Abstract
The aryl hydrocarbon receptor (AhR) is an orphan receptor in the basic-helix-loop-helix PAS family of transcriptional regulators. Although the endogenous regulator of this pathway has not been identified, the AhR is known to bind and be activated by a variety of compounds ranging from environmental contaminants to flavanoids. The function of this receptor is still unclear; however, animal models indicate that the AhR is important for normal development. One hypothesis is that the AhR senses cellular stress and initiates the cellular response by altering gene expression and inhibiting cell cycle progression and that activation of the AhR by exogenous environmental chemicals results in the dysregulation of this normal function. In this review we will examine the role of the AhR in the regulation of genes and proteins involved in cell adhesion and matrix remodeling, and discuss the implications of these changes in development and disease. In addition, we will discuss evidence suggesting that the AhR pathway is responsive to changes in matrix composition as well as cell-cell and cell-matrix interactions.
Keywords: AhR; extracellular matrix; ECM; matrix metalloproteinase; MMP; 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD); development
1. Introduction
The basic helix-loop-helix Per/AhR-Arnt/Sim (bHLH-PAS) family of transcriptional regulators has diverse biological roles regulating the expression of genes involved in development, hypoxia signaling and circadian rhythms (1). The aryl hydrocarbon receptor (AhR) is unique to this family in that it was originally identified as the receptor for environmental contaminants including the polycyclic aromatic hydrocarbons (PAH). However, accumulating evidence demonstrates that the AhR has a number of diverse ligands both endogenous and exogenous (reviewed in (2)). Indeed, the identification of an endogenous ligand or signal for the AhR is of great interest, as evidence from knockout animal models indicates that this pathway has an important role in normal physiology (1). The current hypothesis is that the AhR senses cellular stress and initiates the cellular response by altering gene expression and inhibiting cell cycle and that activation of the AhR by chemicals such as the PAHs results in the dysregulation of this normal function. In this review we will examine the role of the AhR in the regulation of genes and proteins involved in cell adhesion and matrix remodeling and discuss the implications of these changes in development and disease. In addition, we will discuss evidence suggesting that the AhR pathway is responsive to changes in matrix composition as well as cell-cell and cell-matrix interactions.
The molecular mechanisms of the AhR function have been primarily examined using the PAH, 2,3,7,8-tetrachorodibenzo-p-dioxin (TCDD) to bind to the AhR and initiate signaling (reviewed in (3)). TCDD is the unintentional by-product of industrial combustion. The chemical properties of TCDD render it resistant to both environmental and biological degradation, and therefore TCDD accumulates in the lipids of exposed animals, including humans with an estimated half life of 7.6 years. Accidental acute exposure to TCDD is associated with the increase in a variety of human health problems, including immune dysfunction, neurological pathologies, abnormal development, diabetes, and carcinogenesis.
Using TCDD as an activator, data show that the AhR resides in an inactive multiprotein complex in the cytoplasm bound to accessory proteins including two heat shock protein 90 (HSP90) molecules, a HSP-90-interacting co-chaperone p23 and an immunophilin-like protein, ARA9/XAP2/AIP. Ligand binding to the AhR ligand binding domain results in conformational changes that expose the nuclear localization sequence through alterations of XAP2 binding. After nuclear localization and dissociation from HSP-90 and ARA9 the AhR forms a heterodimer with Arnt (AhR nuclear translocator), and functions as a transcriptional activator by binding to specific DNA enhancer element sequences in the 5′ region of AhR-responsive genes termed xenobiotic response elements (XRE: 5′-GCGTG-3′).
Given the role of the AhR in mediating signals from environmental contaminants, it is not surprising that the first target genes identified for this pathway included phase I xenobiotic metabolizing genes, such as members of the cytochrome p450 (CYP450) family of monooxygenase enzymes and included phase II xenobiotic metabolizing enzymes, such as UGT1A1 (UDP-glucuronosyltransferase 1A1), GST-Ya (glutathione S-transferase Ya) subunit and NADPH-quinone-oxido-reductase (reviewed in (1)). However, data now indicate that the AhR-pathway controls the expression of a variety of genes unrelated to xenobiotic metabolism, including genes encoding proteins involved in growth control, such as transforming growth factor-α (4), transforming growth factor-β2 (4), Bax (5) and p27kip1 (6); cytokines interleukin-1β and interleukin-2 (7,8); and nuclear transcription factors such as c-fos, Jun-B, c-Jun and Jun-D (9).
Interestingly, several of the identified targets of AhR activation are also important in the regulation of tissue and matrix remodeling, suggesting that the AhR-pathway has an endogenous role in mediating matrix metabolism and deposition. In addition, data also demonstrate that changes in cell-cell and cell-substratum interactions have an impact on the AhR signaling (Fig. 1). Studies show that cellular suspension of normal human keratinocytes, liver cell lines, and 10T1/2 cells, resulting in the removal of both cell-cell and cell-subtratum interactions, promotes AhR binding to XREs and increases expression of the AhR-target gene cytochrome p450 1A1 (CYP1A1) in the absence of exogenous ligand. (10,11). Recently, using a model of branching morphogenesis in mammary epithelial cells, the expression of AhR, Arnt, CYP1A1 and cytochrome p450 1B1 (CYP1B1) was linked to interactions in culture with specific matrix proteins (12). Matrices that failed to support branching morphogenesis also failed to demonstrate an increase in AhR-pathway gene expression. These data indicate that the AhR pathway can be activated in the absence of ligand by changes in cell –cell and cell-matrix interactions. Taken together, these data indicate that the AhR may function to sense changes in cell-cell/matrix interactions and mediate a cellular response through alterations in gene expression. These findings are further supported by data from a variety of cell types demonstrating that activation of the AhR pathway results in the loss of contact inhibition of cell proliferation (10,13–15)
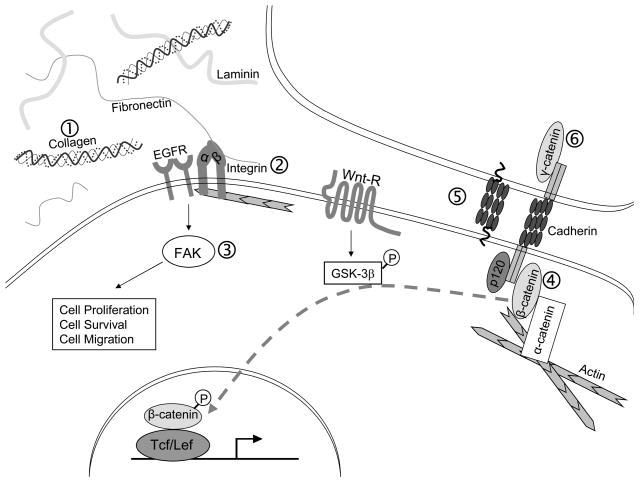
Figure 1. AhR signaling alters cell-cell and cell-matrix interactions.
Activation of the AhR pathway alters cellular adhesion in a variety of model systems. (1) AhR pathway alters the expression of ECM proteins, including types I and IV collagen and fibronectin in fetal heart and marmoset myocardium and thymocytes (17–20). (2) Expression of several integrin subtypes, including the β7 subtype is altered following AhR signaling in cytotoxic T-cells (22), thymic epithelial cells (23), macrophages (24), T-lymphocytes (25) and human amniotic epithelial cells (26). (3) Focal adhesion kinase (FAK) phosphorylation in HUVECs (29). (4) Over-expression of β-catenin results in increased AhR expression in prostate cancer cell lines (36). (5) Expression of T-cadherin is decreased following TCDD exposure in smooth muscle cells in culture and murine fetal hearts (19,32). (6) AhR-activation results in the loss of γ-catenin associated with the adherence complex in rat liver epithelial cells (33).
