Abstract
Type 2 diabetes is a disorder of dysregulated glucose homeostasis. Normal glucose homeostasis is a complex process involving several interacting mechanisms, such as insulin secretion, insulin sensitivity, glucose production, and glucose uptake. The dysregulation of one or more of these mechanisms due to environmental and/or genetic factors, can lead to a defective glucose homeostasis. Hyperglycemia is managed by augmenting insulin secretion and/or interaction with hepatic glucose production, as well as by decreasing dietary caloric intake and raising glucose metabolism through exercise. Although these interventions can delay disease progression and correct blood glucose levels, they are not able to cure the disease or stop its progression entirely. Better management of type 2 diabetes is sorely needed. Advances in genotyping techniques and the availability of large patient cohorts have made it possible to identify common genetic variants associated with type 2 diabetes through genome-wide association studies (GWAS). So far, genetic variants on 19 loci have been identified. Most of these loci contain or lie close to genes that were not previously linked to diabetes and they may thus harbor targets for new drugs. It is also hoped that further genetic studies will pave the way for predictive genetic screening. The newly discovered type 2 diabetes genes can be classified based on their presumed molecular function, and we discuss the relation between these gene classes and current treatments. We go on to consider whether the new genes provide opportunities for developing alternative drug therapies.
Key Words: Type 2 diabetes, drug targets, genetics, personalized medicine.
INTRODUCTION
The last few decades have witnessed a dramatic increase in the prevalence of type 2 diabetes mellitus, due to changes in food intake combined with less physical exercise, a lifestyle often referred to as Western. The long-term consequences of type 2 diabetes are severe and include cardiovascular disease, retinopathy, neuropathy, nephropathy, and diabetic foot disease. While it has been estimated that worldwide around one billion people are obese, over 180 million people suffer from type 2 diabetes and this number is expected to double over the next 25 years. The number of annual deaths due to type 2 diabetes is difficult to estimate because they are often hidden under cardiovascular disease, but the WHO has estimated it to be between 1 and 3 million in 2006. However, since mortality from this disease often occurs many years after its onset, even 3 million deaths is probably an underestimate of the death toll in the near future. Although external factors such as food-intake-related obesity has attracted much attention, the genetic predisposition for diabetes is also important. While the life-time risk for type 2 diabetes in the Western world is around 10%, first-degree relatives of patients have a 20–40% risk for the disease, and concordance rates for identical twins have been estimated to be 57% or higher (up to 90%) for type 2 diabetes in male twins [1]. The endeavor to find the underlying genes was unsuccessful for many years although hundreds of genetic associations have been described, based largely on candidate genes. Unfortunately, replication was only possible for a very few variants. However, the recent advent of genome-wide association studies (GWAS) holds promise, since such studies have now uncovered a number of common genetic variants related to diabetes for which replication is also possible. These genes had not previously been linked to diabetes and these loci are therefore expected to lead to new insights into the disease mechanisms.
GLUCOSE HOMEOSTASIS AND DIABETES
Diabetes is a state of persistent hyperglycemia, leading to irreversible damage in a number of tissues, especially the retina, the kidney glomeruli, neural tissue and blood vessels. Normally plasma glucose levels are kept within a narrow range, a process referred to as glucose homeostasis. This homeostasis is regulated in a complex way and involves several axes.
After eating a meal, a person’s plasma glucose rises temporarily, after which glucose is rapidly taken up by liver and muscle and to a lesser degree by fat tissue, due to the effects of insulin released from the pancreatic β-cells. Under these circumstances, glucose is converted into glycogen, which is then stored in the liver. When plasma glucose levels have dropped, insulin secretion gradually diminishes. In the postprandial state, a minimum level of plasma glucose is assured by glucose production in the liver, partly due to release from glycogen and partly due to new formation from precursors such as lactate and the amino acids alanine and glutamine. Insulin has a number of other effects, including lipid storage in adipocytes, and it even increases DNA replication and protein synthesis. The regulation of insulin release is complex; apart from the prevailing plasma glucose level, release from various peptides from the intestine (e.g., glucagon-like peptide 1, GLP1) after a meal and feedback mechanisms ensure adequate insulin release.
