Abstract
OBJECTIVE
KCNQ1 gene polymorphisms are associated with type 2 diabetes. This linkage appears to be mediated by altered β-cell function. In an attempt to study underlying mechanisms, we examined the effect of four KCNQ1 single nucleotide polymorphisms (SNPs) on insulin secretion upon different stimuli.
RESEARCH DESIGN AND METHODS
We genotyped 1,578 nondiabetic subjects at increased risk of type 2 diabetes for rs151290, rs2237892, rs2237895, and rs2237897. All participants underwent an oral glucose tolerance test (OGTT); glucagon-like peptide (GLP)-1 and gastric inhibitory peptide secretion was measured in 170 participants. In 519 participants, a hyperinsulinemic-euglycemic clamp was performed, in 314 participants an intravenous glucose tolerance test (IVGTT), and in 102 subjects a hyperglycemic clamp combined with GLP-1 and arginine stimuli.
RESULTS
rs151290 was nominally associated with 30-min C-peptide levels during OGTT, first-phase insulin secretion, and insulinogenic index after adjustment in the dominant model (all P ≤ 0.01). rs2237892, rs2237895, and rs2237897 were nominally associated with OGTT-derived insulin secretion indexes (all P < 0.05). No SNPs were associated with β-cell function during intravenous glucose or GLP-1 administration. However, rs151290 was associated with glucose-stimulated gastric inhibitory polypeptide and GLP-1 increase after adjustment in the dominant model (P = 0.0042 and P = 0.0198, respectively). No associations were detected between the other SNPs and basal or stimulated incretin levels (all P ≥ 0.05).
CONCLUSIONS
Common genetic variation in KCNQ1 is associated with insulin secretion upon oral glucose load in a German population at increased risk of type 2 diabetes. The discrepancy between orally and intravenously administered glucose seems to be explained not by altered incretin signaling but most likely by changes in incretin secretion.
Recent genome-wide association (GWA) studies confirmed the significance of established candidate gene regions for type 2 diabetes, i.e., PPARã, KCNJ11, TCF7L2, and WFS1, and also revealed several novel type 2 diabetes susceptibility loci, i.e., SLC30A8, HHEX, CDKAL1, IGF2BP2, and CDKN2A/B, none of which were considered as functional candidates (1–5). Comprehensive metabolic analysis of genotyped cohorts, comprising measurement of insulin sensitivity and insulin secretion with state-of-the-art methods, revealed that the novel variants influence insulin secretion but show little, if any, impact on insulin sensitivity (6–11).
Two recent GWA studies identified KCNQ1 as a novel diabetes susceptibility gene (12–13). Similar to the other novel gene variants that are associated with type 2 diabetes, the KCNQ1 risk alleles for type 2 diabetes also appear to be associated with impaired pancreatic β-cell function as assessed by fasting state– and oral glucose tolerance test (OGTT)-derived indexes of insulin secretion (13).
KCNQ1 contains 19 exons and spans more than 400 kb on chromosome 11p15.5 (14). The KCNQ1 gene encodes the pore-forming α-subunit of the voltage-gated K+ channel (KvLQT1), which plays an important role in controlling the ventricular repolarization process (15). Mutations in KCNQ1 have been associated with inherited cardiac disorders, such as long QT syndrome and familial atrial fibrillation. The long QT syndrome may occur in a recessive form that is associated with deafness (Jervell and Lange-Nielsen syndrome) or in an autosomal dominant variant not associated with deafness (Romano-Ward syndrome) (16). In addition to the heart and cochlea, KCNQ1 is ubiquitously expressed in epithelial cells, including the exocrine and endocrine pancreas (17). KCNQ1 was reported previously to be expressed in insulin-secreting INS-1 cells, and inhibition of this potassium channel by the sulfonamide analog 293B was found to significantly increase insulin secretion in the presence of tolbutamide (18).
The aim of the present study was to investigate the influence of common type 2 diabetes–associated KCNQ1 single nucleotide polymorphisms (SNPs) on insulin secretion kinetics in response to orally and intravenously administered glucose during an OGTT and intravenous glucose tolerance test (IVGTT) as well as a hyperglycemic clamp combined with glucagon-like peptide (GLP)-1 and arginine administration.
RESEARCH DESIGN AND METHODS
We studied 1,578 nondiabetic participants at an increased risk for type 2 diabetes due to family history of diabetes (first-degree relatives of type 2 diabetic patients), history of gestational diabetes, overweight, impaired fasting glucose, or impaired glucose tolerance determined in an OGTT (Table 1). Subjects were recruited from an ongoing study on the pathophysiology of type 2 diabetes (19). A subset of 519 participants was studied by a hyperinsulinemic-euglycemic clamp, 314 participants by an IVGTT, and 102 subjects by a hyperglycemic clamp combined with GLP-1 and arginine stimuli. First-degree relatedness among subjects was less than 1%. Informed written consent for all studies was obtained from all participants, and the local ethics committee approved the protocols.
