Abstract
Chronic obstructive pulmonary disease (COPD) is a complex disease with multifactorial background, based on the interaction of environmental and genetic factors. Environmental factors are clearly related to the development of the disease. However, family and twin studies suggested genetics factors to be one of the important determinants for the development of COPD. Different approaches have been used to identify genes of interest. Genomewide linkage analysis found areas of interest on different chromosomes, with some genes located in this regions being identified and replicated as susceptibility genes. Numerous of candidate genes that could be linked to disease pathogenesis have been implicated in COPD genetics. However, the candidate gene approach is often limited by inconsistent results in other study populations. Recently, a combination of different methods is used giving more evidence for some candidate genes, including TGFß-1, Surfactant, SERPINE2 and microsomal epoxide hydrolase. In the future ongoing exact phenotype definition, combination of several approaches, genome-wide association studies and animal model genetics will lead to new insights into the genetics of COPD, with epigenetic factors needs to be further investigated and considered in concert with genetic findings.
Introduction
Chronic obstructive pulmonary disease (COPD) is a major health problem as it causes increasing mortality and morbidity in all industrial countries (Pauwels et al 2001; Hurd 2005). Progressive irreversible airflow limitation is the central pathogenetic aspect in COPD. However, the disease is not only determined by pulmonary function, but by airway tissue inflammation, airway remodeling and systemic consequences.
The most important risk factor in the etiopathogenesis of COPD is cigarette smoking. Nevertheless, only 10%–20% of all heavy cigarette smokers develop COPD (Fletcher 1976; Redline et al 1987; Sherrill 1990; Global Initiative for Chronic Obstuctive Lung Disease 2001; Mannino 2002, 2003; Mannino et al 2003). There is strong evidence for a relationship between decline in lung function and smoking behavior. However, smoking account only for about 15% of lung function variability (Beck et al 1981). These observations demonstrate that next to smoking other factors seems to be of importance for the development of the disease. Epidemiological data suggest genetics to be one of those factors, as COPD is known to aggregate in families (Larson et al 1970; Tager et al 1976; Lebowitz et al 1984), with a stronger correlation between parents and children or siblings than between spouses (Higgins and Keller 1975; Tager et al 1976; Kauffmann et al 1989). Twin (Redline 1987, 1990) and segregation studies (Givelber 1998) provide evidence that genetic predisposition plays a role in COPD (Hubert et al 1982; Astemborski et al 1985; Redline et al 1987; Rybicki et al 1990; Chen et al 1996; Givelber et al 1998) and had indicated that the genetic background of COPD is composed of several genes with small effects, rather than a single major gene. Despite the general fact that genetic studies in COPD are difficult to conduct due to the variating phenotypes subsume under the disease definition, there are different possible approaches to analyze genetic factors of importance. Basic genetic studies include family and twin studies, as well as segregation studies. In linkage and positional cloning studies susceptibility genes could be identified on the basis of their chromosomal position. The genetic bases of COPD might also be investigated, similar to other diseases with complex and multifactorial etiology, by the analysis of candidate genes – genes that are postulated to play an important role in disease pathogenesis. This review will focus on the candidate gene approach after giving an short overview about the results of linkage studies.
Linkage studies
The genetic bases of lung function and/or COPD had been investigated using linkage or positional cloning studies. Linkage studies investigate haplotypes, or short segments of the genome, conserved between generations by virtue of their size. If a haplotype is found that is passed down along with a disease through a family, it could be speculated that there is a gene within or close to that may have an effect on the disease. To perform linkage analysis phenotypic data and DNA from affected families are needed at least from two generations. From each family all members typed for genetic markers throughout the whole genome, following by determination whether any of these markers are inherited with the disease more often than predicted by chance. In case of a positive correlation the disease is “linked” to that marker on a distinct chromosome. This method provides the possibility to identify novel genes, because linkage of the disease to a genetic marker depends only on the close proximity of that marker with the susceptibility gene, independent of a potential functional correlation. But these studies are not easy to conduct in COPD since the disease has a late onset in life and family members that are needed for analysis may no longer be alive.
Genomewide linkage analysis exploring genetic linkage to lung function was performed in several studies. The genetic determinants of FEV1, FVC and FEV1/FVC ratio had been studied in healthy subjects and patients with COPD. Two population- and one disease-based linkage analysis found strongest evidence of linkage to FEV1/FVC ratio (chromosome 4 [logarithm of the odds favoring genetic linkage (LOD) 3,5], chromosome 2q [LOD 4,12], and chromosome 2 [LOD 4,12]) (Joost et al 2002; Silverman et al 2002; Wilk et al 2003; DeMeo et al 2004). In healthy young adults of the Framingham Heart cohort a genome-wide scan of 330 families with over 1500 members found that loci strongly influencing FEV1 occurred on chromosome 6 [LOD score 2,4] and for FVC on chromosome 21 [LOD 2,6] (Joost et al 2002). In another study including more than 2100 healthy subjects from 391 families who participated in the National Heart, Lung and Blood Institute (NHLBI) Family Heart Study FEV1 and FVC were suggestively linked to regions of chromosome 18 [LOD 2,4]/ [LOD 1.5 / 2,4] (Wilk et al 2003). Interestingly, in a genomewide screen for pulmonary function in families of patients with asthma significant evidence for linkage of pre- and postbronchodilator FEV1%VC was obtained for chromosome 2q (2q32, LOD 4,9/6.03) (Postma 2005). The Boston Early-onset (BEO) studies families with subjects suffering from severe early-onset COPD. A genome-wide scan of 585 family members participating in this study showed the highest LOD score for FEV1 was 1.53 on chromosome 12 and 2.05 for FVC on chromosome 1 (Silverman et al 2002). If only smoking subjects were included, evidence for linkage to FEV1/FVC ratio was increased on chromosome 2 (LOD 4.13) and for FEV1 on chromosome 12p (LOD 3,26). Evidence for linkage of chronic bronchitis or airway obstruction was shown for chromosome 22 (LOD 2.08)/12p (LOD 2,13), respectively. Table 1 summarizes the main results of these linkage studies.