2. The AhR pathway in the regulation of the extracellular matrix and cellular adhesion
Cell-cell and cell-matrix interactions are critical for cellular differentiation, proliferation and migration (reviewed in (16)). The extracellular matrix (ECM) is a dynamic structure that is continually being remodeled. ECM remodeling, its assembly and degradation, is regulated by the three-dimensional environment and cellular tension mediated by the integrins and proteolytic cascades. The ECM consists of a variety of proteins and polysaccharides that form a complex network between cells and within tissues. The composition of the ECM varies between tissues; however, the main components often include fibrous structural proteins (collagens, laminins, fibronectins, vitronectin and elastin); proteoglycans, as well as a variety of specialized proteins (growth factors, small matricellular proteins, and SIBLINGS (small integrin-binding glycoproteins)). Therefore, the ECM not only provides scaffolding for cellular contact, but also acts as a reservoir for growth factors and other signaling molecules. Data indicate that TCDD exposure and activation of the AhR pathway alters the expression of several ECM proteins, including types I and IV collagen and fibronectin (17–20) (Fig. 1). During development, TCDD exposure results in increased expression of collagens 4, 6, 9 13, 18 and tropoelastin in the fetal heart (19). In addition, enzymes related to collagen deposition were also increased including several matrix metalloproteinases (discussed further below), mast cell carboxypeptidase, and endothelin-1 (19). In the marmoset model, TCDD exposure induced expression of types I and IV collagen and fibronectin in the myocardium and in thymocytes (17,18).
Cell-matrix interactions are mediated by two families of membrane associated proteins, the integrins and the syndecans (reviewed in (21)). Members of both these families interact with specific ECM proteins and growth factors and transmit these extracellular signals into the cell through interactions with the contractile cytoskeleton and with other signaling pathways. The integrins are heterodimeric transmembrane receptors made up of α and β subunits that bind extracellularly to specific ECM proteins and intracellularly to actin of the intracellular cytoskeleton via cytoplasmic binding proteins. There are currently 18 α and 8 β integrin subunits identified in mammals, combining to create 24 distinct αβ integrin receptors. Integrin receptor subtype expression is regulated by both the amount of ligand and the composition of the ECM. In addition to their roles in tissue maintenance, integrins also mediate the assembly of fibronectin into fibrils that attach to the cell surface, which, as the first step in ECM assembly, provides a framework for the binding of additional matrix proteins including thrombospondin and type I collagen. Data from several labs demonstrate that several integrin subunits are targets of PAH/TCDD exposure and the AhR pathway and suggest that alterations in integrin subunit expression are critical for TCDD-mediated effects in cytotoxic T-cells (22) (Fig. 1). In the monocytic leukemia THP-1 cell line, mRNA levels of β7 and α5 integrins are increased following AhR activation, whereas expression of α6 is decreased in human thymic epithelial cells (23). The increase in β7 integrin is also observed in primary human macrophages and developing T-lymphocytes (24,25). A variety of other integrin subunits were also altered following PAH exposure, though to a lesser extent in comparison to β7, these include α11, αE, αL, and β8 in primary macrophages (25) and α10 and α2 in human amniotic epithelial cells (26). These studies also demonstrate, using chemical inhibition and siRNA, that the increase in β7 expression requires both the AhR pathway and the binding of the transcription factor c-maf. Taken together these data indicate that PAH/TCDD exposure and activation of the AhR pathway alters integrin subunit expression and cell-matrix interactions and may ultimately alter the physiological processes regulated by these interactions including epithelial/macrophage homing and migration. Further, over-expression of c-maf/β7 integrin is associated with malignant transformation in myelomas and T-lymphomas (27) suggesting that some of the PAH’s carcinogenic effects may be mediated by alterations in integrin expression and cell-matrix interactions. These data indicate that the AhR pathway regulates the composition of the integrin receptors on the cell surface and therefore influences the interactions between the cell and the matrix.
Integrins are part of a large submembranous structure termed the focal adhesion. This large integrin-based multiprotein complex is critical for strong cell-substrate interactions and mediates the bidirectional signaling between the extracellular receptors and the cytoplasm. In addition to the integrins, the focal adhesion also contains integrin-related receptors, growth factor receptors, paxillin, vinculin and signaling proteins including Src, Grb2 and FAK (focal adhesion kinase). Src is known to be rapidly phosphorylated following TCDD exposure in a variety of cell lines and animal models and data indicate that Src is necessary for TCDD-induced pathologies (28). However, signaling through Src does not appear to be critical for TCDD-induced release from contact inhibition (15) suggesting that Src activation may not be sufficient for TCDD-induced changes in cell adhesion.
Recent data suggest that FAK activation may be involved in AhR-pathway induced changes in cell signaling. FAK is a non-receptor protein tyrosine kinase that is involved in the rapid modulation of the integrin signaling cascade (21). FAK is critical for the regulation of cell growth and proliferation, protection from apoptosis, adhesion and migration/invasion. Therefore, it is not surprising that FAK plays an important role in embryogenesis and morphogenesis and that dysregulation of FAK is associated with malignant transformation in addition to other pathologies. Data show that phosphorylation of FAK is inhibited by exposure to the AhR-agonist 3-methylcholanthrene (3-MC) in human umbilical vascular endothelial cells (HUVECs) (29) (Fig. 1) and is associated with the failure of the HUVECs to from tubules on matrigel. Inhibition of FAK phosphoryaltion was demonstrated to be AhR dependant using the AhR-antagonist α-napthoflavone, and was associated with loss of proliferation.. These data suggest that FAK may be an important regulator of AhR-mediated effects on cell-substratum contact and on the loss of contact inhibition.
Cadherins are adhesion proteins critical for modulating cell-cell interactions (30). This large family of cell surface glycoproteins form Ca2+ dependant homotypic interactions with cadherins on adjacent cells and are important for cell recognition, sorting, coordinated cell movement and the induction and maintenance of cell/tissue polarity. Classical cadherins are found in almost all vertebrate tissues and their expression is regulated from methylation-induced repression of gene expression to ubiquination-mediated proteolytic protein turnover of cadherins at the cell surface. Cadherins are found primarily associated with adherens junction, and modulate cell adhesion via dynamic interactions with the actin cytoskeleton. The intracellular region of cadherins interact with specific proteins including α-catenin, β-catinin and plakoglobin (γ-catenin), which are required for cadherin’s adhesive properties, and p120 catenin which appears to be important for regulating the conversion of weak cell-cell adhesion to strong cell-cell adhesion via cadherin clustering (30).
Expression of T-cadherin (cadherin 13/H-cadherin) is decreased following AhR activation (Fig. 1). T cadherin is an atypical cadherin expressed in diverse organs and cell types during development and in adult tissues (30). Although the extracellular domain is similar to that of the classical cadherins, T-cadherin lacks both the transmembrane domain and the cytoplasmic domain typically found in classical cadherins and is instead linked to the cell surface by a GPI moiety. T-cadherin is postulated to be a tumor suppressor as its expression is reduced or lost in a variety of tumor types, mediated by methylation of the T-cadherin promoter (31). In smooth muscle cells, activation of AhR signaling results in decreased expression of T-cadherin (32). Decreased expression of T-cadherin was also observed in the murine fetal heart following TCDD exposure (19).