TABLE 1.
Clinical characteristics of the study population
| Sex (female/male) | 1,044/534 |
| IFG/IGT/IFG and IGT | 164/152/123 |
| Age (years) | 40 ± 13 |
| BMI (kg/m2) | 28.9 ± 8.2 |
| Waist circumference (cm) | 94 ± 17 |
| Fasting glucose (mmol/l) | 5.11 ± 0.55 |
| Glucose: 120-min OGTT (mmol/l) | 6.27 ± 1.66 |
| Fasting insulin (pmol/l) | 63.7 ± 52.9 |
| Insulin: 30-min OGTT (pmol/l) | 493.5 ± 392.7 |
Data are n or means ± SD. IFG, impaired fasting glucose; IGT, impaired glucose tolerance.
Subjects were genotyped for rs151290, rs2237892, rs2237895, and rs2237897 (all located in intron 15) in the KCNQ1 gene. rs2237897 and rs2237895 showed the strongest association with type 2 diabetes in a recent study (12). In the third screening of another study, rs151290 and rs2237895 were found to be most significantly associated with type 2 diabetes (13). As rs2237895 was already included in the SNPs chosen from the study by Unoki et al. (12), the SNP with the strongest association with type 2 diabetes in the replication study by Yasuda et al. (13), rs2237892, was additionally picked.
Genotyping was done using the TaqMan assay (Applied Biosystems, Forster City, CA). The TaqMan genotyping reaction was amplified on a GeneAmp PCR system 7000, and fluorescence was detected on an ABI PRISM 7000 sequence detector (Applied Biosystems). Genotyping for rs2237897 using the TaqMan assay was successful only in major allele carriers. Therefore, in those subjects for whom the TaqMan assay failed, genotypes were directly determined by bidirectional sequencing.
Quality control was performed as described previously (19). Overall, genotyping success rate was 99.9% (100% for rs151290 and rs2237895 and 99.9% for rs2237892 and rs2237897), and the error rate was 0% (3.2% of all samples were regenotyped by bidirectional sequencing). For OGTT, IVGTT, and hyperinsulinemic-euglycemic clamp, the assays were performed as previously described in detail (10).
The hyperglycemic clamp, combined with GLP-1 and arginine administration, was performed as described previously (20). After 120 min of hyperglycemic clamp at 10 mmol/l, a bolus of GLP-1 (4.5 pmol/kg) was given [human GLP-1 (7–36)amide; Poly Peptide, Wolfenbüttel, Germany], followed by a continuous GLP-1 infusion (1.5 pmol · kg−1 · min−1) during the next 80 min. At 180 min, a bolus of 5 g arginine hydrochloride (Pharmacia & Upjohn, Erlangen, Germany) was injected over 45 s while the GLP-1 infusion was continued. Blood for the measurement of glucose, insulin, and C-peptide was taken at −30, −15, 0, 2.5, 5, 7.5, 10, 20, 40, 60, 80, 100, 120, 125, 130, 140, 150, 160, 170, 180, 182.5, 185, 187.5, 190, and 200 min. This clamp allows measurement of different aspects of stimulus-secretion coupling: first and second phases of glucose-induced insulin secretion, GLP-1–induced insulin secretion, and the response to additional arginine administration.
Plasma glucose, insulin, and C-peptide concentrations were measured as described previously (19). GLP-1 and gastric inhibitory polypeptide (GIP) immunoreactivitiy were determined using radioimmunoassays specific for the COOH-terminal of the peptides (21,22). To avoid incretin degradation, venous blood was drawn into chilled tubes containing EDTA and aprotinin (Trasylol; 20,000 kallikrein inhibitor units/ml, 200 μl per 10 ml blood; Bayer, Leverkusen, Germany) and kept on ice. After centrifugation at 4°C, plasma for hormone analyses was kept frozen at −20°C. BMI and waist and hip circumferences were measured as described earlier (19).
First-phase insulin secretion (picomoles per liter), insulin sensitivity from the OGTT (arbitrary units), and clamp-derived insulin sensitivity (arbitrary units) were calculated as reported previously (19). Insulinogenic index was assessed by (insulin at 30 min − insulin at 0 min)/(glucose at 30 min − glucose at 0 min). Insulin secretion during the IVGTT was assessed as the sum of C-peptide levels and insulin levels, respectively, during the first 10 min after glucose administration. Insulin secretion during the hyperglycemic clamp was calculated as reported previously using insulin levels determined during the clamp (20). Fold increase of incretins during OGTT was assessed by the ratio of the 30-min incretin value to the basal incretin value.
Statistical analyses.