Type 2 diabetes is characterized by the combination of disturbances in insulin secretion, and an impairment of the effects of insulin, so-called insulin resistance, which is often related to obesity. Insulin resistance is caused by defects in the signaling pathways that process the insulin signal in its target tissues. It has often been stated that the disease starts with insulin resistance accompanied by raised insulin and glucose levels. At a later stage, we presume β-cells undergo further damage and apoptosis, and are not able to keep up with the demand for insulin release, which then results in higher glucose levels. At least one of the pathways involved in insulin secretion, β-cell regeneration, β-cell survival, or β-cell development are important in determining the vulnerability of the β-cell pool in insulin-resistant conditions. Studies have shown decreases in β-cell function prior to the augmentation of plasma glucose in normal glucose-tolerant first-degree relatives of type 2 diabetes subjects; in these studies these relatives had normal insulin sensitivity [2, 3]. While obesity is a major risk factor for diabetes – around 50% of obese subjects will develop type 2 diabetes at some stage (depending on the age when they became obesity) –, it should be noted that 20% of type 2 diabetes subjects are not obese. It thus seems that obesity is a major risk factor for developing type 2 diabetes, but that it is the vulnerability of the β-cell pool which determines whether obesity in fact triggers type 2 diabetes.
GENETICS OF TYPE 2 DIABETES
Although monogenic forms of diabetes have been found (Table 1) (reviewed in [4]), the majority of cases of type 2 diabetes do not show inheritance as a Mendelian trait, but rather as a genetically complex disorder in which genetic variants predispose individuals to develop the disease. Twin studies have revealed concordance rates of over 50 % (and reported up to 90%) for type 2 diabetes in identical twins [1], which still leaves a substantial role for environmental factors, such as excess food and limited physical activity. The rapid rise in diabetes prevalence over the last few decades strongly suggests that genetic variants involved in type 2 diabetes are interacting with environmental factors.
Table 1.
Genes Associated with Diabetes: Overview of their Target Tissue1, Function2, and Related Medication3
| Diabetes Gene | Proposed Diabetes Target Cell Type / Tissue1 | Monogenic Diabetes4 | Type 2 Diabetes5 | Proposed Function(s) for Gene Product2 | Drug(s) Affecting the Same Pathway as the Diabetes Gene3 |
|---|---|---|---|---|---|
| ABCC8 | Pancreas β-Cell | X | - B-cell ion homeostasis and insulin secretion; ATP-binding cassette transporter that modulates ATP-sensitive potassium channels and insulin release | Sulfonylurea derivatives | |
| ADAMTS9 | Unknown | X | - Cleavage of proteoglycans | Unknown | |
| CDC123 | Pancreas β-Cell | X | - Cell cycle regulation | Unknown | |
| CDKAL1 | Pancreas β-Cell | X | - Growth and development - Proinsulin to insulin conversion |
Unknown | |
| CDKN2A | Pancreas β-Cell | X | - Cell cycle regulation | Unknown | |
| CEL | Unknown | X | - Glycoprotein that is important in regulation of cholesterol metabolism | Unknown | |
| FTO | Hypothalamus | X | - Associated to obesity | Unknown | |
| GCK | Unknown | X | - Catalyzes reaction from glucose to glucose-6-phosphate | Unknown | |
| HHEX | Pancreas β-Cell | X | - Growth and development; transcription factor | Unknown | |
| HNF4α | Pancreas β-Cell | X | - Growth and development; transcription factor | Unknown | |
| IDE | Pancreas β-Cell | X | - Termination of the response to insulin | Unknown | |
| IGF2BP2 | Pancreas β-Cell | X | - Growth and development | Unknown | |
| JAZF1 | Pancreas β-Cell | X | - Cell cycle regulation; transcriptional repressor | Unknown | |
| KCNJ11 | Pancreas β-Cell | X | X | - B-cell ion homeostasis and insulin secretion | Sulfonylurea derivatives |
| KCNQ1 | Pancreas β-Cell | X | - B-cell ion homeostasis and insulin secretion | Sulfonylurea derivatives | |
| KLF11 | Unknown | X | - Unknown | Unknown | |
| NEUROD1 | Pancreas β-Cell | X | - Growth and development; transcription factor that activates several genes including insulin and is important for early β-cell development | Unknown | |
| NOTCH2 | Pancreas β-Cell | X | - Growth and development; transcription factor; receptor for membrane bound ligands | Unknown | |
| PDX1 | Pancreas β-Cell | X | - Growth and development; nuclear protein that acts as a transcriptional activator of several genes including insulin and is important for early β-cell development | Unknown | |
| PPARG | Adipocyte | X | - Nuclear receptor (transcription factor) that regulates adipocyte differentiation | Thiazolidinediones | |
| SLC30A8 | Pancreas β-Cell | X | - B-cell ion homeostasis and insulin secretion; cellular efflux of Zn2+ ions - Proinsulin to insulin conversion |
Sulfonylurea derivatives | |
| TCF1 | Pancreas β-Cell | X | - Growth and development; Transcription factor that forms a complex with the product of TCF2 important for Wnt signaling | Unknown | |
| TCF2 | Pancreas β-Cell | X | X | - Growth and development; transcription factor that forms a complex with the product of TCF1 important for Wnt signaling - Cell cycle regulation |
Unknown |
| TCF7L2 | Pancreas β-Cell | X | - Wnt signaling - Proinsulin to insulin conversion |
Unknown | |
| THADA | Pancreas β-Cell | X | - Apoptosis | Unknown | |
| TSPAN8 | Unknown | X | - Glycoprotein involved in the mediation of signal transduction | Unknown | |
| WFS1 | Pancreas β-Cell | X | X | - Apoptosis; Endoplasmic Reticulum stress pathway activation | Unknown |
Genes included in the list are involved in type 2 diabetes, Maturity Onset Diabetes of the Young (MODY), or Permanent Neonatal Diabetes Mellitus (PNDM). The cut-off p-value for the inclusion of type 2 diabetes genes identified by GWAS is 1 x 10-8 [12, 21-23]. The third and fourth columns of the table show whether a gene is involved in monogenic4 or complexly inherited type 2 diabetes5.