In addition to changes in cadherin expression, the expression of the intracellular proteins that interact with the cadherins is also altered by AhR activation (Fig. 1). In rat liver epithelial cells, TCDD exposure resulted in the loss of γ-catenin/plakoglobin from association with adhesion sites and movement to the cytoplasm, whereas localization of E-cadherin, α-catenin and β catenin remained at the adhesion sites (33). These data further demonstrate that the loss of γ-catenin expression is due to transcriptional inhibition or mRNA destabilization and that the loss of γ-catenin in response to TCDD at the adhesion sites correlated with a loss of contact inhibition and G1 arrest in these cells. γ-catenin is essential for anchoring the E-cadherin in adherens junctions and is associated with desmocollins and desmogleins in desmosomes, both of which are necessary to maintain proper cellular adhesion and growth control (34). Loss of γ-catenin correlates with malignancy and is associated with poor prognosis in several tumor types, including lung (35). These data suggest that one mechanism of the tumor promoting activity of TCDD may be through alterations in γ-catenin expression and reduced stability of cell-cell adhesions.
The above findings demonstrate that cell-cell adhesion is altered following activation of the AhR pathway and suggest that these alterations may have an impact on cellular behavior. Data also show that the AhR is a target of signaling mediated through the cadherin/catenin complex. In prostate cancer cells, over-expression of β-catenin results in increased expression of AhR mRNA and protein (36). β-catenin is a critical part of cadherin-based adhesions; however, it is also an essential co-activator for Wnt-mediated changes in gene expression. Wnt is a secreted lipoglycoprotein that functions as a morphogen and is critical for cell fate and tissue morphogenesis in development (37). Wnt-signaling is mediated by the transcription factor TCF/LEF (T-cell factor/lymphocyte enhancer factor) which requires β-catenin as its co-activator. Interestingly, data also demonstrate that cellular suspension, which activates the AhR pathway, also demonstrate an increase in nuclear β-catenin (10). Taken together these data suggest that β-catenin mediates the changes in AhR signaling observed following loss of adhesion (Fig. 1).
3. AhR pathway and matrix remodeling
ECM degradation is accomplished through the coordinated expression and activity of several proteolytic cascades (Fig. 2). Proteolytic enzymes, including the serine proteases, the matrix metalloproteinases (MMPs) and cysteine proteases contribute to ECM modulation in several ways including the direct degradation of matrix proteins, the release of small bioactive peptides and the release of growth factors stored in the ECM. Members from all three of these protease families have been shown to be targets of the AhR pathway.
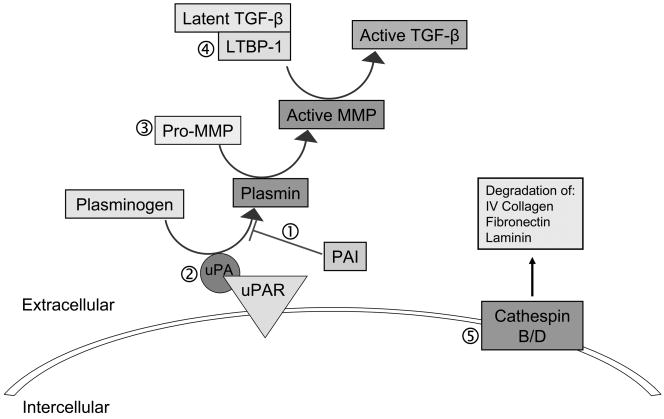
Figure 2. AhR pathway alters matrix metabolism through altering expression and activity of proteases.
(1) AhR pathway activation alters expression of both the uPA protease and PAI-1/2. TCDD induces expression of PAI-1 in mouse hepatoma cells and in the rat ovary (43,44). PAI-2 expression is enhanced by AhR-ligands in human breast epithelial cells, endometrial cells, lymphocytes, as well as in HEPG2, U937 monocytic and SCC-13 keratinocyte cell lines (45–48). (2) Expression of both uPA and tPA (tissue plasminogen activator) are increased by TCDD exposure in a rat model of ovulation (44). (3) Several MMPs are targets of the AhR pathway, including MMP-1 in normal human keratinocytes and A2058 melanoma cells(52,53), MMP-2 and -9 in A2058 melanoma cells (53), MMP-2, -9and -13 in the murine fetal heart (19,20) as well as in the regenerating zebrafish tailfin (54). TCDD exposure also induced MMP-1, 3 and 13 in U937 macrophages and correlated with increased cell migration (55), and in human endometrial cells (56), and MMP-9 expression is induced in prostate cancer cells (57). Further, in AhR−/− mouse embryo fibroblasts, MMP-2 activity is reduced (58). (4) AhR regulates TGF-β levels as demonstrated in AhR−/− MEF where the total TGF-β levels were elevated resulting in decrease proliferation and increased apoptosis respectively. It has been recently determined that LTBP-1 (latent TGF-β binding protein) mediates the increase in TGF-β1 in AhR−/− MEF cells and is also responsible for activity of two proteases involved in TGF-β release, plasmin and elastase. Reduction in LTBP-1 increases the MMP-2 protease. (5) Two cathespins are down regulated in response to AhR activation. Cathespin B expression is reduced by TCDD in porcine thyrocytes (39). Both TCDD exposure and a constitutively active AhR inhibits estrogen-induced cathespin D expression in MCF-7 cells.(40,41).
The cathespins are cysteine proteases primarily localized to the endolysosomal vesicles (38). However, recent findings indicate that these proteases also have activity extracellularly and they have been implicated in promoting tumor progression. The two cathespins identified to be targets of the AhR pathway are cathespin B and cathespin D (Fig. 2). Cathespin B expression is down-regulated by TCDD in porcine thyrocytes (39). And although the physiological effect of this down-regulation is unclear, it is known that cathespin B can degrade purified type IV collagen, laminin and fibronectin in vitro. In MCF-7 cells, both TCDD exposure and a constitutively active AhR both inhibits estrogen-induced cathespin D expression (40,41). This inhibition is mediated through the AhR which inhibits the binding of transactivators, including the estrogen receptor α (ERα), to the major late promoter element in the cathespin D enhancer (40).
The serine proteases are also targets of the AhR pathway. Urokinase plasminogen activator (uPA) uPA binds to cell surface receptor uPAR (uPA receptor) and is then converted to an active enzyme via the action of cysteine proteases. uPA activates plasmin, which cleaves ECM proteins, activates several pro-MMPs and may also be involved in the cleavage of collagen via endocytosis (42). The plasminogen activator inhibitors -1 and -2 (PAI-1, -2) are direct and rapid inhibitors of uPA. PAI-1 also binds to vitronectin and blocks its binding to the integrin receptor αVβ3. Both these inhibitors will effectively block the proteolytic cascades, although data also suggest that depending on cell type, conformation and concentration, PAI-1 can either enhance or inhibit cell migration and adhesion. Data demonstrate that the activation of the AhR pathway alters expression of both the uPA protease and PAI-1/2 (Fig. 2). TCDD induces expression of PAI-1 in mouse hepatoma cells and in the rat ovary (43,44). Expression of PAI-2 is induced by AhR-ligands in human breast epithelial cells, endometrial cells, lymphocytes, as well as in HEPG2, U937 monocytic and SCC-13 keratinocyte cell lines (45–48). Data using human endometrial cells suggest that AhR-mediated PAI-2 expression is regulated post-transcriptionally (46). Interestingly, expression of both uPA and tPA (tissue plasminogen activator) are also increased by TCDD exposure in a rat model of ovulation (44). Data suggest that TCDD increases uPA mRNA expression in rat liver cells by inducing the binding of a 50kDA protein to the 3′ untranslated region and stabilizing the message (49). Taken together, these findings indicate that AhR pathway signaling is important for maintaining the proper regulation of the plasminogen proteolytic cascade.
