Abstract
With the publishing of the first complete, whole genome of a human cancer and its paired normal, we have passed a key milestone in the cancer genome sequencing strategy. The generation of such data will, thanks to technical advances, soon become commonplace. As a significant number of proof-of-concept studies have been published, it is important to analyze now the likely implications of this data and how it might frame cancer research in the near future. The diversity of genes mutated within individual tumor-types, the most striking feature of all studies reported to date, challenges gene-centric models of tumorigenesis. While cancer genome sequencing will revolutionize certain aspects of personalized care, the value of these studies in facilitating the development of new therapies, their primary goal, appears less promising. Most significantly, however, the cancer genome sequencing strategy, as currently applied, fails to characterize the most relevant genomic features of cancer – the mutational heterogeneity within individual tumors.
Keywords: Cancer Genome, Mutations, Sequencing, Molecular Heterogeneity
Cancer is a genomic disease associated with the accumulation of mutations (1). The prevailing models of tumorigenesis stress tumor progression as the result of sequential mutations in key cancer genes, each mutation driving a new round of clonal proliferation (2,3). By cataloging all clonal mutations found in a variety of cancers, it is hoped that it will be possible to identify cancer's key mutated genes and, in so doing, revolutionize basic and clinical cancer research (4). A series of studies have now been undertaken to annotate the cancer genomes of a number of different tumor types and it is timely to summarize these results and to evaluate the implications of these findings.
Initial results on sequencing of tumor genomes
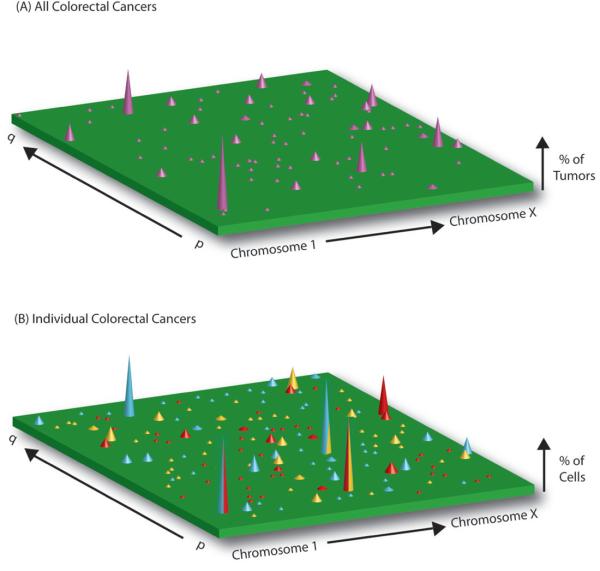
Early attempts at systematically identifying multiple somatic mutations within individual cancers yielded few prevalent clonal alterations (5-7). The relatively limited sequence coverage of these initial studies spurred on attempts at more comprehensive screens of the cancer genome. The first complete sequencing of all likely coding exons of the human cancer genome, conducted in breast and colon cancers (8,9), concluded that these cancers contain a median of 84 and 76 clonal mutations, respectively, which alter protein function (Table 1). While almost ten percent of the 18,197 genes studied were detectably mutated in at least one specimen, each tumor displayed a unique and diverse profile of mutated genes and, other than those previously known (e.g. TP53, APC), no new prevalently mutated genes were identified. The authors of this study proposed that the cancer genome landscape is composed of a handful of commonly mutated gene “mountains” but dominated by a vastly larger number of infrequently mutated gene “hills” (Fig. 1a); a view consistent with a large number of mutations, each providing a small fitness advantage, driving tumor progression (9). It is this clonal mutational heterogeneity, the authors concluded, which underlies the wide variation in tumor behavior and responsiveness to therapy.
Table 1.