Candidate-Based Association Studies
Several candidate gene association studies have been performed to identify genes involved in type 2 diabetes. These studies test some of the genetic variants in a gene that is a strong candidate for being involved in the disease. Although a few genes have been identified in this way, in general these studies have not been very successful because the results could not be replicated and the p-values for association of the genetic variants were moderate.
Obviously, the few diabetes genes that were identified and replicated in candidate-based association studies had already been linked to a diabetic phenotype, since the tested genes were mainly selected on the basis of their function and their potential relationship with diabetes. The genes identified by candidate gene approaches were found to be involved in rare, monogenic forms of diabetes. The TCF2 gene was linked to maturity-onset diabetes of the young (MODY) 5 [5], while WFS1 gene mutations led to Wolfram (or DIDMOAD) syndrome, a rare, lethal, neurodegenerative disorder that includes diabetes insipidus, diabetes mellitus, optic nerve degeneration, and inner ear deafness [6]. The KCNJ11 gene is related to permanent neonatal diabetes mellitus (PNDM), a rare form of diabetes starting before the age of 6 months [7]. Finally, mutations in PPARγ can lead to insulin resistance, hypertension, and lipodystrophy [8]. Although the molecular impact of mutations in these genes is not yet fully understood, it is likely that mutations in them will have differing effects and that mutations involved in type 2 diabetes will have a milder impact on human physiology than those involved in monogenic diabetes.
Genome-Wide Association Studies (GWAS)
Recent advances in genotyping techniques and the collection of large, type 2 diabetes patient cohorts have made it possible to perform hypothesis-free genome-wide association studies (GWAS) to identify common genetic variants that increase susceptibility to type 2 diabetes. The human genome harbors around 3 billion base-pairs, which contain at least 3 million common single nucleotide polymorphisms (SNPs) according to the International HapMap Project. However, alleles represented by SNPs that are close together have often stayed on the same chromosome in further generations, forming a so-called haplotype. This implies that when one variant has been typed, we know the genotype of a set of other variants that surround the initial genetic marker; in genetic terminology it is said that the SNPs that stayed together on a chromosome are in high linkage disequilibrium. It has been estimated that, in a Caucasian population, assessing 500,000 SNPs will detect around 80% of the common genetic variation. GWAS typically involve the assessment of such numbers of SNPs determined in large case-control studies for association with a disease or with a so-called “phenotypic trait”, i.e., type 2 diabetes. It should be noted that GWAS identify association of a genetic locus, and not directly of a gene. It is likely that the most associated SNPs in a genomic region are not the causal variant, but that the disease-producing SNP is in high linkage disequilibrium with the “associated” SNPs. It is also possible that a SNP, even when located in a gene, can influence the expression of a nearby gene located several thousand base-pairs or more away [9]. It is therefore difficult to determine which gene is responsible for the association signal in a GWAS with full certainty. A detailed description of a GWAS is reviewed by McCarthy et al. [10].
The genome-wide approach has been very successful for type 2 diabetes, leading to the identification of over a dozen common genetic variants associated with the disease lying near genes that had not previously been associated with a diabetic phenotype (Table 1) [11-24]. In the near future this number will probably increase and a large part of the genetic variation that confers susceptibility to type 2 diabetes will be unveiled. At present, the total effect of the identified genetic variants explains only a small part of the total estimated genetic variation which is mainly due to the small effect from each of the identified variants (with odds ratios around 1.2 for each gene, which implies a mere 20% extra risk for the disease) [25, 26]. Identifying more genetic variants - by including more cases to increase the statistical power of GWAS, performing GWAS in non-Caucasian populations, improving the genomic coverage of the GWAS, and performing meta-analyses of the studies published so far - could overcome these problems. Moreover, thorough analysis of the associated region by deep sequencing could lead to variants with higher odds ratios being identified (see section 5).