Table 1.
Linkage analysis: genomewide scans
| Study | Population | Main results | LOD score§ |
|---|---|---|---|
| Framingham Heart Study
(Joost et al 2002) |
Healthy Subjects:
1578 members of 330 families |
Chromosome 6: FEV1 | 2.4 |
| Chromosome 21: FVC | 2.6 | ||
| National Heart, Lung and Blood Institute Family Heart Study
(Wilk et al 2003) |
Healthy Subjects:
2178 members of 391 families |
Chromosome 4: FEV1/FVC | 3.5 |
| Chromosome 18: FEV1 | 2.5 | ||
| Chromosome 18: FVC | 2.9 | ||
| Boston Early-onset Study
(Silverman et al 2002) |
Patients with COPD + Pedigrees:
585 members of 72 families |
Chromosome 2q: FEV1/FVC | 4.13 |
| Chromosome 12p: FEV1 | 3.26* | ||
| Chromosome 19q: Chronic bronchitis | 3.30* | ||
| Chromosome 12p: Airway Obstruction | 3.14* | ||
| Netherland Asthma Study
(Postma et al 2005) |
Patients with Asthma + Pedigrees:
200 families |
Chromosome 2q: Pre- and postbronchodilator FEV1%VC | 4.9/6.03** |
LOD > 3 denotes significant evidence for linkage; LOD < 3 denotes suggestive evidence;
only smoker;
pre- and postbronchodilator FEV1%VC was obtained for chromosome 2q (2q32, LOD 4,9/6.03).
Instance areas of interest from linkage studies included SERPINE2 and IL-8 as potential candidate genes (both on chromosome 2), as well as microsomal GST1 and TGFß genes (chromosome 12 and 19) in terms of the severity of pulmonary function in COPD. However, there is no direct evidence for the significance of linkage studies for COPD pathogenesis.
Candidate genes – association studies
Variations, or polymorphism, within genes are classified by means of a change in DNA alone or effect on the protein it codes for. Sigle nucleotide polymorphism consists of a sigle base change are the most common in the human genome. They could either be functional or not. Candidate gene analysis compare the frequency of gene variations or polymorphism in groups with the given disease and control individuals. In diseases with complex and multifactorial etiology, such as COPD, the use of candidate genes may help to identify genetic factors of potential importance. In the candidate gene approach genes are tested directly for their involvement in disease process. Genes that are selected are those that could be postulated to be functional related to the disease. The candidate gene approach is limited on the one hand by the fact that only known genes can be examined, controls and patients are difficult to match and the need of a very exact phenotype definition. On the other hand COPD is a multifactorial disease that is influenced by genetic as well as environmental factors resulting in a complex genotype-environment interaction. However, a reasonable large number of successful association studies had analyzed candidate genes selected on the base of COPD pathophysiology.
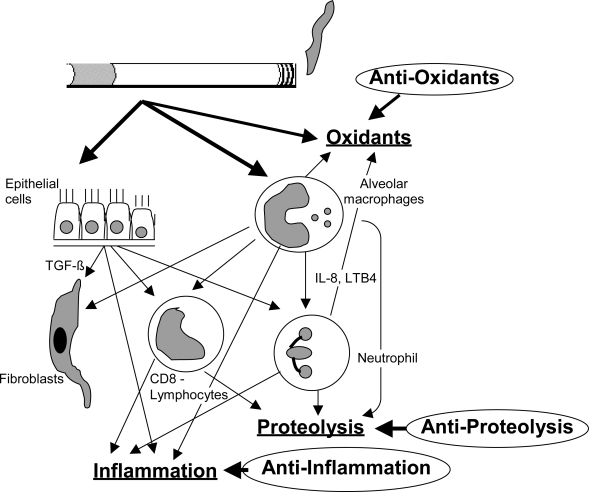
COPD is characterized by a progressive decline in lung function due to increasing irreversible airway obstruction. This results from ongoing chronic airway inflammation and loss of lung elastic recoil due to parenchymal destruction. Many inflammatory cells, mediators, enzymes and maybe infectious agents are involved. The genes that are implicated in the pathogenesis of COPD are involved in proteolysis and antiproteolysis (Shapiro and Senior 1999; Stockley 2001), oxidants and anti-oxidants (MacNee 2000), inflammation (O’Donnell et al 2006), metabolism of toxic substances, airway hyperresponsiveness, mucus homöostasis and host defense mechanisms (O’Donnell et al 2006). Figure 1 show a graphical depiction of cells and mediator systems involved in COPD pathogenesis.