Data are means ± SD. Log-transformation of metabolic variables was performed before simple and multivariate linear regression analyses. Distribution was tested for normality using the Shapiro-Wilk W test. The secretion indexes were compared using multivariate regression models. In these models, the trait was the dependent variable, whereas age, sex, BMI, insulin sensitivity, and genotype were the independent variables. To account for the number of SNPs tested and the number of independent traits analyzed (anthropometrics, insulin sensitivity, and insulin secretion) in the OGTT study, a Bonferroni-corrected α-level of P < 0.00425 was considered statistically significant. Given that the IVGTT study, the hyperglycemic clamp, and measurement of incretin levels were hypothesis driven, we considered only the number of SNPs tested resulting in a Bonferroni-corrected α-level of P = 0.0127. The statistical software package JMP 7.0 (SAS Institue, Cary, NC) was used. In the dominant model, dependent on the SNP tested, the OGTT study was sufficiently powered (1-β > 0.8) to detect effect sizes as small as 0.13–0.24 (one-tailed t test), the hyperinsulinemic-euglycemic clamp 0.23–0.49, the IVGTT study 0.29–0.62, and the combined hyperglycemic clamp 0.53–0.98. Power calculation was performed using G*power software available at http://www.psycho.uni-duesseldorf.de/aap/projects/gpower. Hardy-Weinberg equilibrium was tested using the χ2 test.
RESULTS
Characterization and genotyping of a German population at increased risk for type 2 diabetes.
We genotyped 1,578 nondiabetic subjects from the southwest of Germany whose clinical characteristics are presented in Table 1. Of these subjects, 68.1% had a family history of diabetes, i.e., at least one second-degree relative with type 2 diabetes. The observed minor allele frequency (MAF) and the MAF published by HapMap were 0.208 and 0.217, respectively, for rs151290, 0.064 and 0.075 for rs2237892, and 0.037 and 0.051 for rs2237897. Whereas the observed MAF for rs2237895 was 0.427, an MAF for this SNP was not published by HapMap. All allele frequencies were in Hardy-Weinberg equilibrium (χ2 test, P > 0.05).
Association of genetic variation in KCNQ1 with anthropometric and metabolic data.
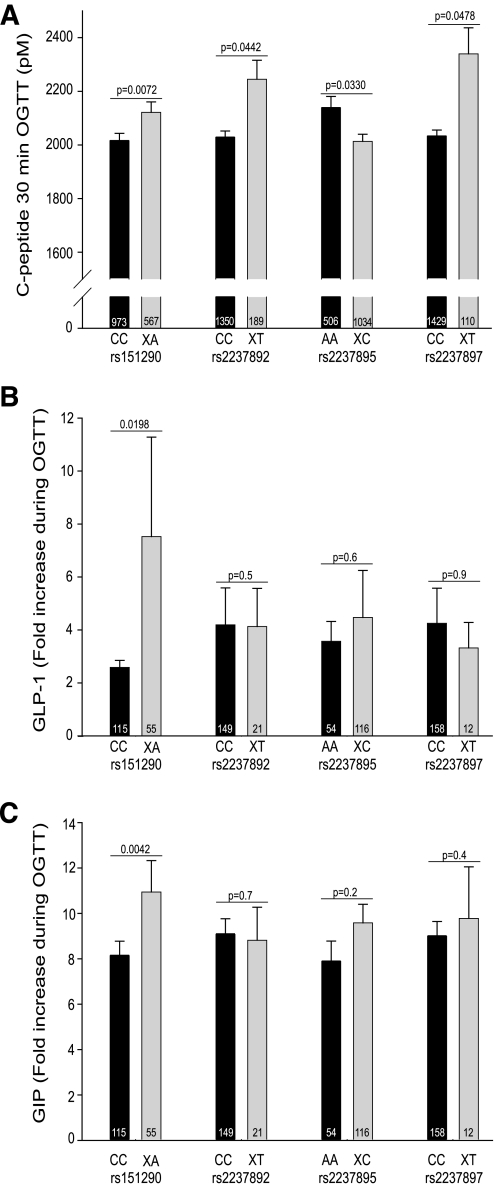
The four SNPs were not associated with anthropometric data, such as BMI, waist circumference, and body fat content, except for a nominal association between rs2237895 and BMI in the additive model only (P = 0.0252; Table 2). rs151290 was nominally associated with 30-min C-peptide levels during OGTT, first-phase insulin secretion, and the insulinogenic index (P = 0.0072, P = 0.0072, and P = 0.0104, respectively) after adjustment for sex, age, BMI, and insulin sensitivity (Table 2 and Fig. 1A). rs2237892 was significantly associated with 30-min insulin levels during OGTT (P = 0.0010) and nominally with 30-min C-peptide concentrations during OGTT and the insulinogenic index (P = 0.0330 and 0.0472, respectively) after adjustment for sex, age, BMI, and insulin sensitivity in the dominant model. rs2237895 was nominally associated with 30-min C-peptide levels during OGTT, first-phase insulin secretion, and the insulinogenic index (P = 0.0442, P = 0.0410, and P = 0.0409, respectively) after adjustment for sex, age, BMI, and insulin sensitivity in the dominant model. rs2237897 was nominally associated with 30-min C-peptide levels during OGTT (P = 0.0478) after adjustment for sex, age, BMI, and insulin sensitivity in the dominant model. Whereas indexes of insulin secretion were improved in minor allele carriers of rs151290, rs2237892, and rs2237897, minor allele carriers of rs2237895 depicted reduced insulin secretion. Nominal associations were found between rs2237897 and fasting insulin and OGTT-derived insulin sensitivity (P = 0.0388 and 0.0340, respectively) after adjustment for age, sex, and BMI in the dominant model. rs2237895 was also nominally associated with OGTT-derived insulin sensitivity (P = 0.0245) after adjustment for age, sex, and BMI in the dominant model. rs151290 was nominally associated with OGTT-derived insulin sensitivity (P = 0.0330) after appropriate adjustment in the additive model. However, such an association was not found in the dominant model (P = 0.4). rs2237892 was not associated with OGTT-derived insulin sensitivity (P ≥ 0.3).