Functions of Type 2 Diabetes Genes
As discussed above, type 2 diabetes is a disease characterized by impaired β-cell secretion of insulin, in combination with resistance to insulin in its target tissues. Both insulin secretion and insulin sensitivity are influenced by genetic and environmental factors [27]. Genes that harbor variants associated with diabetes can thus be expected to exert their effect through one of these pathways.
Most genes that play a role in monogenic diabetes are important for pancreatic β-cell growth and/or function (Table 1). Genetic variants in three (KCNJ11, TCF2, WFS1) out of four replicated genes for type 2 diabetes found by candidate-based genetic association studies are also related to decreased β-cell function. The best characterized gene is KCNJ11 which encodes a member of a β-cell potassium channel involved in insulin secretion [28]. Mutations in TCF2 are associated with insulin secretion [29], while knock-out mutations in WFS1 have been found to lead to Endoplasmic Reticulum (ER) stress and subsequent apoptosis of pancreatic β-cells [30]. The protein product of the fourth gene identified by a candidate gene approach, PPARγ, is involved in adipocyte differentiation and function (reviewed in [31]). Since rare mutations in this gene lead to insulin resistance and lipodystrophy, variations in this gene are likely to contribute to type 2 diabetes susceptibility through altered insulin effects in adipose tissue.
To explore the functions of the newly discovered genes, various studies have investigated their roles in determining sub-phenotypes of type 2 diabetes, such as peripheral insulin sensitivity and β-cell insulin secretion, and the genetic variants identified in GWAS (except KCNQ1 because variants in this gene have only recently been identified) [32-36]. In general, these studies indicate that the genetic variants in the genes identified by association studies act through interference with insulin secretion and not with peripheral insulin sensitivity, and are thus similar to the genes identified through candidate gene approaches and in monogenic diabetes. However, insulin secretion and insulin sensitivity are related through complex and poorly understood mechanisms. Expression studies have revealed that most of the newly discovered type 2 diabetes genes are expressed in multiple cell types throughout the body and that the expression of some of these genes under diabetic conditions is unaltered in the pancreatic islets whereas it is different in other tissues (reviewed in [37]). Functional studies of the molecular mechanisms by which most genetic variants lead to impaired insulin secretion and the use of animal models have not yet been published but are sorely needed.
Since only a small percentage of the variability of the genetic influence on diabetes risk has been uncovered so far, it is still possible that there are mutations in genes that affect other axes of the underlying mechanisms of diabetes. It has been proposed that multiple hits in several diabetes pathways generally occur in subjects destined to develop type 2 diabetes, and that disturbances in β-cell function are ultimately decisive for the actual development of the hyperglycemic state.
Genetic Subgroups
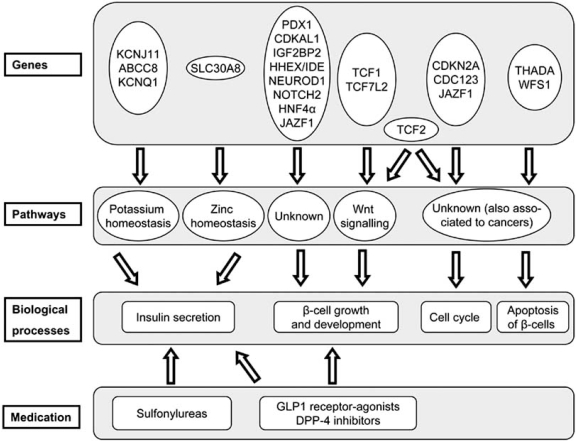
It can be proposed that some of the diabetes genes can be grouped into four subgroups based on what is known about their molecular function (see Fig. 1). First, KCNJ11, KCNQ1, ABCC8, and SLC30A8 gene products are involved in cellular ion homeostasis and insulin secretion. A second group of genes – TCF7L2, TCF1, TCF2, HHEX / IDE, IGF2BP2, CDKAL1, NEUROD1, PDX1, and NOTCH2 – is likely to be involved in the growth and development of the pancreas. Of this group TCF7L2, TCF1, and TCF2 are important in Wnt signaling (reviewed in [38]). A third group is related with cell cycle events: JAZF1 and TCF2 have both been associated with prostate cancer and these genes might play a role in the regulation of the cell cycle [39-41]. CDKN2A and, presumably, CDC123 also play a role in the cell cycle. Finally, THADA and WFS1 are thought to be involved in the apoptosis of β-cells. Genes involved in the cell cycle and apoptosis of β-cells might play a role in diabetes by dysregulating the response of β-cells when a higher insulin output is called for when insulin resistance is present.