Figure 1.
Graphical depiction of cells and mediator systems involved in COPD pathogenesis. Oxidative stress, inflammation and proteolysis and their opponents are the important components influencing disease pathogenesis.
Reactive oxygen species (ROS) are induced as a direct result of cigarette smoke inhalation and/or increased production by activated inflammatory cells and activation of the xantine oxidase pathway. These oxidants may inhibit by anti-oxidative enzymes, such as α1-antitrypsin. Parenchymal damage may be due to several mechanisms, including oxidative stress, inflammation and apoptosis of vascular endothelial cells.
Numerous candidate genes have been studied in COPD case-control association studies and several candidate genes have been reported to be significantly associated with COPD phenotypes (see Table 1). However, the evidence supporting these associations have been inconsistent in most of the cases due to failure in replication by other studies. The inconsistent replication might be due to the study populations (differences in phenotype definition, control subjects or COPD subtypes and/or racial differences), population stratification, multiple testing, random error or small case numbers. However, combination of different study methods, as performed in recent studies, seems to provide more replicatable results. This review will focus on some candidate genes and recent data.
Proteolysis and antiproteolysis
α1-antitrypsin
Severe alpha1-antitrypsin (AAT) deficiency is the one proven genetic risk factor for COPD. AAT is an acute phase protein that provides the major defense against neutrophil elastase. There are four main variants of AAT, classified by their speed of movement in gel electrophoresis: F = fast, M = medium, S = slow and Z = very slow (Fagerhol and Laurell 1970). In 1963 Laurell and Eriksson demonstrated that individuals with extremely low levels of AAT had an increased prevalence of emphysema. In the following time it was shown that AAT deficiency was usually associated with the Z isoform of AAT (Kueppers et al 1964; Eriksson 1965). The AAT variants are inherited in a co-dominant fashion, with the PiM allele being the wild-type and most prevalent. Homozygosity of the Z variant (Glu342Lys) results in severe AAT deficiency due to a phenotype with reduced plasma AAT levels to 10% compared to normal AAT levels (for review see Needham and Stockley (2004)). However, the risk for COPD among PiMZ heterozygous at the α1-antitrypsin locus remains controversial. A recent meta-analysis compared COPD cases versus control subjects and found an increased risk for COPD among PiMZ subjects, whereas population based studies reported usually similar pulmonary function values (FEV1) in subjects with PiMZ and PiM (Hersh et al 2004). There is significant genotype-environment interaction between the Pi Type and pack years of smoking (Sandford et al 1999). However, this interaction could not explain the complete phenotype variations (Silverman et al 1992), suggesting gene-gene interaction and/or confounding other factors next to AAT deficiency to be of importance.
Matrix metalloproteinases (MMP’s) and tissue inhibitor of MMP’s (TIMP)
Matrix Metalloproteinases comprise a family of more than 20 related proteolytic enzymes that play an important role in tissue remodeling and repair in development and inflammation. These enzymes degrade collagen, inactivate AAT and activate tumor necrosis factor-α (TNF-α). A functional SNP of the MMP9 gene promoter region had been studied in COPD and emphysema and had been associated in both, Chinese and Japanese populations (Minematsu et al 2001; Zhou et al 2004) However, this polymorphim was not associated with COPD in the Lung health Study (Joos et al 2002).
An insertion in the promoter region of MMP1 (G → GG, − 1607) that increases transcription of MMP1 by creating an additional transcription factor binding site, was negatively associated with rapid decline of FEV1 (Joos et al 2002), as well as a haplotype containing the MMP1 together with an MMP12 SNP (Asn357Ser) (Joos et al 2002).
There have been four tissue inhibitors of matrix metalloproteinases (TIMP) described, that inhibit active forms of MMP’s, with different affinities to each MMP. MMP2 is inhibited by TIMP-2 and this system seems to play an important role in emphysema (Ohnishi et al 1998). Two SNP’s of the TIMP2 gene have been associated with COPD in a Japanese population (Hirano et al 2001). The authors speculate that a linked gene may be associated with genetic susceptibility and probably not TIMP itself.
α1-antichymotrypsin (SERPINA 3)
α1-antichymotrypsin (SERPINA3) is capable of inhibiting cathepsin G and mast cell chymase in a reversible fashion. In the SERPINA3 gene two functional SNPs have been associated with low α1-antichymotrypsin levels and COPD (characterized by airway resistance) in a Swedish population (Poller et al 1990, 1993). The association was not replicated in a Japanese Population. However, a third gene variation (non-synonymous mutation with affects to the signal peptide region) was found to be increased in the COPD group. Non of these polymorphims showed differences in an Italian study of patients with airflow obstruction, though next to COPD, the patient group also included subjects with bronchiectasis (Benetazzo et al 1999).