TABLE 2.
Associations of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with anthropometric and metabolic traits
| rs151290 (0.208) |
rs2237892 (0.064) |
rs2237895 (0.427) |
rs2237897 (0.037) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CA | AA | Padd | Pdom | CC | AA | TT | Padd | Pdom | AA | AC | CC | Padd | Pdom | CC | CT | TT | Padd | Pdom | |
| n | 995 | 508 | 75 | — | — | 1,384 | 183 | 10 | — | — | 521 | 765 | 292 | — | — | 1,463 | 111 | 3 | — | — |
| Age (years) | 39 ± 13 | 40 ± 14 | 38 ± 14 | 0.5 | 0.7 | 40 ± 13 | 39 ± 14 | 34 ± 12 | 0.4 | 0.5 | 39 ± 13 | 40 ± 13 | 39 ± 14 | 0.0091 | 0.05 | 40 ± 13 | 38 ± 15 | 37 ± 15 | 0.2 | 0.11 |
| BMI (kg/m2) | 28.7 ± 8.1 | 29.1 ± 8.5 | 29.1 ± 7.3 | 0.6 | 0.4 | 28.8 ± 8.3 | 29.1 ± 7.7 | 31.2 ± 6.6 | 0.3 | 0.3 | 29.2 ± 8.1 | 29.0 ± 8.5 | 27.8 ± 7.6 | 0.0252 | 0.08 | 28.8 ± 8.2 | 29.6 ± 8.1 | 34.7 ± 6.8 | 0.16 | 0.15 |
| Waist circum-ference (cm) | 93 ± 17 | 95 ± 18 | 95 ± 17 | 0.4 | 0.17 | 94 ± 17 | 94 ± 17 | 99 ± 15 | 0.4 | 0.4 | 94 ± 17 | 94 ± 18 | 92 ± 17 | 0.09 | 0.10 | 94 ± 17 | 95 ± 17 | 103 ± 23 | 0.2 | 0.13 |
| Body fat (%) | 30.7 ± 10.8 | 31.4 ± 11.0 | 31.9 ± 11.0 | 0.2 | 0.3 | 30.8 ± 10.9 | 32.0 ± 10.7 | 32.8 ± 8.7 | 0.17 | 0.2 | 31.6 ± 10.8 | 30.8 ± 11.0 | 30.5 ± 10.5 | 0.17 | 0.07 | 30.9 ± 10.8 | 32.6 ± 11.0 | 38.3 ± 5.2 | 0.19 | 0.4 |
| Fasting glucose (mmol/l) | 5.11 ± 0.56 | 5.09 ± 0.52 | 5.15 ± 0.54 | 0.5 | 0.6 | 5.10 ± 0.55 | 5.12 ± 0.52 | 5.05 ± 0.52 | 0.0457 | 0.7 | 5.09 ± 0.54 | 5.10 ± 0.53 | 5.13 ± 0.61 | 0.9 | 0.8 | 5.11 ± 0.55 | 5.07 ± 0.54 | 5.45 ± 0.71 | 0.6 | 0.7 |
| Glucose: 120-min OGTT (mmol/l) | 6.32 ± 1.66 | 6.14 ± 1.64 | 6.52 ± 1.64 | 0.0035 | 0.0303 | 6.26 ± 1.65 | 6.31 ± 1.66 | 6.96 ± 2.20 | 0.0121 | 0.14 | 6.18 ± 1.63 | 6.27 ± 1.66 | 6.41 ± 1.70 | 0.5 | 0.6 | 6.26 ± 1.65 | 6.31 ± 1.72 | 7.39 ± 2.60 | 0.7 | 0.9 |
| Fasting insulin (pmol/l) | 62.8 ± 48.8 | 64.2 ± 59.0 | 72.6 ± 61.3 | 0.13 | 0.5 | 62.6 ± 51.4 | 70.8 ± 62.54 | 82.7 ± 67.0 | 0.18 | 0.7 | 65.4 ± 51.4 | 63.3 ± 55.7 | 61.8 ± 47.9 | 0.06 | 0.0198 | 62.9 ± 52.8 | 73.6 ± 54.4 | 101 ± 48.5 | 0.11 | 0.0388 |
| Insulin: 30-min OGTT (pmol/l) | 477 ± 369 | 512 ± 416 | 584 ± 508 | 0.18 | 0.2 | 480 ± 379 | 589 ± 473 | 565 ± 433 | 0.0027 | 0.0010 | 528 ± 424 | 487 ± 380 | 449 ± 362 | 0.18 | 0.08 | 484 ± 384 | 616 ± 486 | 592 ± 368 | 0.12 | 0.09 |
| First-phase insulin secretion (pmol/l) | 1,235 ± 782 | 1,322 ± 906 | 1,433 ± 1,085 | 0.0203 | 0.0072 | 1,245 ± 811 | 1,470 ± 1,024 | 1,445 ± 900 | 0.12 | 0.11 | 1,343 ± 899 | 1,250 ± 824 | 1,205 ± 771 | 0.09 | 0.0410 | 1,253 ± 824 | 1,532 ± 1,027 | 1,536 ± 526 | 0.2 | 0.10 |