Fig. (1) Classification of diabetes genes with a potential role in β-cell function.
Diabetes genes involved in either type 2 diabetes, Maturity Onset Diabetes of the Young (MODY), or Permanent Neonatal Diabetes Mellitus (PNDM) are included. These genes are assigned to pathways and biological process as described in the text. Medications as shown are known to affect a subset of β-cell functions.
MANAGEMENT OF TYPE 2 DIABETES AND HOW IT IS RELATED TO GENETICS
The corner stone for diabetes management still lies in diet and exercise [42, 43]. There is also a slowly expanding list of drugs being used to treat type 2 diabetes, all of which act through one of the pathways important in diabetes pathophysiology. However, neither changes in lifestyle nor the use of medication are sufficient to cure diabetes, although both interventions can delay the progression of disease. There is therefore an urgent need to develop new medications or strategies to counter the huge increase in cases expected in the future. Since the management of type 2 diabetes with either lifestyle changes, medication or both, is more effective when started at an early stage, improving the techniques for early diagnosis and the opportunities for early intervention will greatly improve the effects of current ways of managing type 2 diabetes.
Pathways important for the function, growth and development of pancreatic β-cells provide obvious drug targets and are already being used, since defects in insulin secretion play a central role in diabetes. Sulfonylurea derivatives are widely used and act through improving β-cell function by closing β-cell potassium channels and thereby enhancing insulin secretion. Some of the genes associated with both monogenic (ABCC8 and KCNJ11) and complex diabetes (KCNJ11, KCNQ1) encode subunits of these potassium channels and accordingly, monogenic diabetes of this type can be well managed with sulfonylureas. SLC30A8 encodes a zinc transporter protein in the β-cell and the function of this gene product might be related to that of the potassium channel genes by regulating ion homeostasis in the β-cell [44].
The actions of various other drugs involve provoking different effects rather than augmenting insulin secretory function: thiazolidinediones enhance insulin activity by acting on adipose tissue, metformin lowers hepatic glucose output, and pramlintidine delays gastric emptying and inhibits the release of glucagons. GLP1 receptor-agonists or DPP-4 inhibitors have combined actions on food intake by regulating satiety and enhancing insulin secretion in the short term, and β-cell neogenesis and proliferation in the long term (reviewed in [45]). The exact physiological mechanisms that underlie these improvements are, however, unclear. Except for PPARγ and genes involved in pancreas growth and survival, there is no evidence that any of the other genes identified by GWAS are involved in any of these processes. The protein product of PPARγ is likely to be involved in the mechanism targeted by thiazolidinediones because these compounds are ligands of PPARγ receptors [46]. Although it seems likely that some of the newly discovered diabetes genes act in a pathway that is targeted by GLP1 receptor-agonists or DPP-4 inhibitors, more research into both the mechanisms of the medication and the function of the genes is needed.
IMPLICATIONS FOR PREVENTION AND TREATMENT
Genetic Screening for Prediction and Prevention
The effectiveness of current type 2 diabetes management is greatly improved when it is started at an early stage of the disease. If genetic testing could be used to predict type 2 diabetes, preventive measures could be taken and diabetes could potentially be managed more easily. However, the variants associated with type 2 diabetes that have been identified so far only explain a small percentage of the total genetic variation that is thought to be present [25, 26]. It is therefore not yet possible to perform accurate predictive genetic testing but, in the near future, research should provide more insight into the opportunities for such testing. Firstly, it is expected that many more common genetic variants will be identified by performing GWAS in other populations and by improving their power and genomic coverage (i.e., more patients included in studies and more SNPs tested). Secondly, performing thorough analyses of the genomic regions that show association to the disease by resequencing large numbers of patients and controls may identify genetic variants that have higher odds ratios than the common genetic variants identified so far. In a GWAS it is unlikely that the functional variant will be identified, but rather one or more common variants to which the disease variant is in linkage disequilibrium, i.e., that the actual causative mutation(s) is in close vicinity to the tested variant. If the frequency of the causal disease variant is lower than the tested common SNP, we can expect the odds ratio of the causal SNP to be higher and this would make the causal SNP a better candidate to use for genetic testing. One drawback of genetic testing is that many variants have to be tested for each subject, which is laborious and costly at present, but improved genotyping and resequencing technologies will make screening for such variants feasible in the near future.
