplementary Material
Acknowledgments
This work was supported in part by the National Science Foundation, Grant SBIR II (0450262), awarded to MicroSurfaces, Inc.
Footnotes
Note Added after ASAP Publication. The version of this paper published on April 12, 2008, was missing the Supporting Information paragraph. The version published on May 7, 2008 has the correct information.
Supporting Information Available: Details on the XPS analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ellis C. Nat Rev Drug Discovery. 2004;3:577. [Google Scholar]
- 2.Huang CJ, Harootunian A, Maher MP, Quan C, Raj CD, McCormack K, Numann R, Negulescu PA, González JE. Nat Biotechnol. 2006;24:439. doi: 10.1038/nbt1194. [DOI] [PubMed] [Google Scholar]
- 3.Lerner RA. Angew Chem, Int Ed. 2006;45:8106. doi: 10.1002/anie.200603381. [DOI] [PubMed] [Google Scholar]
- 4.Hollinger P, Hudson PJ. Nat Biotechnol. 2005;23:1126. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 5.Kiessling LL, Gestwicki JE, Strong LE. Curr Opin Chem Biol. 2000;4:696. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari M. Nat Rev Cancer. 2005;5:161. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 7.Brian AA, McConnell HM. Proc Natl Acad Sci USA. 1984;81:6159. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka M, Sackmann E. Nature. 2005;437:656. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 9.Groves JT, Boxer SG. Acc Chem Res. 2002;35:149. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- 10.Castellana ET, Cremer PS. Surf Sci Rep. 2006;61:429. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriksen J, Rowat AC, Brief E, Hsueh YW, Thewalt JL, Zuckermann MJ, Ipsen JH. Biophys J. 2006;90:1639. doi: 10.1529/biophysj.105.067652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simons K, Vaz WLC. Annu Rev Biophys Biomol Struct. 2004;33:269. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 13.Buck B, Zamoon J, Kirby TL, DeSilva TM, Karim C, Thomas D, Veglia G. Protein Expression Purif. 2003;30:253. doi: 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- 14.Metcalfe EE, Traaseth NJ, Veglia G. Biochemistry. 2005;44:4386. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 15.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Biochemistry. 2006;45:13827. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 16.Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Proc Natl Acad Sci USA. 2007;104:14676. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoll W, Frank CW, Heibel C, Naumann R, Offenhausser A, Ruhe J, Schmidt EK, Shen WW, Sinner A. Rev Mol Biotechnol. 2000;74:137. doi: 10.1016/s1389-0352(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 18.Cremer PS, Boxer SG. J Phys Chem B. 1999;103:2554. [Google Scholar]
- 19.Cheng Y, Boden N, Bushby RJ, Clarkson S, Evans SD, Knowles PF, Marsh A, Miles RE. Langmuir. 1998;14:839. [Google Scholar]
- 20.Jeuken LJC, Daskalakis NN, Han X, Sheikh K, Erbe A, Bushby RJ, Evans SD. Sens Actuators, B. 2007;124:501. [Google Scholar]
- 21.Plant AL. Langmuir. 1999;15:5128. [Google Scholar]
- 22.Meuse CW, Krueger S, Majkrzak CF, Dura JA, Fu J, Connor JT, Plant AL. Biophys J. 1998;74:1388. doi: 10.1016/S0006-3495(98)77851-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross EE, Rozanski LJ, Spratt T, Liu S, O’Brien DF, Saavedra SS. Langmuir. 2003;19:1752. [Google Scholar]
- 24.Holden MA, Jung SY, Yang T, Castellana ET, Cremer PS. J Am Chem Soc. 2004;126:6512. doi: 10.1021/ja048504a. [DOI] [PubMed] [Google Scholar]
- 25.Albertorio F, Diaz AJ, Yang T, Chapa VA, Kataoka S, Castellana ET, Cremer PS. Langmuir. 2005;21:7476. doi: 10.1021/la050871s. [DOI] [PubMed] [Google Scholar]
- 26.Albertorio F, Chapa VA, Chen X, Diaz AJ, Cremer PS. J Am Chem Soc. 2007;129:10567. doi: 10.1021/ja0731266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowe JH, Crowe LM. Nat Biotechnol. 2000;18:145. doi: 10.1038/72580. [DOI] [PubMed] [Google Scholar]
- 28.Groves JT, Ulman N, Boxer SG. Science. 1997;275:651. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki V, Sirenko O, Schafer RJ, Nguyen L, Gutsmann T, Brade L, Groves JT. BMC Biotechnol. 2005;5:18. doi: 10.1186/1472-6750-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, Frutos AG, Lahiri J. J Am Chem Soc. 2002;124:2394. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 31.McBee TW, Saavedra SS. Langmuir. 2005;21:3396. doi: 10.1021/la047646f. [DOI] [PubMed] [Google Scholar]
- 32.Schon A, Freire E. Biochemistry. 1989;28:5019. doi: 10.1021/bi00438a017. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki V, Sirenko O, Schafer RJ, Groves JT. J Am Chem Soc. 2005;127:2826. doi: 10.1021/ja042430l. [DOI] [PubMed] [Google Scholar]
- 34.Lauer S, Goldstein B, Nolan RL, Nolan JP. Biochemistry. 2002;41:1742. doi: 10.1021/bi0112816. [DOI] [PubMed] [Google Scholar]
- 35.Mammen M, Choi SK, Whitesides GM. Angew Chem, Int Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.