| Insulinogenic index (pmol/mmol) | 137 ± 398 | 157 ± 174 | 160 ± 142 | 0.0246 | 0.0104 | 140 ± 350 | 174 ± 160 | 136 ± 68 | 0.14 | 0.0472 | 171 ± 263 | 126 ± 413 | 144 ± 153 | 0.06 | 0.0409 | 145 ± 323 | 136 ± 443 | 139 ± 42 | 0.4 | 0.3 |
| C-peptidesum 0–10 min IVGTT (pmol/l)* | 8,357 ± 3,544 | 8,355 ± 3,321 | 9,508 ± 7,534 | 0.9 | 0.8 | 8,299 ± 3,423 | 9,189 ± 5,624 | — | 0.9 | 0.9 | 8,776 ± 4,646 | 8,208 ± 3,313 | 8,432 ± 3,426 | 0.5 | 0.4 | 8,286 ± 3,399 | 10772 ± 7,904 | — | 0.4 | 0.4 |
| ISI: OGTT (U) | 16.3 ± 10.7 | 16.6 ± 10.9 | 14.3 ± 9.6 | 0.0330 | 0.4 | 16.6 ± 10.9 | 14.5 ± 9.2 | 15.0 ± 14.5 | 0.3 | 1.0 | 16.1 ± 10.7 | 16.3 ± 10.8 | 16.6 ± 10.7 | 0.08 | 0.0245 | 16.5 ± 10.8 | 14.4 ± 10.5 | 9.3 ± 8.2 | 0.10 | 0.0340 |
| ISI: clamp (U)† | 0.089 ± 0.060 | 0.080 ± 0.047 | 0.068 ± 0.027 | 0.3 | 0.14 | 0.087 ± 0.055 | 0.075 ± 0.056 | 0.134 | 0.4 | 0.3 | 0.088 ± 0.055 | 0.080 ± 0.056 | 0.095 ± 0.050 | 0.4 | 0.3 | 0.086 ± 0.054 | 0.082 ± 0.064 | — | 0.5 | 0.5 |
Data are means ± SD unless otherwise indicated. For statistical analysis, data were log transformed. Anthropometric data were adjusted for sex and age. Indexes of insulin sensitivity were adjusted for sex, age, and BMI. Indexes of insulin secretion were adjusted for sex, age, BMI, and insulin sensitivity. ISI, insulin sensitivity index; Padd, additive model; Pdom, dominant model.
*IVGTT data were available from 314 subjects.
†ISI (clamp) data were available from 519 subjects.
FIG. 1.
A: Associations of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with insulin secretion. Insulin secretion was assessed by C-peptide levels at 30 min during an OGTT. Unadjusted data from 1,578 subjects are presented. B: Association of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with increase of GLP-1 levels during an OGTT. C: Association of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with increase of GIP levels during OGTT. Incretin increase was assessed by the ratio of levels at 30 min during OGTT to fasting levels. Unadjusted data from 170 subjects are presented. Before multivariate linear regression analysis in the dominant model, non–normally distributed data were log–transformed. C-peptide levels were adjusted for sex, age, BMI, and insulin sensitivity. Incretin increase was adjusted for sex, age, and BMI. P values are given above the columns. Sample sizes are given at the bottom of the columns.