Oxidative stress and antioxidants
An imbalance between oxidative noxes and antioxidants resulting in oxidative stress is postulated to be one of the important mechanisms in the development of COPD. Airway epithelium and lung matrix could be damaged by oxidative stress and enhance lung inflammation with up-regulation of pro-inflammatory cytokines (MacNee 2000). Cigarette smoke is a major source of free radicals, nitric oxide and other oxidants. Moreover, during inflammatory processes radicals are released by leucocytes, that are known to be increased in the lung of patients with COPD. The lung is protected by antioxidative enzymes, including gluthathione-S-transferase, superoxide dismutase and catalase (Young et al 2006). Therefore, functional genetic variants may be of importance by altering antioxidative capacity of the lung.
Glutathione-S-transferase (GST)
The family of glutathione-S-transferase comprises enzymes that can detoxify some noxes of tobacco smoke. There are functional polymorphisms, including one SNP in GSTP1 (A313G → Ile105Val) and three alleles of GSTM1 each affecting enzyme activity.
The 105Ile variant of the GSTP1 gene was associated with airflow obstruction in a Japanese population and with rapid decline of FEV1 as well as low baseline lung function in the Lung Health Study (He et al 2002; He 2004).
The GSTM1 alleles include one null allele, having no detectable GSTM1 activity in case of homozygosity. This genotype had been previously linked to emphysema and COPD (Baranova et al 1997; Harrison et al 1997). However, there are negative studies concerning airflow obstruction and rapid decline in lung function (Yim et al 2000; He et al 2002).
Superoxide dismutase (SOD)
Superoxide dismutase belongs to three superoxides that can scavenger reactive oxygen species (ROS). They are part of the protection system of the lung, particularly during inflammation. A functional SNP (C760G) in the SOD3 gene was found to have a protective effect in Caucasian COPD populations (Juul et al 2006; Young et al 2006), probably dependent on gene-smoking interaction, as the genetic variant of the SOD3 gene may confer protection from adverse affects of cigarette smoke.
Microsomal epoxide hydrolase
Microsomal epoxide hydrolase (mEH) is expressed in human bronchial epithelial cells and detoxify highly reactive intermediates present in cigarette smoke (Bartsch et al 1992). There are two functional polymorphisms that affect enzyme activity that account for a modest change in activity level. For individuals carrying both His variants (exon3: Tyr113His; exon4: His139Arg) a higher risk for development COPD and emphysema were demonstrated (Yoshikawa et al 2000). SNPs in the microsomal epoxide hydrolase (mEH) gene have been associated with severe COPD cases, emphysema and COPD phenotypes (Smith and Harrison 1997; Koyama and Geddes 1998; Yoshikawa et al 2000; Park et al 2005), as well as with rapid decline in FEV1 in smokers (Sandford et al 2001; He et al 2002). However, the association failed to be replicated in a Corean population (Yim et al 2000).
Inflammation and inflammatory mediators
COPD is characterized by chronic airflow obstruction due to chronic inflammation based on an abnormal inflammatory response (Celli and MacNee 2004). Different mediators and cells have been implicated in the pathogenesis of COPD, including TNFα, IL8 and TGF-ß. This extends beyond the lung to systemic manifestations, with circulating mediators and/or cytokines in patients serum (Sevenoaks and Stockley 2006). Cigarette smoke activates macrophages to release TNF-α, LTB4, IL-8 and other neutrophil chemotactic factors, as well as antiproteases. TNF-α promotes further IL-8 release from other cells in the respiratory tract by NF-κB mediated effects on gene transcription. This increases local neutrophilic inflammation, and hence the release of proteases. Epithelial cells also stimulate fibroblasts via TGFß, a key-player in the pathogenesis of fibrosis.
Tumor necrosis factor – alpha (TNF-α)
There is one SNP in the promoter region of the TNF-α gene, that is known to be functional by directly affecting gene regulation (Wilson et al 1997). Heterogeneity of study results demonstrate a relevance for the SNP – 308 in Asian but not in Caucasian populations, as in Taiwanese and Japanese collectives there is an increased prevalence in COPD compared to controls (Huang et al 1997). Additionally, airflow obstruction without chronic bronchitis, and severity of emphysema had been associated with this polymorphism in Japanese subjects (Sakao et al 2001, 2002). However, these results could not be confirmed in Caucasian collectives (Sandford et al 2001; Hersh et al 2005; Seifart et al 2005), maybe due to variation in genotype frequencies between races, or by linkage disequilibrium with HLA alleles, seen previously in the Caucasian population (Wilson et al 1993).
Transforming growth factor-beta (TGFß)
TGFß regulates extra-cellular matrix production, cell growth and differentiation, tissue repair and some immune responses (Blobe et al 2000). Animal experiments suggests that disordered activation of TGFß relates to the pathogenesis of COPD.