Data also indicate that the expression and activity of the MMPs are targets of the AhR pathway. MMPs are a family of zinc and calcium dependent enzymes that together are able to degrade all the components of the ECM. There are currently 23 MMPs characterized in humans and are classified based upon their substrate specificity, sequence similarity and domain organization (reviewed in (50)). MMPs are mainly regulated at the level of transcription and activation and are essential for tissue remodeling events such as wound healing and embryogenesis. Not surprisingly, loss of control of MMPs is a hallmark of a variety of diseases, including in cancer, arthritis, atherosclerosis, aneurysms, nephritis ulceration and fibrosis. MMP transcription is induced by various growth factors and cytokines including interleukin-1 (IL-1) and tumor necrosis factor-α (TNF-α) (51). Other factors such as retinoic acid and glucocorticoids are known to inhibit MMP expression (51).
Activation of the AhR pathway has been shown to modulate the expression and activity of the MMPs in a variety of cell types. MMP-1 expression is increased in normal human keratinocytes and A2058 melanoma cells following exposure to TCDD (52,53). Expression and activity of MMP-2 and MMP-9 are also up-regulated in response to TCDD in the A2058 melanoma cells and this increase is associated with increased in vitro invasion (53). AhR-mediated changes in MMP expression are not limited to cell types of the skin. Increased expression of MMP-2, -9and -13 is observed in the murine fetal heart following in utero exposure to TCDD (19,20) as well as in the regenerating zebrafish tailfin (54). TCDD exposure also induced MMP-1, 3 and 13 in U937 macrophages and correlated with increased cell migration (55) TCDD-induced MMP expression is also observed in human endometrial cells (56) and it is postulated to disrupt normal progesterone-mediated MMP expression and contribute to endometriosis (56). MMP-9 expression is also induced in prostate cancer cells in response to TCDD (57). Further support that the AhR pathway is important for MMP activity comes from AhR−/− mouse embryo fibroblasts, where loss of AhR results in a reduction in MMP-2 activity (58). However, in this context, there was no discernable change in MMP-2 mRNA expression indicating that the AhR pathway may regulate MMPs differently depending on cellular context. Considering the interplay between the protease cascades, it is possible that in this case the AhR is modifying the proteolytic activation of the MMPs, perhaps via alterations in the serine proteases (Fig. 2).
4. AhR pathway in development
Data from animal model systems demonstrate that activation/inhibition of the AhR pathway results in pathological lesions that result from aberrant matrix metabolism. Embryonic development of several organ systems is disrupted in AhR−/− murine models demonstrating evidence of uncoordinated tissue/matrix remodeling (59,60). For example, AhR−/− animals have smaller livers as a result of massive portosystemic shunting and exhibit a failure in the closure of the ductus venosus (61), a process that requires extensive matrix remodeling. Mammary morphogenesis, which requires the remodeling of the local ECM to facilitate ductal branching, is disrupted in AhR−/− females, displaying a 50% reduction in estrous-induced terminal end buds in comparison to wild-type animals (62). In mammary gland development, the expression of MMP-2, -3, -11 and TIMP-1 are localized to the mammary ducts and mediate ECM remodeling (63). TIMP-1 and MMP-3 are also important for proper murine mammary development in that the down regulation of TIMP-1 or over-expression of MMP-3 results in enhanced budding and growth of mammary ducts (64). These data suggest that one of the endogenous functions of AhR signaling may be to coordinate tissue morphogenesis in mammals.
Activation of the AhR pathway by exogenous ligands also results in the deregulation of tissue morphogenesis. Postnatal development of the seminal vesicles, including vesicle branching and differentiation, a process that requires coordinated matrix remodeling, is reduced in rats exposed to TCDD in utero (65). Similar phenotypes are observed in C57BL/6 mice following in utero and lactational exposure to TCDD and also display a loss of AhR expression (66). Mammary gland morphogenesis is also altered following exposure to TCDD similar to that observed in the AhR knockout animals. In weaning rats, TCDD exposure results in decreased tubule branching (67) and TCDD exposure of pregnant mice impairs mammary gland development and lactation (68). Mice exposed to TCDD in utero also develop cleft palate, which is thought to result from insufficient differentiation of the epithelial cells of the palatal shelves (69). Cleft clitoris and incomplete vaginal opening are also observed in female rats exposed in utero to TCDD (70,71).
AhR-mediated developmental alterations in matrix remodeling is not limited to mammalian models: development in zebrafish (Danio rerio) is also disrupted by exposure to TCDD. In zebrafish, TCDD toxicity is mediated through the zebrafish AhR2 and Arnt1, similar to mammalian systems (72). Exposure of zebrafish embryos to TCDD results in a variety of defects including craniofacial malformations, pericardial and yolk sac edema, uninflated swim bladders, cardiovascular dysfunction and abnormal heart looping (73). Interestingly, TCDD embryonic exposure of zebrafish results in a failure of the common cardinal vein to migrate resulting in heart elongation (74). This TCDD-induced lesion is dependant on the AhR and is reminiscent of the abnormal fetal vascular structures observed in AhR knockout mice (75).
In addition to effects on ECM remodeling during the development, the AhR pathway also appears to be important for wound repair and regeneration, processes that require extensive remodeling. The zebrafish has been an extremely useful model in studying epimorphic regeneration, as zebrafish are capable of fully regenerating their tailfins if amputated. Regeneration occurs in three distinct phases: 1) wound healing and the formation of an epithelial wound layer at the site of injury, 2) blastema formation directly underneath the wound cap and the 3) regenerative outgrowth phase that progresses with the proliferation, differentiation, and apoptosis of blastemal cells and are dependant on angiogenesis and nerve innervation. The whole process occurs within 14 days and results in a completely regenerated tailfin, and this process depends on proper protease expression and activity (reviewed in (76)). It is therefore not surprising that proper ECM remodeling is requisite for normal regeneration to occur. For example, the plasminogen activator/plasmin system has been recognized as a key player in wound healing by regulating the expression of MMP’s, TGF-β, TIMP-1, fibronectin, collagen, and uPA. In the zebrafish, MMP-2, MT1-MMP, and TIMP-2 were found to be expressed and activated in the blastema and wound epithelium of the regenerating tailfin; inhibition of MMPs or MT1-MMP specifically, results in the impairment of blastema and epithelial wound layer formation (77).
Since TCDD exposure and corresponding AhR activation result in the inhibition of tailfin regeneration, AhR has been implicated to be a modulator of this dynamic regenerative process by conducting the misregulation of ECM remodeling, thus hindering the maturation of the ECM, as evidenced by microarray analysis of TCDD exposed fish during tailfin regeneration (54). Both zfAhR2 and zfARNT1 mediate TCDD induced tailfin inhibition, since morpholino knockdown of zfAhR2 or zfARNT1 in TCDD treated larval zebrafish restored tailfin regeneration (78). Further investigations based on the previous microarray analysis identified deficiencies in collagen and proteoglycan localization and reduced neovascularization and neuronal regeneration to be potential physiological manifestations of TCDD induced, AhR mediated, ECM deregulation (79). AhR activation in regenerating tailfins also resulted in the activation of the Wnt signaling pathway, suggesting possible crosstalk between the AhR and Wnt pathways (80). Wnt/β-catenin signaling activation has been previously shown to be a negative regulator of zebrafish tailfin regeneration and is essential for all stages of regeneration (81). Upregulation of R-Spondin1, a Wnt coreceptor, and LRP6, the Wnt core-receptor and potential target for R-Spondin1, was determined to mediated TCDD induced inhibition of tailfin regeneration, as either R-Spondin1 or LRP6 knockdown restored tailfin regeneration in the presence of TCDD despite AhR activation (80).