Selected details of Cancer Genome Sequencing Studies.
| Study | Cancer type | Total number of genes analyzed |
No. of mutated genes |
Total no. of mutations (non-silent) |
Average no. of mutations per tumour |
|---|---|---|---|---|---|
| Sjoblom et al.(7) & Wood et al. (8) |
Breast (n=11)* |
18,191 | 1,137 | 1,243 | 84 |
| Colorectal (n=11)* |
848 | 942 | 76 | ||
| Greenman et al. (9) | Diverse (n=210) |
518 | 581 | 798 | NA |
| Jones et al. (10) | Pancreatic (n=24) |
20,661 | 1,007 | 1,163 | 48 |
| Parson et al. (11) | Glioblastoma (n=21)# |
20,661 | 685 | 748# | 47 |
| TCGA (12) | Glioblastoma (n=91) |
601 | 223 | 453 | NA |
| Ding et al. (13) | Lung (n=188) | 623 | 348 | 1,013 | NA |
| Ley et al. (14) | Acute Myeloid Leukemia (n=1) |
Whole Genome |
10 | 500-1,000 | NA |
Abbreviations for Table 1
NA: Not applicable
Discovery phase of study only
Excludes post-therapy samples and MMR deficient samples
Figure 1. The Mutational Landscape of the Cancer Genome.
(a) The cancer genome landscape of colorectal cancer as proposed by Wood et al. (9) demonstrates the mutational heterogeneity between individual tumors. The height of each violet peak represents the percentage of colorectal tumors found to carry a clonal mutation in a particular gene. The landscape comprises a small number of “mountains”, genes which are mutated in a large number of cancers [e.g. TP53 (23)], and a significantly larger number of “hills”–genes mutated in only a small number of individual tumors. While there may be 50 or more genes clonally mutated in a given tumor, any given gene is rarely mutated in more than a few tumors. (b) The mutational heterogeneity within individual tumors not detected by the cancer genome sequencing studies. The mutational landscape of three tumors are shown; each tumor is represented by a set of differently colored peaks (blue, red and yellow). The height of each peak reflects the number of cells within a single tumor that have a particular mutated gene. Split colored peaks represent the small number of mutated genes shared in common between more than one tumor. Within an individual neoplasm, a few mutations are present throughout the population (tall peaks), a greater number are present in minority subclones (short peaks) and the majority are found in only one or a few cells (invisibly distributed throughout the green background.)
Subsequent studies on diverse cancer types (Table 1) have generated the most extensive catalogs of somatic point mutations yet undertaken (10-14). The primary conclusion to be drawn from these studies is that most cancer types display substantial heterogeneity at the genetic level. Individual solid organ tumors harbor on average more than 50 non-silent mutations in the coding regions of different genes (Colorectal≈Breast>>Pancreatic>GBM), and although a few of these genes are mutated in a high proportion of tumors, the prevalence at which the majority are mutated among different tumors of the same cancer type is low. Additionally, these studies focused exclusively on identifying mutations in exons of known protein-coding genes. With the ever-increasing recognition that so-called “junk” DNA and intronic sequences contain functional elements, including regulatory regions and non-coding RNAs, the total number of clonal mutations with functional consequences cannot be easily estimated.
The characterization of the first hematopoietic cancer genome represents an important methodological milestone in cancer genome sequencing – truly unbiased whole genome sequencing (15). Prior attempts at re-sequencing the tyrosine kinase gene family in acute myeloid leukemia (AML) yielded few mutations (16,17). By exhaustively interrogating the cancer and paired normal genomes of a single AML patient using massively parallel sequencing, Ley et al. (15) identified 500-1000 non-synonymous somatic mutations throughout the cancer genome of which 10 mutations were in protein-coding genes. While the overall number of protein coding mutations is considerably fewer than that described for solid organ tumors, similar to the mutational diversity of other tumor-types, the genes identified are mutated in only a small fraction of AML cases. Surprisingly, none of the 8 newly identified genes were found to be mutated in a further 187 cases of AML.