Two recent studies, with new study designs using a combination of different methods, strongly suggests TGFß1 to be of importance for COPD phenotypes. On the basis of the linkage analysis in the Boston Early-Onset COPD study that revealed suggestive linkage between an area on chromosome 19q – containing the TGFß1 gene – and FEV1 (Celedon et al 2004), TGFß was genotyped in subjects of the Boston Early-Onset COPD study. Out of five, three SNPs, one in the TGFß1 promotor region and two SNPs in the 3’ genomic region of TGFß1, had been significantly associated with pre- and post-bronchodilator FEV1. Again, among 304 cases from the National Emphysema Treatment Trail (NETT) and smoking controls, this association was replicated for two of the SNPs (Hersh et al 2006), who linked them both to subjective measures of dyspnoea, though not objective exercise capacity. This apparent discordance may be important when defining phenotypes within COPD. The latter two SNPs have an effect on TGFß1 levels, as the one in the promoter region C-509T enhances promoter function, thus increasing levels of TGFß (Grainger et al 1999). While the second is a C -> T change at position 613, that leads to an amino acid substitution (Leu -> Pro) and higher production of TGFß1 (Suthanthiran et al 2000). If both of these SNPs are implicated in COPD subjects, it suggests that TGFß may have a protective role.
Recently, the Pro allele was found to be less common in COPD subjects relative to resistant smokers (OR: 0.59, p = 0.01) and controls (OR: 0.62, p = 0.005) (Wood and Stockley 2006).
Vitamin D binding protein
Vitamin D binding protein is a precursor of macrophage activating factor (MAF) and increases the neutrophil chemotactic properties of different peptides. This function could be prevented by neutrophile elastase inhibitors pointing out to a potential relationship between the protease-antiprotease pathway and inflammation.
Two non-synonymous SNPs had been identified in the Vitamin D binding protein (GC2 and GC1S alleles), with the GC2 allele seemed to be protective in studies of Caucasian subjects (Horne et al 1990; Schellenberg et al 1998). The GC1S allele has not been shown to have a significant association with COPD.
The GC1F allele has been associated with an increased risk of developing airflow obstruction, emphysema and rapid decline of FEV1 in Japanese subjects. However, the results for homozygous individuals concerning an increased risk of developing COPD in Caucasian populations are conflicting (Horne et al 1990; Schellenberg et al 1998). Furthermore, the allele failed to be linked to rapid decline in lung function (Sandford et al 2001). The study heterogeneity might be due to difference in allele frequency between the racial groups.
Interleukin 13
Mice overexpressing IL-13 develop cathepsine and matrix metalloproteinase dependent emphysema (Zheng et al 2000).
A functional polymorphism in the promoter region (C-1055T) of the IL-13 gene leads to increased IL-13 production (van der Pouw Kraan et al 1999). The T allele had been demonstrated to be more common in COPD patients (van der Pouw Kraan et al 2002), but this association has not been replicated yet.
Surfactant and SERPINE2
Surfactant
Surfactant proteins are important for normal lung function and play a role in the innate host defense and control of inflammation in the lung. Genetic heterogeneity of the surfactant proteins may explain protein and mRNA content in the lung.
Surfactant protein B is important for the formation of the active surfactant surface film (Schurch et al 1998) and is essential for normal lung function (Robertson et al 1991). SP-B knock out mice show disruption of surfactant film and function and die from respiratory failure (Clark et al 1995, 1997). Furthermore, those mice are more susceptible to oxidative lung injury (Tokieda et al 1999).
A gene variation within intron 4 of the surfactant protein B (SP-B) gene had been associated with respiratory failure in COPD (Seifart et al 2002). One of the known SNPs (B1580 C/T = Thr131Ile) in the SP-B gene was associated with COPD in the Boston Early-Onset study and was confirmed in a following family study (Hersh et al 2005). The same polymorphism, had been previously associated as a smoking dependent risk factor in a case-control study in a Mexican population (Guo et al 2001). When gene-environment interaction was taking into account, the same polymorphism was associated with dyspnoea score and exercise capacity in the NETT cohort (Hersh et al 2006). Thus, surfactant protein B polymorphisms seemed to be of importance in the genetic background of COPD, probably especially in fact of gene-smoking or gene-environment interaction.
SERPINE 2
Prior linkage analysis in the Boston Early Onset COPD study demonstrated significant linkage of a region on chromosome 2q with COPD. Candidate gene selection in this region were done by integration of the linkage results with results from murine lung development and gene-expression studies of human COPD tissue (Demeo et al 2006). This procedure identified SERPINE2 as a positional candidate susceptibility gene for COPD. Significant association of 18 SNPs in the SERPINE2 gene with COPD phenotypes were demonstrated in a family-based study, while five of these could be confirmed in a case control analysis with cases from the National Emphysema Treatment Trail.
SERPINE2 is known to be an inhibitor of thrombin, urokinase and plasmin. There are no studies investigating a separate different function of SERPINE2 in the lung so far. Nevertheless, SERPINE2 is postulated to be a COPD – susceptibility gene that is likely influenced by gene-by-smoking interaction (Demeo et al 2006).
Recent study approach
There are recent studies combining several study methods analysing candidate gene to obtain more reliable data. This include the combination of family-based and case-control studies, emerging data from linkage analysis to identify possible candidate genes and including DNA microarray data (Celedon et al 2004; Hersh et al 2005; Demeo et al 2006; Hersh et al 2006a, 2006b).