None of the four SNPs were associated with IVGTT-derived indexes of insulin secretion (all P ≥ 0.4), and insulin sensitivity measured with the clamp technique was not affected by any of the genotypes (all P ≥ 0.14). The discrepancy between OGTT- and IVGTT-derived insulin secretion pointed to an influence of common genetic variation in the KCNQ1 gene on incretin production or incretin signaling. Recently, the two diabetes susceptibility loci TCF7L2 and WFS1 were found to be associated with impaired GLP-1–induced insulin secretion (23,24). Therefore, we also studied the influence of the four KCNQ1 variants on a hyperglycemic clamp combined with GLP-1 administration. However, no associations were found between the KCNQ1 variants and glucose-, GLP-1–, and arginine-induced insulin secretion during the hyperglycemic clamp after appropriate adjustment (all P > 0.05; supplementary Table 1, available in the online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1589/DC1). To test the influence of genetic variation in KCNQ1 on incretin secretion, in a subset GLP-1 and GIP levels were measured during OGTT. rs151290 was significantly associated with the glucose-stimulated GIP increase and nominally associated with the GLP-1 increase after adjustment for sex, age, and BMI in the dominant model (P = 0.0042 and P = 0.0198, respectively; Fig. 1B and C). The reason for the large SEM values of the fold increase of GLP-1 during OGTT appears to be an outlier with an extremely high 200-fold increase. After exclusion of this outlier, the difference between homozygous major allele carriers and risk allele carriers remains nominally significant (CC 2.6 ± 0.3 vs. XA 3.9 ± 0.7; P < 0.05). No associations were detected between the other three SNPs and basal or stimulated incretin levels (all P ≥ 0.05; Table 3).
TABLE 3.
Associations of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with the incretins GLP-1 and GIP
| rs151290 (0.208) |
rs2237892 (0.064) |
rs2237895 (0.427) |
rs2237897 (0.037) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CA | AA | Padd | Pdom | CC | CT | TT | Padd | Pdom | AA | AC | CC | Padd | Pdom | CC | CT | TT | Padd | Pdom | |
| n | 116 | 47 | 8 | — | — | 150 | 21 | 0 | — | — | 55 | 92 | 24 | — | — | 159 | 12 | 0 | — | — |
| GLP-1 (pmol/l) | ||||||||||||||||||||
| 0 min | 18.1 ± 9.1 | 15.0 ± 7.1 | 15.9 ± 8.6 | 0.2 | 0.10 | 17.2 ± 8.7 | 16.8 ± 9.0 | 0.9 | 0.9 | 16.1 ± 8.2 | 17.8 ± 8.9 | 16.8 ± 8.8 | 0.4 | 0.3 | 17.3 ± 8.8 | 15.5 ± 6.8 | 0.9 | 0.9 | ||
| 30 min | 35.4 ± 19.1 | 41.7 ± 35.2 | 95 ± 17 | 0.7 | 0.5 | 36.9 ± 25.6 | 36.5 ± 14.9 | 0.4 | 0.4 | 31.9 ± 14.6 | 40.3 ± 28.7 | 34.5 ± 23.1 | 0.3 | 0.3 | 37.0 ± 25.0 | 34.1 ± 15.7 | 1.0 | 1.0 | ||
| 120 min | 30.4 ± 13.5 | 29.0 ± 13.4 | 28.6 ± 9.3 | 0.4 | 0.18 | 29.7 ± 13.7 | 29.8 ± 10.9 | 0.5 | 0.5 | 26.3 ± 12.3 | 31.4 ± 13.9 | 31.0 ± 13.0 | 0.15 | 0.05 | 29.8 ± 13.7 | 29.3 ± 8.5 | 0.7 | 0.7 | ||
| GIP (pmol/l) | ||||||||||||||||||||
| 0 min | 15.5 ± 11.3 | 12.5 ± 7.3 | 12.1 ± 4.6 | 0.3 | 0.12 | 14.8 ± 10.7 | 12.1 ± 5.7 | 0.6 | 0.6 | 16.4 ± 14.2 | 13.9 ± 7.8 | 12.5 ± 6.6 | 0.2 | 0.09 | 14.7 ± 10.5 | 12.4 ± 5.2 | 0.8 | 0.8 | ||
| 30 min | 84.5 ± 30.3 | 101.2 ± 47.8 | 71.8 ± 28.4 | 0.10 | 0.10 | 89.0 ± 37.1 | 84.7 ± 33.9 | 0.8 | 0.8 | 87.6 ± 31.7 | 90.1 ± 41.3 | 84.1 ± 27.4 | 0.8 | 0.5 | 88.1 ± 37.3 | 93.6 ± 27.1 | 0.5 | 0.5 | ||
| 120 min | 71.4 ± 29.1 | 76.1 ± 35.3 | 60.1 ± 26.0 | 0.8 | 0.9 | 72.0 ± 31.1 | 73.3 ± 29.5 | 0.9 | 0.9 | 74.7 ± 34.2 | 70.2 ± 30.3 | 73.7 ± 24.8 | 0.7 | 0.8 | 71.8 ± 31.2 | 76.2 ± 26.9 | 0.5 | 0.5 | ||
Data are means ± SD unless otherwise indicated. For statistical analysis, data were log transformed. Incretin data were adjusted for sex, age, and BMI. Padd, additive model; Pdom, dominant model.
DISCUSSION
Two recent GWA studies showed that common genetic variation in KCNQ1 is associated with type 2 diabetes (12,13). One SNP, rs2237892, has been found to be associated with a fasting parameter of insulin secretion (homeostasis model assessment of β-cell function) in a Japanese population and with an OGTT-derived insulin secretion parameter (corrected insulin response) in a European cohort (13).