New Opportunities for Intervention
Even if the exact functions of the majority of the genes associated with type 2 diabetes are still elusive, they can be broadly grouped into several classes (see Fig. 1). When certain genes are involved in the same molecular pathway or physiological process, not only these genes but the entire pathway or process would be an obvious target for new anti-diabetes drugs. In combination with genetic screening, such information could be used to optimize diabetes management by prescribing drugs that act on those pathways that are affected in a patient, and vice versa, it is also possible that altering the activity, be it stimulation or inhibition, of undisturbed processes could improve glucose homeostasis in selected patients. Improved drug treatment in diabetes can be seen in the current management of various MODY subtypes. As described above, mutations in genes involved in β-cell signaling and/or growth in the pancreas have been found to be responsible for MODY subtypes. After these mutations were identified, the management of these diseases was greatly improved by the use of sulfonylurea derivatives, which enhances insulin secretion instead of exogenous insulin [44]. The good response of MODY patients to sulfonylurea treatment serves as an excellent example of personalized therapy based on genetic screening.
Evidence that genes identified by GWAS can represent targets for managing type 2 diabetes is provided by the association to this disease of variants in the KCNQ1 and KCNJ11 genes. Both these genes encode proteins that are members of β-cell potassium channels important for insulin secretion and these channels are targeted by sulfonylurea derivates which are already widely used anti-diabetic drugs [44].
Another gene, found through a candidate-based association study and likely to act in the pathway targeted by thiazolidinediones, is PPARγ. On the physiological level no relationships are known between current medications and all the other genetic variants identified in either GWAS or candidate gene association studies. Therefore all these genes represent new potential targets for intervention.
Type 2 diabetes genes that are presumed to play a role in cell cycle regulation (JAZF1, CDKN2A, and CDC123), apoptosis (THADA and WFS1), or pancreas development and growth (TCF7L2, TCF2, HHEX / IDE, IGF2BP2, CDKAL1, and NOTCH2) have not yet been studied well enough to predict whether interference with their products could be used in managing diabetes. Studies are needed in order to better classify these genes.
Common variants in type 2 diabetes genes relating to cell cycle events and apoptosis, and representing different alleles than those associated to type 2 diabetes, are also associated with various cancers. In addition, the risks of developing diabetes and prostate cancer are correlated in a complex way: overall diabetes risk and prostate cancer risk are inversely correlated, but while diabetes risk and the risk of developing aggressive prostate cancer show no correlation, these two risks are positively correlated in lean men and in men who undertake vigorous physical activity [47]. Systemic administration of agents interfering with the gene products of diabetes genes that are also associated with cancers could, therefore, be beneficial in treating diabetes, but they might be potentially carcinogenic. Tissue-specific drug targeting or additive medication to overcome the carcinogenic side-effects of such medication needs to be developed to overcome this problem. Hence, cell-cycle related genes are probably not the most promising targets for developing new diabetes treatments, unless cell-type-specific targeting of drugs becomes a reality.
Although the genes containing variants associated with type 2 diabetes that seem to be involved in pancreatic growth and development have not been well studied yet, we can speculate on several possibilities for intervention with such genes. Because most genes in this category are not associated with other diseases, they represent promising intervention targets. Variants in these genes might affect the regenerative or proliferative potential of the β-cell population when there is an increased demand for insulin secretion. In this case it would be possible to use such genes, or the molecular pathways they are involved in, as a target for intervention, in an attempt to correct the poor response of the β-cells and ultimately aiming to improve insulin secretion.
Another possibility is that variants in these genes cause pancreatic damage, which would lead to endocrine malfunction of the pancreas at an early stage of the diabetic development. The possibilities for therapeutic intervention would largely depend on the severity and reversibility of such a malfunction.
Genetic variants in three genes identified (or confirmed) by GWAS (TCF7L2, CDKAL1, and SLC30A8) are associated with impairment of proinsulin-to-insulin conversion, whereas variants in other genes identified by GWAS are not associated to this [48]. Even if the affected proinsulin-to-insulin conversion is not a primary but a secondary effect of genetic variants in one of the three genes, reconstituting this process would enhance insulin secretion and thereby enhance β-cell function. Although the conversion of proinsulin-to-insulin has long been known to be involved in type 2 diabetes [49], the mechanisms that regulate this process are still undetermined and there is no current therapy that acts by interfering with this process. Association of TCF7L2, CDKAL1, and SLC30A8 to impaired proinsulin-to-insulin conversion has provided evidence that these genes are somehow related to this process and this clears the way for functional research and development of new medication.