Hersh et al (2005) genotyped selected candidate genes previously reported to be important in COPD in a family based study, followed by a case-control study. Only one polymorphism in the surfactant protein B gene (SFTPB Thr131Ile) and one in the heme oxygenase gene were replicated across both studies. A similar approach was used in 80 markers in 22 positional or biological plausible candidate genes with exact phenotype definition including COPD related phenotypes. The analysis revealed SNPs in the epoxide hydrolase, latent transforming growth factor ß binding protein-4 (LTBP4), TGF-ß1 and again SP-B to be significant associated with COPD related phenotypes (Hersh et al 2006).
Data obtained from previous linkage analysis were used in two recent studies. One integrated additional results from gene expression profiling (Demeo et al 2006), and both were followed by association analysis in a family based samples and a case control study. One identified SERPINE2 to be a COPD-susceptibility gene, likely influenced by gene-smoking interaction, while the other associated TGFß1 gene variants with the pathogenesis of COPD among smokers (Celedon et al 2006).
Gene expression profiling
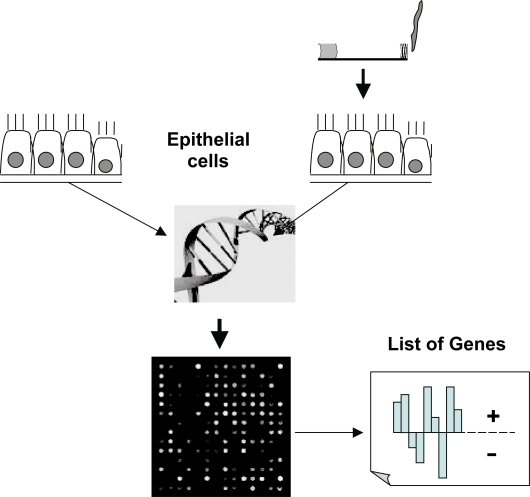
Gene expression profiling using cDNA microarrays may identify the expression of several thousand genes simultaneous. The method point a “photograph” of the current genetic activity in one tissue in comparison to another. Figure 2 shows graphically the principle of the method. Gene expression profiling using cDNA microarrays may provide the possibility to identify candidate genes involved in the pathogenesis of emphysema, though lung tissue represents an end-stage picture of gene expression. Linkage of the obtained gene expression profiles to chromosomal regions that were previously associated with COPD in linkage analyses will focus enclose potential candidate genes. cDNA microarrays are capable of profiling gene expression patterns of thousands of genes and/or gene clusters. It could be used to detect variations in transcriptional programs, including signal transduction and regulatory systems.
Figure 2.
Shows graphically the principle of gene expression profiling using cDNA microarrays. The activity of up to 20000 genes could be simultaneous evaluated. Usually two tissues or cell populations are compared and the final result shows the different genetic activity of the sample of interest compared to a reference control.
One study using microarray technology in lung tissue from patients with severe emphysema and controls, with one group of patients that had been previously diagnosed as AAT-deficient, showed surprisingly only modest differences between individuals with normal lung function and patients with end-stage emphysema (Golpon et al 2004). However, 150 genes were significantly different expressed between normal and emphysematous lung tissue samples. Expression of about 30–50 genes differ between emphysema, emphysema due to AAT-deficiency and controls, with a reduction in protein biosynthesis capacity as a clear characteristic of the lung gene expression pattern of patients with AAT deficiency-related emphysema. Significant heterogeneity within the individuals of each group emphasize the multifactorial nature of the disease. In another study, that compares lung tissue of smokers with normal to mild or severe emphysema 102 genes distinguished between the different types of smoking related lung tissue (Spira et al 2004). A total of 76 genes were upregulated in the lungs of smokers with severe emphysema. These include extracellular matrix (ECM)-related genes, whereas many immune and cell signaling-related genes were downregulated in these lungs. Overrepresented gene categories comprises ECM constituents and stress-related genes with oxidoreductase, isomerase, and complement activity. The study revealed distinct molecular subclasses of severe emphysema with the body mass index being the only clinical variable that differed between the groups.
The future
There had been some progress in the last years in genetics of chronic obstructive pulmonary disease. However, there are many areas that require more investigation. Not at least, epigenetic needs have to be considered.
Future genetic studies will include additional linkage analysis with and without combination of gene expression profiling. Focusing on novel phenotypes, defined by molecular or clinical means, will be essential in identifying genetic determinants for COPD. Animal model genetics may help in identifying several pathways of importance and provide more insight in some aspects of pathogenesis.
Recently, analyses using the genomewide association analysis approach (Hirschhorn 2005; Hirschhorn and Daly 2005) – an aproach that involves genotyping of 300,000–5,000,000 SNPs across the whole genome – is expected to provide new insights into COPD pathogenesis, though it still has limitations concerning statistical calculation, number of investigated SNPs and costs of the studies.