Angiogenesis, the outgrowth of new blood vessels from existing vasculature, is a process dependant on matrix remodeling (reviewed in (82)). In both AhR knockout mice and TCDD-exposed zebrafish, fetal vascular structures are improperly remodeled, resulting in abnormal heart and liver development (74,75). Altered vascular remodeling is also observed following TCDD exposure in the chick embryo resulting in reduced number and size of differentiated coronary arteries (83) and decreased expression and secretion of cardiac vascular endothelial growth factor (VEGF). Interestingly, the AhR agonist 3-methylcholanthrene inhibits angiogenesis of human umbilical vascular endothelial cells (29). Taken together, these findings strongly suggest a role for AhR signaling in inhibiting the angiogenic pathway through alterations in matrix remodeling.
5. Molecular mechanisms of AhR-induced changes in adhesion and matrix remodeling
At this time, the mechanism(s) of TCDD-induced changes in ECM remodeling is not well-understood, although data indicate that these processes are dependent on the AhR-signaling pathway. In some cases, the AhR has a direct effect on gene transcription through binding to XREs in the promoter, where as in other cases AhR-alterztions in gene expression occur through stabilization of the mRNA (49). In other cases the mechanism is not direct, and involves the interaction with other signaling pathways. Many of the pathways identified as interacting with the AhR also have an impact on matrix remodeling. Therefore, these pathway interactions may be critical for modulating AhR-induced changes in matrix remodeling, and may contribute to cell-type specific differences in gene expression.
5.1 The estrogen receptor pathway
Interactions between the AhR and ER pathway are well documented and suggest that crosstalk between these two pathways is critical for AhR-induced effects in estrogen-sensitive tissues (reviewed in (84) (Fig. 3). The effect of AhR and ER pathway interaction on matrix remodeling has been extensively studied in the developing mammary gland. During the development of the mammary gland, high levels of AhR protein are present in C57BL/6J mice during estrous-stimulation. AhR null mice display a reduced number of terminal end buds (TEBs) and altered shape of the mammary glands as compared to wild-type littermates (62). Similarly, the ERα knockout mice exhibit severely underdeveloped glands lacking TEB formation and tubule branching demonstrating that development of the mammary gland is also dependent on ERα (85). It is interesting to note that in mammary gland organ culture, AhR protein has been shown to be concentrated around developing lobules implicating AhR’s involvement in proper mammary development (62). Interestingly, similar TEB phenotypes are observed in mouse mammary glands where MMP expression was impaired (86) suggesting that the ultimate function for both these pathways may be to regulate the appropriate temporal and spatial expression of the MMPs. Taken together, the mammary phenotype observed in the AhR null mouse and the AhR localization around developing lobules suggests that ER-mediated mammary development is dependent on AhR signaling.
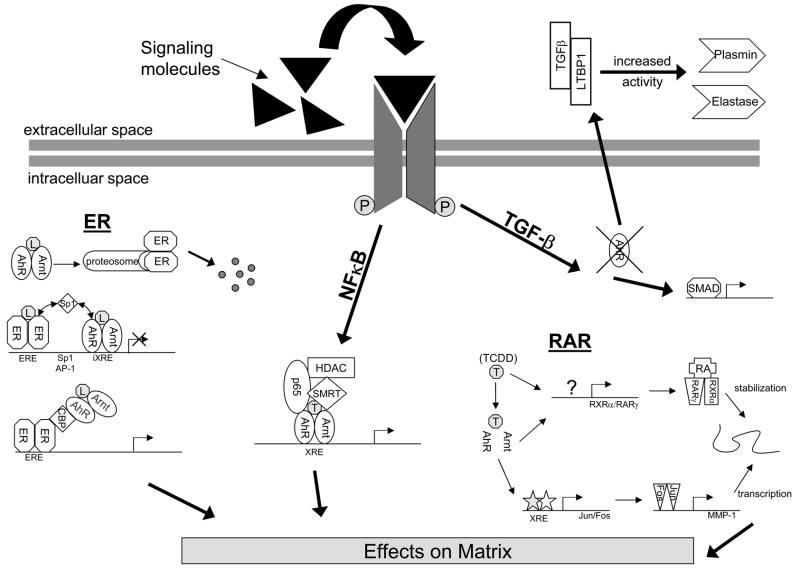
Figure 3. The AhR pathway can alter matrix metabolism and deposition through interactions with other signaling pathways.
Depicted are some of the processes known to take place between the AhR pathway; NFκB, Estrogen Receptor (ER), Retinoic Acid Receptor (RAR) and Transforming Growth Factor β (TGFβ) pathways.
The direct interaction of ERα/AhR in MCF-7 cells offers a feasible theory for the effects seen above in TEB formation. It has been shown that in mesengial cells, an increase in 17β-estradiol (E2)-induced MMP-9 expression and activity is associated with increases in both ERα and ERβ expression (87). Further, E2 treatment increases the expression activity of MMP-2 in human retinal pigment epithelial cells (88). E2 exposure along with over-expression of ERα in endometrial carcinoma cells resulted in increased expression of MMP-1, -7 and -9 and invasiveness (89). These findings demonstrate a direct link between estrogen treatment and modulation of MMP expression while also suggesting that estrogen receptors may be required for this action.
5.2 The NF-kB pathway
Nuclear factor-κB (NF-κB) is a member of a family of transcription factors defined by a common Rel homology domain (RHD). Members of the NF-κB family dimerize through the RHD to form either transcriptionally active or inactive dimers. RelB, c-Rel and p65 are mammalian family members that can form transcriptionally active dimers, while p50 and p52 lack a transactivation domain. NF-κB dimers are cytoplasmic until activation occurs resulting in translocation into the nucleus (reviewed in (90)). The NF-κB family had been shown to regulate inflammatory response, apoptosis, immune responses, development, environmental stress response, osteogenesis and cell growth and is activated in a variety of disease that involve inappropriate MMP activity, such as arthritis, neurodegenerative diseases and cancer.
The NF-κB pathway has been shown to interact with the AhR pathway in many different studies, some of which are highlighted below (reviewed in ())(Fig. 3). Benzo-α-pyrene (BAP), an AhR agonist known to inhibit osteoclast differentiation and bone resorption, caused enhanced co-immunoprecipitation of AhR and p65 in a mouse macrophage cell line (RAW 264.7) furthering indicating that interactions of AhR and NFκ-B may control remodeling events.
NF-κB regulation of MMPs is a key regulatory step in ECM degradation in response to cytokines, growth factors and oxidative stress (91). Interleukin-1 (IL-1) and tumor necrosis factor-α (TNF-α) are two well studied stimulators of MMP production and IL-1-induced MMP-1 expression was recently seen to require both ERK and NF-kB signaling (51,92). Since activation of AhR has been shown to require ERK signaling in some cells and NF-κB has been shown to be directly associated with AhR in certain cell types, it is possible that AhR may also be involved in IL-1 and TNF-α stimulation of MMPs (93).
5.3 The retinoic acid signaling pathway
Data from a variety of animal and cell culture models have demonstrated interactions between the AhR and retinoic acid signaling pathways (reviewed in (94)) (Fig. 3). Dietary derived all-trans RA (atRA) is the main signaling retinoid in the body and is vital for biological functions such as embryogenesis, growth and differentiation and reproduction which require extensive matrix remodeling.