In a German population at increased risked for type 2 diabetes, we detected nominal associations of KCNQ1 SNPs rs151290, rs2237892, rs2237895, and rs2237897 with several OGTT-derived indexes of insulin secretion, including C-peptide at 30 min during OGTT, first-phase insulin secretion, and insulinogenic index. Whereas insulin secretion was lower in homozygous major allele carriers of rs151290 (CC), rs2237892 (CC), and rs2237897 (CC), β-cell function was improved in homozygous major allele carriers of rs2237895 (AA). Thus, our data confirm the previous study reporting an association between rs2237892 and indexes of insulin secretion (13). Furthermore, our findings are in agreement with the two previous studies that identified the C allele as the type 2 diabetes risk allele for rs151290, rs2237892, rs2237895, and rs2237897 (12,13).
None of the SNPs was associated with insulin secretion during IVGTT, pointing to an influence of common genetic variation in KCNQ1 on incretin secretion or incretin signaling. Recently, we found that SNPs of the two diabetes susceptibility genes TCF7L2 and WFS1 were associated with impaired GLP-1 signaling that contributed to the pathogenetic mechanism (23,24). In contrast, none of the KCNQ1 variants were associated with GLP-1–induced insulin secretion. However, we found an association between rs151290, the SNP with the most prominent effect on insulin secretion after an oral glucose load, and glucose-stimulated GLP-1 and GIP levels. These results may indicate that altered incretin secretion after food intake provides a potential link between KCNQ1 gene variants and impaired β-cell function. In line with this assumption, KCNQ1 is expressed along the entire gastrointestinal tract (25) and is involved in transport mechanisms in gastrointestinal epithelia (26).
It is worth noting that associations with alterations of glucose-stimulated incretin secretion were found only for rs151290, though rs2237892, rs2237895, and rs2237897 were also associated with indexes of insulin secretion during OGTT. The reason for these inconsistent results could be either that the effects of rs2237892, rs2237895, and rs2237897 on incretin secretion may be too small to be detected in our limited sample size or that these KCNQ1 variants regulate insulin secretion differently than rs151290.
We are aware that the SNPs presented are located within intronic noncoding regions and that, therefore, the mechanisms of their actions remain elusive. The NCBI Reference Sequence (RefSeq) of KCNQ1 contains 14 missense mutations, two frame-shift mutations, one nonsense mutation, and one SNP in the 5′-untranslated region. Only 4 of these 18 mutations are captured by the HapMap data. None of these SNPs are in linkage disequilibrium with any of the three chosen SNPs rs151290, rs2237892, and rs2237897. SNP rs2237895 is also not captured by the HapMap data. However, we cannot rule out that the chosen SNPs may be in linkage disequilibrium with a functional candidate that is not captured by the HapMap data. Alternatively, given that none of the chosen SNPs are located in coding regions, common genetic variants in KCNQ1 may affect gene expression and not the function of the gene product.
The present study has certain limitations that need to be taken into account. First, our study comprised subjects at an increased risk for type 2 diabetes, which may affect the phenotype of incretin secretion or mask some other effects of KCNQ1 SNPs. Second, we were not able to detect effect sizes smaller than 53% with sufficient power (80%) in the combined hyperglycemic clamp study. Thus, effects sizes of KCNQ1 SNPs below 53% possibly remained undetected in this study. Therefore, we cannot rule out that genetic variation in KCNQ1 may, in addition to its effects on glucose-stimulated incretin secretion, also alter GLP-1–induced insulin secretion.
In summary, common genetic variation in the KCNQ1 gene is associated with β-cell function in our German population at increased risk of type 2 diabetes, confirming previous data in Japanese and European cohorts. The discrepancy between orally and intravenously administered glucose seems to be explained not by altered incretin signaling but most likely by changes in incretin secretion.
Supplementary Material
Acknowledgments
This study was supported in part by grants from the German Research Foundation (Fr 1561/5-1, Ga 386/9-1, and Heisenberg-Grant STE 1096/1-1).
The study was also supported by Merck Sharp & Dohme. No other potential conflicts of interest relevant to this article were reported.