References
- Astemborski JA, Beaty TH, et al. Variance components analysis of forced expiration in families. Am J Med Genet. 1985;21:741–53. doi: 10.1002/ajmg.1320210417. [DOI] [PubMed] [Google Scholar]
- Baranova H, Perriot J, et al. Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum Genet. 1997;99:822–6. doi: 10.1007/s004390050455. [DOI] [PubMed] [Google Scholar]
- Bartsch H, Castegnaro M, et al. Expression of pulmonary cytochrome P4501A1 and carcinogen DNA adduct formation in high risk subjects for tobacco-related lung cancer. Toxicol Lett. 1992:64–65. doi: 10.1016/0378-4274(92)90222-6. Spec No:477–83. [DOI] [PubMed] [Google Scholar]
- Beck GJ, Doyle CA, et al. Smoking and lung function. Am Rev Respir Dis. 1981;123:149–55. doi: 10.1164/arrd.1981.123.2.149. [DOI] [PubMed] [Google Scholar]
- Benetazzo MG, Gile LS, et al. alpha 1-antitrypsin TAQ I polymorphism and alpha 1-antichymotrypsin mutations in patients with obstructive pulmonary disease. Respir Med. 1999;93:648–54. doi: 10.1016/s0954-6111(99)90105-1. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, et al. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Celedon JC, Lange C, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13:1649–56. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Resp J. 2004;23:932–46. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- Chen Y, Horne SL, et al. Segregation analysis of two lung function indices in a random sample of young families: the Humboldt Family Study. Genet Epidemiol. 1996;13:35–47. doi: 10.1002/(SICI)1098-2272(1996)13:1<35::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Clark JC, Weaver TE, et al. Decreased lung compliance and air trapping in heterozygous SP-B-deficient mice. Am J Respir Cell Mol Biol. 1997;16:46–52. doi: 10.1165/ajrcmb.16.1.8998078. [DOI] [PubMed] [Google Scholar]
- Clark JC, Wert SE, et al. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci USA. 1995;92:7794–8. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMeo DL, Celedon JC, et al. Genome-wide linkage of forced midexpiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:1294–301. doi: 10.1164/rccm.200404-524OC. [DOI] [PubMed] [Google Scholar]
- DeMeo DL, Mariani TJ, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet. 2006;78:253–64. doi: 10.1086/499828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S. Studies in alpha 1-antitrypsin deficiency. Acta Med Scand Suppl. 1965;432:1–85. [PubMed] [Google Scholar]
- Fagerhol MK, Laurell CB. The Pi system-inherited variants of serum alpha 1-antitrypsin. Prog Med Genet. 1970;7:96–111. [PubMed] [Google Scholar]
- Fletcher CM. Letter: Natural history of chronic bronchitis. Br Med J. 1976;1:1592–3. doi: 10.1136/bmj.1.6025.1592-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givelber RJ, Couropmitree NN, et al. Segregation analysis of pulmonary function among families in the Framingham Study. Am J Respir Crit Care Med. 1998;157:1445–51. doi: 10.1164/ajrccm.157.5.9704021. [DOI] [PubMed] [Google Scholar]
- Givelber RJ, Couropmitree NN, Gottlieb DJ, et al. Segregation analysis of pulmonary function among families in the Framingham Study. American Journal of Respiratory and Critical Care Medicine. 1998;157:1445–51. doi: 10.1164/ajrccm.157.5.9704021. [DOI] [PubMed] [Google Scholar]
- Global Initiative for Chronic Obstuctive Lung Disease 2001. NHLBI/WHO Workshop report. National Institutes of Health. Publication number 2701.
- Golpon HA, Coldren CD, et al. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol. 2004;31:595–600. doi: 10.1165/rcmb.2004-0008OC. [DOI] [PubMed] [Google Scholar]
- Grainger DJ, Heathcote K, et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8:93–7. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- Guo X, Lin HM, et al. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. 2001;18:482–90. doi: 10.1183/09031936.01.00043401. [DOI] [PubMed] [Google Scholar]
- Harrison DJ, Cantlay AM, et al. Frequency of glutathione S-transferase M1 deletion in smokers with emphysema and lung cancer. Hum Exp Toxicol. 1997;16:356–60. doi: 10.1177/096032719701600703. [DOI] [PubMed] [Google Scholar]
- He JQ, Ruan J, et al. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med. 2002;166:323–8. doi: 10.1164/rccm.2111059. [DOI] [PubMed] [Google Scholar]
- Hersh CP, Dahl M, et al. Chronic obstructive pulmonary disease in alpha1-antitrypsin PI MZ heterozygotes: a meta-analysis. Thorax. 2004;59:843–9. doi: 10.1136/thx.2004.022541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh CP, Demeo DL, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol. 2005;33:71–8. doi: 10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh CP, Demeo DL, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006a;173:977–84. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh CP, Demeo DL, et al. Genetic determinants of functional impairment in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006b;3:476. doi: 10.1513/pats.200603-039MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M, Keller J. Familial occurrence of chronic respiratory disease and familial resemblance in ventilatory capacity. J Chronic Dis. 1975;28:239–51. doi: 10.1016/0021-9681(75)90053-3. [DOI] [PubMed] [Google Scholar]