CONCLUSIONS
GWAS are an important tool for identifying genetic variation and they have been very successful in finding 19 genetic loci involved in type 2 diabetes. Genotyping techniques will continue to improve and patient cohorts will become larger, which will lead to more genetic variants being identified in the near future. Although the true causal SNPs are often not known, it is anticipated they will be uncovered by deep sequencing of the genomic regions containing association signals. These advances will help elucidate a higher percentage of the total genetic variation, and improve opportunities for genetic screening and personalized diabetes care. Functional studies and animal models harboring specific gene deletions will be needed to study the role of such mutations, while genetic studies using sharply defined endophenotypes will greatly enhance our knowledge about the type 2 diabetes genes identified by GWAS.
At present, it is too early to expect results from GWAS to lead to new therapies. Nevertheless, it is clear that genetic studies are crucial to dissecting the mechanisms underlying the biological processes and to finding ways to intervene with them, as they provide important clues for directing the focus of functional research. The ongoing avalanche of genetic information about type 2 diabetes will thus pave the way to further research in the field and has already yielded important information to aid the development of new therapeutic strategies.
ACKNOWLEDGEMENTS
We thank the Dutch Diabetes Foundation (project grant 541741) for financial support.
We thank J.L. Senior for critically reading the manuscript. There are no conflicts of interest.
REFERENCES
- 1.Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763–768. doi: 10.1007/BF00275741. [DOI] [PubMed] [Google Scholar]
- 2.van Haeften TW, Dubbeldam S, Zonderland ML, Erkelens DW. Insulin secretion in normal glucose-tolerant relatives of type 2 diabetic subjects. Assessments using hyperglycemic glucose clamps and oral glucose tolerance tests. Diabetes Care. 1998;21:278–282. doi: 10.2337/diacare.21.2.278. [DOI] [PubMed] [Google Scholar]
- 3.Pimenta W, Korytkowski M, Mitrakou A, Jenssen T, Yki-Jarvinen H, Evron W, Dailey G, Gerich J. Pancreatic beta-cell dysfunction as the primary genetic lesion in NIDDM. Evidence from studies in normal glucose-tolerant individuals with a first-degree NIDDM relative. Jama. 1995;273:1855–1861. [PubMed] [Google Scholar]
- 4.Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr. Rev. 2008;29:254–264. doi: 10.1210/er.2007-0024. [DOI] [PubMed] [Google Scholar]
- 5.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, Bell GI. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat. Genet. 1997;17:384–385. [Google Scholar]
- 6.Strom TM, Hortnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, Gerbitz KD, Meitinger T. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum. Mol. Genet. 1998;7:2021–2028. doi: 10.1093/hmg/7.13.2021. [DOI] [PubMed] [Google Scholar]
- 7.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, Edghill EL, Frayling TM, Temple IK, Mackay D, Shield JP, Sumnik Z, van Rhijn A, Wales JK, Clark P, Gorman S, Aisenberg J, Ellard S, Njolstad PR, Ashcroft FM, Hattersley AT. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 8.Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O'Rahilly S. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 9.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 11.Florez JC, Manning AK, Dupuis J, McAteer J, Irenze K, Gianniny L, Mirel DB, Fox CS, Cupples LA, Meigs JB. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes. 2007;56:3063–3074. doi: 10.2337/db07-0451. [DOI] [PubMed] [Google Scholar]
- 12.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 13.Hanson RL, Bogardus C, Duggan D, Kobes S, Knowlton M, Infante AM, Marovich L, Benitez D, Baier LJ, Knowler WC. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- 14.Hayes MG, Pluzhnikov A, Miyake K, Sun Y, Ng MC, Roe CA, Below JE, Nicolae RI, Konkashbaev A, Bell GI, Cox NJ, Hanis CL. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56:3033–3044. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- 15.Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, Zhang L, Zhao Y, Mitchell BD, O'Connell J, Shuldiner AR. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 16.Salonen JT, Uimari P, Aalto JM, Pirskanen M, Kaikkonen J, Todorova B, Hypponen J, Korhonen VP, Asikainen J, Devine C, Tuomainen TP, Luedemann J, Nauck M, Kerner W, Stephens RH, New JP, Ollier WE, Gibson JM, Payton A, Horan MA, Pendleton N, Mahoney W, Meyre D, Delplanque J, Froguel P, Luzzatto O, Yakir B, Darvasi A. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am. J. Hum. Genet. 2007;81:338–345. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 18.