Interactions between the RA and AhR signaling pathways include changes in the availability of atRA in the liver and extrahepatic tissues by AhR-mediated regulation of atRA synthesis and metabolism, as well as on storage and transport (94). Further, these two signaling pathways directly impact each other through alterations in receptor availability and modulation of transcriptional regulation. Although it appears that their intersection may be mediated by specific co-activator and co-repressor proteins, the exact mechanism is yet undefined. However, it is clear that a portion of toxicity related to TCDD and related congeners are mediated through their effect on RA homeostasis and on the atRA-signaling pathway.
Data from our laboratory demonstrate that co-treatment of normal human keratinocytes with atRA and TCDD results in enhanced of MMP-1 expression over exposure to TCDD alone (52). atRA and TCDD co-treatment also resulted in the enhanced expression of PAI-2, indicating that atRA/TCDD co-activation is not limited to MMPs. The induction of MMP-1 by co-treatment with atRA and TCDD does not rely on transcriptional interaction between the RARs and AhR, but instead is mediated through two distinct mechanisms: TCDD-induced transcription of MMP-1 and atRA-enhancement of MMP-1 mRNA stability. It is interesting to note that TCDD exposure in normal human keratinocytes does result in an increase in RARγ and RXRα expression and may facilitate the effects of atRA in these cells (52). Expression of a dominant negative RARα mutant resulted in enlarged terminal end buds, increased branching and an accompanying increase in MMP-3 expression in developing murine mammary glands (95), similar to the phenotype observed in AhR knockout animals, suggesting that TCDD/AhR alterations in the RA-signaling pathway may be involved in the abnormal mammary development through disruption of the MMPs.
5.4 The TGF-β pathway
Recently, the theory of the AhR interacting with various different molecular pathways has lead to the theory of the AhR as a modulator of cellular processes instead of solely a ligand-activated transcription factor acting upon downstream targets. Transforming growth factor β (TGF-β) belongs to a family of cytokines that are involved in proliferation, growth and matrix deposition and turnover (96). Latent TGF-β is bound to TGF-β binding protein-1 (LTBP-1) and is sequestered in ECM matrix until released by proteases (96). Recent data indicate that there is an interaction between the AhR pathway and TGF-β signaling (reviewed in(97)). TGF-β levels are elevated in mouse embryonic fibroblasts from AhR null mice (AhR−/− MEF) and recent data show that LTBP-1 is essential for the increased TGF-β 1 in AhR−/− MEF cells. Fruthermore, it is also responsible for activity of two proteases involved in TGF-β release, plasmin and elastase. Over-expression of AhR in AhR−/− MEFcells resulted in increased proliferation, independent of ligand activation and TGF-β 1 levels were reduced in the media of the over expressing cells as compared to the AhR−/− MEF cells. (Fig. 3).
Future Considerations
Despite the variety of ligands for the AhR, it is essentially an orphan receptor since its endogenous ligand(s) has not been definitively identified (2). Although this status has not diminished enthusiasm for study in the AhR pathway, the apparent lack of an endogenous ligand, and of an endogenous function, has stalled our understanding of this pathway in its normal context, and therefore in disease. Data from knockout mice suggest that the AhR pathway is involved in mediating functional development in several tissues, as discussed above. In addition, data also suggest that inappropriate activation of the AhR influences disease progression, particularly in the case of tumor progression (98). The findings outlined in this review demonstrating that the AhR alters adhesion and matrix degradation and enhances migration and invasion suggest that the AhR contributes to tumor progression, at least in part, by altering cell-cell and cell-matrix interactions. This is supported by the findings that invasive tumor cell types express higher levels of AhR than their untransformed counterparts (53,99), and that the inhibitor of the AhR pathway, the AhR repressor (AhRR) was recently identified as a tumor suppressor in several tumor types (100).
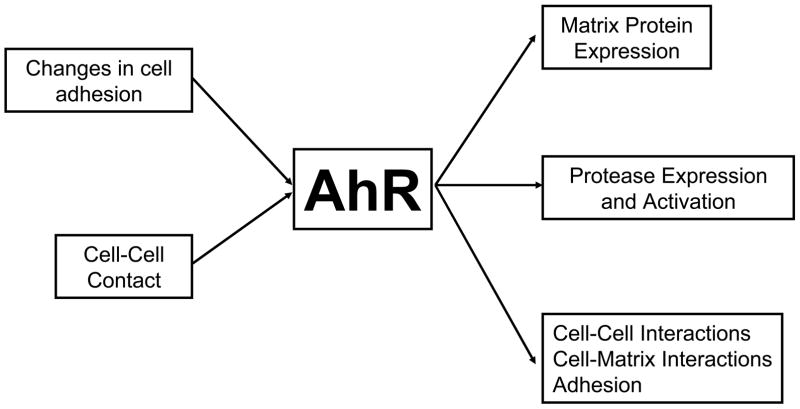
One important consideration for understanding the effect of the AhR pathway in disease is that this pathway is also a target for signaling events induced by alterations in cell-cell and cell-matrix interactions. Therefore, we propose that the AhR is a central modifying pathway in the cellular response to changes in cell adhesion (Fig. 4). This hypothesis is supported by AhR knockout animal models demonstrating altered structural development, and by cell culture demonstrating the direct impact of the AhR pathway on the expression of proteins involved in cell adhesion and matrix metabolism. Therefore, disruption and activation of this pathway by exogenous agents will influence cellular behavior and interactions, which will modify cellular response to other signals.
Figure 4. The AhR as a central regulator of cell adhesion and matrix metabolism.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barouki R, Coumoul X, Fernandez-Salguero PM. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen LP, Bradfield CA. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hankinson O. Annual Review of Pharmacology & Toxicology. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 5.Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, Sherr DH, Tilly JL. Nat Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- 6.Kolluri SK, Weiss C, Koff A, Gottlicher M. Genes Dev. 1999;13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H, Li Y, Sutter TR. Experimental & Clinical Immunogenetics. 1994;11:128–135. doi: 10.1159/000424203. [DOI] [PubMed] [Google Scholar]
- 8.Jeon MS, Esser C. J Immunol. 2000;165:6975–6983. doi: 10.4049/jimmunol.165.12.6975. [DOI] [PubMed] [Google Scholar]
- 9.Hoffer A, Chang CY, Puga A. Toxicology & Applied Pharmacology. 1996;141:238–247. doi: 10.1006/taap.1996.0280. [DOI] [PubMed] [Google Scholar]
- 10.Cho YC, Zheng W, Jefcoate CR. Toxicol Appl Pharmacol. 2004;199:220–238. doi: 10.1016/j.taap.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Sadek CM, Weitzel MA, Allen-Hoffmann BL. Radiation Oncology Investigations 1996 [Google Scholar]