We thank all study participants for their cooperation. We thank the International HapMap Consortium for the public allocation of genotype data. We acknowledge the excellent technical assistance of Anna Bury, Heike Luz, Alke Guirguis, Melanie Weisser, and Roman Werner.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Sladek R, Rocheleau G, Rung J, et al. : A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445: 881– 885 [DOI] [PubMed] [Google Scholar]
- 2.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, Novartis Institutes of BioMedical Research, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007; 316: 1331– 1336 [DOI] [PubMed] [Google Scholar]
- 3.Zeggini E, Weedon MN, Lindgren CM, et al. : Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007; 316: 1336– 1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott LJ, Mohlke KL, Bonnycastle LL: A genomewide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007; 316: 1341– 1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hoek M, Dehghan A, Witteman JC, et al. : Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes 2008; 57: 3122– 3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grarup N, Rose CS, Andersson EA, et al. : Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes 2007; 56: 3105– 3111 [DOI] [PubMed] [Google Scholar]
- 7.Staiger H, Machicao F, Stefan N, et al. : Polymorphisms within novel risk loci for type 2 diabetes determine beta-cell function. PLoS ONE 2007; 2: e832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchhoff K, Machicao F, Haupt A, et al. : Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia 2008; 51: 597– 601 [DOI] [PubMed] [Google Scholar]
- 9.Boesgaard TW, Zilinskaite J, Vänttinen M, et al. : The common SLC30A8 Arg325Trp variant is associated with reduced firstphase insulin release in 846 non-diabetic offspring of type 2 diabetes patients: the EUGENE2 study. Diabetologia 2008; 51: 816– 820 [DOI] [PubMed] [Google Scholar]
- 10.Staiger H, Stancáková A, Zilinskaite J, et al. : A candidate type 2 diabetes polymorphism near the HHEX locus affects acute glucose-stimulated insulin release in European populations: results from the EUGENE2 study. Diabetes 2008; 57: 514– 517 [DOI] [PubMed] [Google Scholar]
- 11.Stancáková A, Pihlajamäki J, Kuusisto J, et al. : SNP rs7754840 of CDKAL1 is associated with impaired insulin secretion in nondiabetic offspring of type 2 diabetic subjects (the EUGENE2 study) and in a large sample of men with normal glucose tolerance. J Clin Endocrinol Metab 2008; 93: 1924– 1930 [DOI] [PubMed] [Google Scholar]
- 12.Unoki H, Takahashi A, Kawaguchi T, et al. : SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008; 40: 1039– 1040 [DOI] [PubMed] [Google Scholar]
- 13.Yasuda K, Miyake K, Horikawa Y, et al. : Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008; 40: 1092– 1097 [DOI] [PubMed] [Google Scholar]
- 14.Neyroud N, Richard P, Vignier N, et al. : Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long-QT syndrome. Circ Res 1999; 84: 290– 297 [DOI] [PubMed] [Google Scholar]
- 15.Barhanin J, Lesage F, Guillemare E, et al. : K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 1996; 384: 78– 80 [DOI] [PubMed] [Google Scholar]
- 16.Towbin JA, Vatta M: Molecular biology and the prolonged QT syndromes. Am J Med 2001; 110: 385– 398 [DOI] [PubMed] [Google Scholar]
- 17.Thévenod F: Ion channels in secretory granules of the pancreas and their role in exocytosis and release of secretory proteins. Am J Physiol Cell Physiol 2002; 283: C651– C672 [DOI] [PubMed] [Google Scholar]
- 18.Ullrich S, Su J, Ranta F, et al. : Effects of I(Ks) channel inhibitors in insulin-secreting INS-1 cells. Pflugers Arch 2005; 451: 428– 436 [DOI] [PubMed] [Google Scholar]
- 19.Stefan N, Machicao F, Staiger H, et al. : Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia 2005; 48: 2282– 2291 [DOI] [PubMed] [Google Scholar]
- 20.Fritsche A, Stefan N, Hardt E, et al. : A novel hyperglycaemic clamp for characterization of islet function in humans: assessment of three different secretagogues, maximal insulin response and reproducibility. Eur J Clin Invest 2000; 30: 411– 418 [DOI] [PubMed] [Google Scholar]
- 21.Deacon CF, Johnsen AH, Holst JJ: Degradation of glucagon-like peptide-1 by human plasma in vitro yields an N-terminally truncated peptide that is a major endogenous metabolite in vivo. J Clin Endocrinol Metab 1995; 80: 952– 957 [DOI] [PubMed] [Google Scholar]
- 22.Deacon CF, Nauck MA, Meier J, et al. : Degradation of endogenous and exogenous gastric inhibitory polypeptide in healthy and in type 2 diabetic subjects as revealed using a new assay for the intact peptide. J Clin Endocrinol Metab 2000; 85: 3575– 81 [DOI] [PubMed] [Google Scholar]
- 23.Schäfer SA, Tschritter O, Machicao F, et al. : Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia 2007; 50: 2443– 2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäfer SA, Müssig K, Staiger H, et al. : A common genetic variant in WFS1 determines impaired glucagon-like peptide-1-induced insulin secretion. Diabetologia 2009; 52: 1075– 1082 [DOI] [PubMed] [Google Scholar]
- 25.Dedek K, Waldegger S: Colocalization of KCNQ1/KCNE channel subunits in the mouse gastrointestinal tract. Pflugers Arch 2001; 442: 896– 902 [DOI] [PubMed] [Google Scholar]
- 26.Vallon V, Grahammer F, Volkl H, et al. : KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proc Natl Acad Sci U S A 2005; 102: 17864– 17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.