While the absence of prevalently mutated genes is disappointing from a targeted theraputic perspective, the major shortcoming of all the above studies is not in the complexity of the problem they are attempting to address, but in the fact that, by design, they do not analyze the deeper heterogeneity within individual tumors. A primary tumor is itself genomically heterogeneous, with each cell having a unique mutational signature. This genetic variation within a cancer cell population reflects the history and dynamics of clonal evolution and, importantly, serves as a reservoir of genetic diversity from which therapy-resistant clones may arise (18). Analysis of disseminated single cells in minimal residual disease has demonstrated that there is a high level of genomic heterogeneity within individual lesions as well as between primary tumors and metastatic cells (19, 20). While current “next-generation” sequencers can detect multiple rare sub-clones with frequencies as low as 1 in 5,000 (21), analyzing rarer sub-clones awaits an, as yet, undeveloped future generation of DNA sequencing methodologies. It is this intra-specimen diversity, not detected by the cancer genome sequencing studies, that may be clinically most relevant and responsible for much of the diverse spectrum of clinical phenotypes. The question arises of whether there is, at a clinically meaningful level, a single consensus cancer genome per tumor?
We posit that a complete description of the cancer genome must capture the mutational heterogeneity within individual neoplasms as well as between different tumors. In addition to the clonal genomic landscape proposed, comprising “mountains” and “hills” of frequently and infrequently mutated genes within a given tumor-type, additional mutational diversity exists within individual tumors themselves, with a large number of sub-clonal mutations being present in only one or a few cells (Fig. 1b). It is this mutational landscape, overlooked by the cancer genome sequencing studies to date, that is the basis for the wide variations in tumor behavior and responsiveness to therapy, and represent the clinically most important features of the cancer genome.
New DNA sequencing technologies are undoubtedly important for guiding biological and medical research, however many limitations exist. To accurately quantify the sub-clonal diversity of a single tumor, it may be necessary to sequence millions of individual genome equivalents per tumor. Present sequencing efforts only detect mutations that are present in the majority of a neoplasm's cells. Until more advanced technologies become available or existing ones are retooled, a critical feature of the genomic landscape will continue to go unexplored.
The mutational heterogeneity of cancer, which has been recognized for many years (22) and now confirmed by DNA sequencing, underlies the complexity of the cancer genome landscape. Currently, the heterogeneity within individual tumor types alone confounds many of the cancer genome strategy's original expectations. A future of multiple targeted therapies and patient stratification, based on a mutational signature of defined key genes for each cancer type, appears less hopeful than initially anticipated. While ongoing studies will continue to uncover the remarkable heterogeneity inherent in tumorigenesis, without addressing the deeper complexity of each individual cancer genome, we may not significantly impact patient care.
Acknowledgments
Grant Support: NIH grants CA102029, CA78885 (L.A. Loeb), AG033485 (J.J. Salk) and NCI/Health Research Board of Ireland Joint Research Fellowship (E.J. Fox).
Footnotes
The authors declared no potential conflicts of interest.
References
- 1.Weinberg RA. The Biology of Cancer. Garland Science. 2006 [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–28. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 4.Pollack A. New York Times. 2005 March 28; Sect. A1. [Google Scholar]
- 5.Wang TL, Rago C, Silliman N, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci U S A. 2002;99:3076–80. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 8.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 9.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 10.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasson MH, Xiang Z, Walgren R, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loriaux MM, Levine RL, Tyner JW, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–96. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci U S A. 2003;100:776–81. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoecklein NH, Hosch SB, Bezler M, et al. Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell. 2008;13:441–53. doi: 10.1016/j.ccr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Klein CA, Schmidt-Kittler O, Schardt JA. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl. Acad. Sci. USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell PJ, Pleasance ED, Stephens PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–21. [PubMed] [Google Scholar]
- 23.Harris CC. p53: at the crossroads of molecular carcinogenesis and cancer risk assessment. 1993;262:1980–1981. doi: 10.1126/science.8266092. [DOI] [PubMed] [Google Scholar]