- 12.Larsen MC, Brake PB, Pollenz RS, Jefcoate CR. Toxicol Sci. 2004;82:46–61. doi: 10.1093/toxsci/kfh242. [DOI] [PubMed] [Google Scholar]
- 13.Weiss C, Faust D, Schreck I, Ruff A, Farwerck T, Melenberg A, Schneider S, Oesch-Bartlomowicz B, Zatloukalova J, Vondracek J, Oesch F, Dietrich C. Oncogene. 2008;27:2198–2207. doi: 10.1038/sj.onc.1210859. [DOI] [PubMed] [Google Scholar]
- 14.Andrysik Z, Vondracek J, Machala M, Krcmar P, Svihalkova-Sindlerova L, Kranz A, Weiss C, Faust D, Kozubik A, Dietrich C. Mutat Res. 2007;615:87–97. doi: 10.1016/j.mrfmmm.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Hoelper P, Faust D, Oesch F, Dietrich C. Arch Toxicol. 2005;79:201–207. doi: 10.1007/s00204-004-0624-6. [DOI] [PubMed] [Google Scholar]
- 16.Daley WP, Peters SB, Larsen M. J Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 17.Riecke K, Grimm D, Shakibaei M, Kossmehl P, Schulze-Tanzil G, Paul M, Stahlmann R. Arch Toxicol. 2002;76:360–366. doi: 10.1007/s00204-002-0338-6. [DOI] [PubMed] [Google Scholar]
- 18.Nottebrock C, Riecke K, Kruse M, Shakibaei M, Stahlmann R. Toxicology. 2006;226:197–207. doi: 10.1016/j.tox.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Thackaberry EA, Jiang Z, Johnson CD, Ramos KS, Walker MK. Toxicol Sci. 2005;88:231–241. doi: 10.1093/toxsci/kfi301. [DOI] [PubMed] [Google Scholar]
- 20.Aragon AC, Kopf PG, Campen MJ, Huwe JK, Walker MK. Toxicol Sci. 2008;101:321–330. doi: 10.1093/toxsci/kfm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan MR, Humphries MJ, Bass MD. Nat Rev Mol Cell Biol. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prell RA, Kerkvliet NI. J Immunol. 1997;158:2695–2703. [PubMed] [Google Scholar]
- 23.Riecke K, Schmidt A, Stahlmann R. Arch Toxicol. 2003;77:358–364. doi: 10.1007/s00204-003-0445-z. [DOI] [PubMed] [Google Scholar]
- 24.McMillan BJ, McMillan SN, Glover E, Bradfield CA. J Biol Chem. 2007;282:12590–12597. doi: 10.1074/jbc.M611446200. [DOI] [PubMed] [Google Scholar]
- 25.Monteiro P, Gilot D, Le Ferrec E, Lecureur V, N’Diaye M, Le Vee M, Podechard N, Pouponnot C, Fardel O. Biochem Biophys Res Commun. 2007;358:442–448. doi: 10.1016/j.bbrc.2007.04.111. [DOI] [PubMed] [Google Scholar]
- 26.Abe Y, Sinozaki H, Takagi T, Minegishi T, Kokame K, Kangawa K, Uesaka M, Miyamoto K. Reprod Biol Endocrinol. 2006;4:27. doi: 10.1186/1477-7827-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morito N, Yoh K, Fujioka Y, Nakano T, Shimohata H, Hashimoto Y, Yamada A, Maeda A, Matsuno F, Hata H, Suzuki A, Imagawa S, Mitsuya H, Esumi H, Koyama A, Yamamoto M, Mori N, Takahashi S. Cancer Res. 2006;66:812–819. doi: 10.1158/0008-5472.CAN-05-2154. [DOI] [PubMed] [Google Scholar]
- 28.Backlund M, Ingelman-Sundberg M. Cell Signal. 2005;17:39–48. doi: 10.1016/j.cellsig.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Juan SH, Lee JL, Ho PY, Lee YH, Lee WS. Eur J Pharmacol. 2006;530:1–8. doi: 10.1016/j.ejphar.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Halbleib JM, Nelson WJ. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 31.Yan Q, Zhang ZF, Chen XP, Gutmann DH, Xiong M, Xiao ZY, Huang ZY. Int J Oncol. 2008;32:1057–1063. [PubMed] [Google Scholar]
- 32.Niermann T, Schmutz S, Erne P, Resink T. Biochem Biophys Res Commun. 2003;300:943–949. doi: 10.1016/s0006-291x(02)02970-4. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich C, Faust D, Moskwa M, Kunz A, Bock KW, Oesch F. Int J Cancer. 2003;103:435–439. doi: 10.1002/ijc.10830. [DOI] [PubMed] [Google Scholar]
- 34.Yin T, Green KJ. Semin Cell Dev Biol. 2004;15:665–677. doi: 10.1016/j.semcdb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Kanazawa Y, Ueda Y, Shimasaki M, Katsuda S, Yamamoto N, Tomita K, Tsuchiya H. Anticancer Res. 2008;28:655–664. [PubMed] [Google Scholar]
- 36.Chesire DR, Dunn TA, Ewing CM, Luo J, Isaacs WB. Cancer Res. 2004;64:2523–2533. doi: 10.1158/0008-5472.can-03-3309. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, He X. Curr Opin Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palermo C, Joyce JA. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Pocar P, Klonisch T, Brandsch C, Eder K, Frohlich C, Hoang-Vu C, Hombach-Klonisch S. Toxicol Sci. 2006;89:408–414. doi: 10.1093/toxsci/kfj042. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Samudio I, Safe S. Mol Cell Endocrinol. 2001;172:91–103. doi: 10.1016/s0303-7207(00)00379-8. [DOI] [PubMed] [Google Scholar]
- 41.Kohle C, Hassepass I, Bock-Hennig BS, Walter Bock K, Poellinger L, McGuire J. Arch Biochem Biophys. 2002;402:172–179. doi: 10.1016/S0003-9861(02)00076-0. [DOI] [PubMed] [Google Scholar]
- 42.Qiu D, Owen K, Gray K, Bass R, Ellis V. Biochem Soc Trans. 2007;35:583–587. doi: 10.1042/BST0350583. [DOI] [PubMed] [Google Scholar]
- 43.Son DS, Rozman KK. Arch Toxicol. 2002;76:404–413. doi: 10.1007/s00204-002-0354-6. [DOI] [PubMed] [Google Scholar]
- 44.Mizuyachi K, Son DS, Rozman KK, Terranova PF. Reprod Toxicol. 2002;16:299–307. doi: 10.1016/s0890-6238(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 45.Gohl G, Lehmkoster T, Munzel PA, Schrenk D, Viebahn R, Bock KW. Carcinogenesis. 1996;17:443–449. doi: 10.1093/carcin/17.3.443. [DOI] [PubMed] [Google Scholar]
- 46.Yang JH. Biochem Biophys Res Commun. 1999;257:259–263. doi: 10.1006/bbrc.1999.0451. [DOI] [PubMed] [Google Scholar]
- 47.Ahn NS, Hu H, Park JS, Kim JS, An S, Kong G, Aruoma OI, Lee YS, Kang KS. Mutat Res. 2005;579:189–199. doi: 10.1016/j.mrfmmm.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 48.Gaido KW, Maness SC. Toxicol Appl Pharmacol. 1995;133:34–42. doi: 10.1006/taap.1995.1124. [DOI] [PubMed] [Google Scholar]
- 49.Shimba S, Hayashi M, Sone H, Yonemoto J, Tezuka M. Biochem Biophys Res Commun. 2000;272:441–448. doi: 10.1006/bbrc.2000.2789. [DOI] [PubMed] [Google Scholar]
- 50.Visse R, Nagase H. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Mol Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 52.Murphy KA, Villano CM, Dorn R, White LA. J Biol Chem. 2004;279:25284–25293. doi: 10.1074/jbc.M402168200. [DOI] [PubMed] [Google Scholar]
- 53.Villano CM, Murphy KA, Akintobi A, White LA. Toxicol Appl Pharmacol. 2006;210:212–224. doi: 10.1016/j.taap.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Andreasen EA, Mathew LK, Tanguay RL. Toxicol Sci. 2006;92:254–269. doi: 10.1093/toxsci/kfj118. [DOI] [PubMed] [Google Scholar]