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 20.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 21.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jorgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 23.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lango H, Palmer CN, Morris AD, Zeggini E, Hattersley AT, McCarthy MI, Frayling TM, Weedon MN. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Hoek M, Dehgan A, Witteman JC, van Duijn CM, Uitterlinden AG, Oostra BA, Hofman A, Sijbrands EJ, Janssens AC. Predicting type 2 diabetes based on polymorphisms from genome wide association studies: a population-based study. Diabetes. 2008;57:3122–3128. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 28.Inagaki N, Gonoi T, Clement JPt, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Coffinier C, Thomas MK, Gresh L, Eddu G, Manor T, Levitsky LL, Yaniv M, Rhoads DB. Selective deletion of the Hnf1beta (MODY5) gene in beta-cells leads to altered gene expression and defective insulin release. Endocrinology. 2004;145:3941–3949. doi: 10.1210/en.2004-0281. [DOI] [PubMed] [Google Scholar]
- 30.Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, Tokita A, Satake C, Tashiro F, Katagiri H, Aburatani H, Miyazaki J, Oka Y. WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum. Mol. Genet. 2006;15:1600–1609. doi: 10.1093/hmg/ddl081. [DOI] [PubMed] [Google Scholar]
- 31.Stumvoll M, Haring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002;51:2341–2347. doi: 10.2337/diabetes.51.8.2341. [DOI] [PubMed] [Google Scholar]
- 32.Grarup N, Andersen G, Krarup NT, Albrechtsen A, Schmitz O, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57:2534–2540. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, Clausen JO, Rasmussen SS, Jorgensen T, Sandbaek A, Lauritzen T, Schmitz O, Hansen T, Pedersen O. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–3111. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 34.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, Weedon MN, Mari A, Hattersley AT, McCarthy MI, Frayling TM, Walker M. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 35.Staiger H, Machicao F, Kantartzis K, Schafer SA, Kirchhoff K, Guthoff M, Silbernagel G, Stefan N, Fritsche A, Haring HU. Novel meta-analysis-derived type 2 diabetes risk loci do not determine prediabetic phenotypes. PLoS ONE. 2008;3:e3019. doi: 10.1371/journal.pone.0003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staiger H, Machicao F, Stefan N, Tschritter O, Thamer C, Kantartzis K, Schafer SA, Kirchhoff K, Fritsche A, Haring HU. Polymorphisms within novel risk loci for type 2 diabetes determine beta-cell function. PLoS ONE. 2007;2:e832. doi: 10.1371/journal.pone.0000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin T. The WNT signalling pathway and diabetes mellitus. Diabetologia. 2008;51:1771–1780. doi: 10.1007/s00125-008-1084-y. [DOI] [PubMed] [Google Scholar]
- 39.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 40.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 41.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 42.Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N. Engl. J. Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 43.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hattersley AT, Pearson ER. Minireview: pharmacogenetics and beyond: the interaction of therapeutic response, beta-cell physiology, and genetics in diabetes. Endocrinology. 2006;147:2657–2663. doi: 10.1210/en.2006-0152. [DOI] [PubMed] [Google Scholar]
- 45.Drucker DJ. Glucagon-like peptides: regulators of cell proliferation, differentiation, and apoptosis. Mol. Endocrinol. 2003;17:161–171. doi: 10.1210/me.2002-0306. [DOI] [PubMed] [Google Scholar]
- 46.Elbrecht A, Chen Y, Cullinan CA, Hayes N, Leibowitz M, Moller DE, Berger J. Molecular cloning, expression and characterization of human peroxisome proliferator activated receptors gamma 1 and gamma 2. Biochem. Biophys. Res. Commun. 1996;224:431–437. doi: 10.1006/bbrc.1996.1044. [DOI] [PubMed] [Google Scholar]
- 47.Leitzmann MF, Ahn J, Albanes D, Hsing AW, Schatzkin A, Chang SC, Huang WY, Weiss JM, Danforth KN, Grubb RL, 3rd Andriole GL. Diabetes mellitus and prostate cancer risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Cancer Causes Control. 2008;19:1267–1276. doi: 10.1007/s10552-008-9198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchhoff K, Machicao F, Haupt A, Schafer SA, Tschritter O, Staiger H, Stefan N, Haring HU, Fritsche A. Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia. 2008;51:597–601. doi: 10.1007/s00125-008-0926-y. [DOI] [PubMed] [Google Scholar]
- 49.Jain K, Logothetopoulos J. Secretion of insulin in a perifusion system and conversion of proinsulin to insulin by pancreatic islets from hyperglycemic rats. Diabetes. 1977;26:650–656. [PubMed] [Google Scholar]