- 55.Vogel CF, Sciullo E, Matsumura F. Cardiovasc Toxicol. 2004;4:363–373. doi: 10.1385/ct:4:4:363. [DOI] [PubMed] [Google Scholar]
- 56.Igarashi TM, Bruner-Tran KL, Yeaman GR, Lessey BA, Edwards DP, Eisenberg E, Osteen KG. Fertil Steril. 2005;84:67–74. doi: 10.1016/j.fertnstert.2005.01.113. [DOI] [PubMed] [Google Scholar]
- 57.Haque M, Francis J, Sehgal I. Cancer Lett. 2005;225:159–166. doi: 10.1016/j.canlet.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 58.Santiago-Josefat B, Mulero-Navarro S, Dallas SL, Fernandez-Salguero PM. J Cell Sci. 2004;117:849–859. doi: 10.1242/jcs.00932. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez FJ, Fernandez-Salguero P, Lee SS, Pineau T, Ward JM. Toxicol Lett. 1995;82–83:117–121. doi: 10.1016/0378-4274(95)03548-6. [DOI] [PubMed] [Google Scholar]
- 61.Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Proc Natl Acad Sci U S A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hushka LJ, Williams JS, Greenlee WF. Toxicol Appl Pharmacol. 1998;152:200–210. doi: 10.1006/taap.1998.8508. [DOI] [PubMed] [Google Scholar]
- 63.Lee PP, Hwang JJ, Mead L, Ip MM. J Cell Physiol. 2001;188:75–88. doi: 10.1002/jcp.1090. [DOI] [PubMed] [Google Scholar]
- 64.Fata JE, Leco KJ, Moorehead RA, Martin DC, Khokha R. Dev Biol. 1999;211:238–254. doi: 10.1006/dbio.1999.9313. [DOI] [PubMed] [Google Scholar]
- 65.Hamm JT, Sparrow BR, Wolf D, Birnbaum LS. Toxicol Sci. 2000;54:424–430. doi: 10.1093/toxsci/54.2.424. [DOI] [PubMed] [Google Scholar]
- 66.Lin TM, Ko K, Moore RW, Simanainen U, Oberley TD, Peterson RE. Toxicol Sci. 2002;68:479–487. doi: 10.1093/toxsci/68.2.479. [DOI] [PubMed] [Google Scholar]
- 67.Brown NM, Lamartiniere CA. Environ Health Perspect. 1995;103:708–713. doi: 10.1289/ehp.95103708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vorderstrasse BA, Fenton SE, Bohn AA, Cundiff JA, Lawrence BP. Toxicol Sci. 2004;78:248–257. doi: 10.1093/toxsci/kfh062. [DOI] [PubMed] [Google Scholar]
- 69.Takagi TN, Matsui KA, Yamashita K, Ohmori H, Yasuda M. Teratog Carcinog Mutagen. 2000;20:73–86. doi: 10.1002/(sici)1520-6866(2000)20:2<73::aid-tcm3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 70.Flaws JA, Sommer RJ, Silbergeld EK, Peterson RE, Hirshfield AN. Toxicol Appl Pharmacol. 1997;147:351–362. doi: 10.1006/taap.1997.8295. [DOI] [PubMed] [Google Scholar]
- 71.Gray LE, Jr, Ostby JS. Toxicol Appl Pharmacol. 1995;133:285–294. doi: 10.1006/taap.1995.1153. [DOI] [PubMed] [Google Scholar]
- 72.Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Toxicol Sci. 2002;68:403–419. doi: 10.1093/toxsci/68.2.403. [DOI] [PubMed] [Google Scholar]
- 73.Antkiewicz DS, Burns CG, Carney SA, Peterson RE, Heideman W. Toxicol Sci. 2005;84:368–377. doi: 10.1093/toxsci/kfi073. [DOI] [PubMed] [Google Scholar]
- 74.Bello SM, Heideman W, Peterson RE. Toxicol Sci. 2004;78:258–266. doi: 10.1093/toxsci/kfh065. [DOI] [PubMed] [Google Scholar]
- 75.Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. Mol Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- 76.Gill SE, Parks WC. Int J Biochem Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bai S, Thummel R, Godwin AR, Nagase H, Itoh Y, Li L, Evans R, McDermott J, Seiki M, Sarras MP., Jr Matrix Biol. 2005;24:247–260. doi: 10.1016/j.matbio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 78.Mathew LK, Andreasen EA, Tanguay RL. Mol Pharmacol. 2006;69:257–265. doi: 10.1124/mol.105.018044. [DOI] [PubMed] [Google Scholar]
- 79.Andreasen EA, Mathew LK, Lohr CV, Hasson R, Tanguay RL. Toxicol Sci. 2007;95:215–226. doi: 10.1093/toxsci/kfl119. [DOI] [PubMed] [Google Scholar]
- 80.Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguay RL. Faseb J. 2008 doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 82.Rundhaug JE. J Cell Mol Med. 2005;9:267–285. doi: 10.1111/j.1582-4934.2005.tb00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ivnitski I, Elmaoued R, Walker MK. Teratology. 2001;64:201–212. doi: 10.1002/tera.1065. [DOI] [PubMed] [Google Scholar]
- 84.Safe S, Wormke M, Samudio I. J Mammary Gland Biol Neoplasia. 2000;5:295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- 85.Bocchinfuso WP, Korach KS. J Mammary Gland Biol Neoplasia. 1997;2:323–334. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- 86.Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, Werb Z. J Cell Biol. 2003;162:1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Potier M, Elliot SJ, Tack I, Lenz O, Striker GE, Striker LJ, Karl M. J Am Soc Nephrol. 2001;12:241–251. doi: 10.1681/ASN.V122241. [DOI] [PubMed] [Google Scholar]
- 88.Marin-Castano ME, Elliot SJ, Potier M, Karl M, Striker LJ, Striker GE, Csaky KG, Cousins SW. Invest Ophthalmol Vis Sci. 2003;44:50–59. doi: 10.1167/iovs.01-1276. [DOI] [PubMed] [Google Scholar]
- 89.Mizumoto H, Saito T, Ashihara K, Nishimura M, Tanaka R, Kudo R. Int J Cancer. 2002;100:401–406. doi: 10.1002/ijc.10504. [DOI] [PubMed] [Google Scholar]
- 90.Tian Y. Biochemical Pharmacology. 2008;X:XX–XX. [Google Scholar]
- 91.Ghosh S, May MJ, Kopp EB. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 92.Fan Z, Yang H, Bau B, Soder S, Aigner T. Rheumatol Int. 2006;26:900–903. doi: 10.1007/s00296-006-0114-7. [DOI] [PubMed] [Google Scholar]
- 93.Tan Z, Chang X, Puga A, Xia Y. Biochem Pharmacol. 2002;64:771–780. doi: 10.1016/s0006-2952(02)01138-3. [DOI] [PubMed] [Google Scholar]
- 94.Murphy KA, Quadro L, White LA. Vitam Horm. 2007;75:33–67. doi: 10.1016/S0083-6729(06)75002-6. [DOI] [PubMed] [Google Scholar]
- 95.Wang YA, Shen K, Wang Y, Brooks SC. Dev Dyn. 2005;234:892–899. doi: 10.1002/dvdy.20570. [DOI] [PubMed] [Google Scholar]
- 96.Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J. Microsc Res Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 97.Fernandez-Salguero P. Biochemical Pharmacology. 2008;XX:XX–XX. [Google Scholar]
- 98.Safe S. Toxicol Lett. 2001;120:1–7. doi: 10.1016/s0378-4274(01)00301-0. [DOI] [PubMed] [Google Scholar]
- 99.Yang X, Liu D, Murray TJ, Mitchell GC, Hesterman EV, Karchner SI, Merson RR, Hahn ME, Sherr DH. Oncogene. 2005;24:7869–7881. doi: 10.1038/sj.onc.1208938. [DOI] [PubMed] [Google Scholar]
- 100.Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martinez A, Narayan G, Kirsch I, Franklin W, Hirsch F, Birrer M, Cuttitta F. J Clin Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]