- Hirano K, Sakamoto T, et al. Tissue inhibitor of metalloproteinases-2 gene polymorphisms in chronic obstructive pulmonary disease. Eur Respir J. 2001;18:748–52. doi: 10.1183/09031936.01.00102101. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN. Genetic approaches to studying common diseases and complex traits. Pediatr Res. 2005;57:74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Horne SL, Cockcroft DW, et al. Possible protective effect against chronic obstructive airways disease by the GC2 allele. Hum Hered. 1990;40:173–6. doi: 10.1159/000153926. [DOI] [PubMed] [Google Scholar]
- Huang SL, Su CH, et al. Tumor necrosis factor-alpha gene polymorphism in chronic bronchitis. Am J Respir Crit Care Med. 1997;156:1436–9. doi: 10.1164/ajrccm.156.5.9609138. [DOI] [PubMed] [Google Scholar]
- Hubert HB, Fabsitz RR, et al. Genetic and environmental influences on pulmonary function in adult twins. Am Rev Respir Dis. 1982;125:409–15. doi: 10.1164/arrd.1982.125.4.409. [DOI] [PubMed] [Google Scholar]
- Hurd S. Global impact of COPD. Exp Lung Res. 2005;31(Suppl 1):57–62. [PubMed] [Google Scholar]
- Joos L, He JQ, et al. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum Mol Genet. 2002;11:569–76. doi: 10.1093/hmg/11.5.569. [DOI] [PubMed] [Google Scholar]
- Joost O, Wilk JB, et al. Genetic loci influencing lung function: a genome-wide scan in the Framingham Study. Am J Respir Crit Care Med. 2002;165:795–9. doi: 10.1164/ajrccm.165.6.2102057. [DOI] [PubMed] [Google Scholar]
- Juul K, Tybjaerg-Hansen A, et al. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:858–64. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- Kauffmann F, Tager IB, et al. Familial factors related to lung function in children aged 6–10 years. Results from the PAARC epidemiologic study. Am J Epidemiol. 1989;129:1289–99. doi: 10.1093/oxfordjournals.aje.a115248. [DOI] [PubMed] [Google Scholar]
- Koyama H, Geddes DM. Genes, oxidative stress, and the risk of chronic obstructive pulmonary disease. Thorax. 1998;53(Suppl 2):S10–14. doi: 10.1136/thx.53.2008.s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueppers F, Briscoe WA, et al. Hereditary deficiency of serum alpha-L-antitrypsin. Science. 1964;146:1678–9. doi: 10.1126/science.146.3652.1678. [DOI] [PubMed] [Google Scholar]
- Larson RK, Barman ML, et al. Genetic and environmental determinants of chronic obstructive pulmonary disease. Ann Intern Med. 1970;72:627–32. doi: 10.7326/0003-4819-72-5-627. [DOI] [PubMed] [Google Scholar]
- Lebowitz MD, Knudson RJ, et al. Family aggregation of pulmonary function measurements. Am Rev Respir Dis. 1984;129:8–11. doi: 10.1164/arrd.1984.129.1.8. [DOI] [PubMed] [Google Scholar]
- MacNee W. Oxidants/antioxidants and COPD. Chest. 2000;117(5 Suppl 1):303S–17S. doi: 10.1378/chest.117.5_suppl_1.303s-a. [DOI] [PubMed] [Google Scholar]
- Mannino DM. COPD: epidemiology, prevalence, morbidity and mortality, and disease heterogeneity. Chest. 2002;121(5 Suppl):121S–126S. doi: 10.1378/chest.121.5_suppl.121s. [DOI] [PubMed] [Google Scholar]
- Mannino DM. Chronic obstructive pulmonary disease: definition and epidemiology. Respir Care. 2003;48:1185–91. discussion 1191–3. [PubMed] [Google Scholar]
- Mannino DM, Buist AS, et al. Lung function and mortality in the United States: data from the First National Health and Nutrition Examination Survey follow up study. Thorax. 2003;58:388–93. doi: 10.1136/thorax.58.5.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minematsu N, Nakamura H, et al. Genetic polymorphism in matrix metalloproteinase-9 and pulmonary emphysema. Biochem Biophys Res Commun. 2001;289:116–19. doi: 10.1006/bbrc.2001.5936. [DOI] [PubMed] [Google Scholar]
- Needham M, Stockley RA. Alpha 1-antitrypsin deficiency. 3: Clinical manifestations and natural history. Thorax. 2004;59:441–5. doi: 10.1136/thx.2003.006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell R, Breen D, et al. Inflammatory cells in the airways in COPD. Thorax. 2006;61:448–54. doi: 10.1136/thx.2004.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Takagi M, et al. Matrix metalloproteinase-mediated extra-cellular matrix protein degradation in human pulmonary emphysema. Lab Invest. 1998;78:1077–87. [PubMed] [Google Scholar]
- Park JY, Chen L, et al. Polymorphisms for microsomal epoxide hydrolase and genetic susceptibility to COPD. Int J Mol Med. 2005;15:443–8. [PMC free article] [PubMed] [Google Scholar]
- Pauwels RA, Buist AS, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–76. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]
- Poller W, Faber JP, et al. A leucine-to-proline substitution causes a defective alpha 1-antichymotrypsin allele associated with familial obstructive lung disease. Genomics. 1993;17:740–3. doi: 10.1006/geno.1993.1396. [DOI] [PubMed] [Google Scholar]
- Poller W, Meisen C, et al. DNA polymorphisms of the alpha 1-antitrypsin gene region in patients with chronic obstructive pulmonary disease. Eur J Clin Invest. 1990;20:1–7. doi: 10.1111/j.1365-2362.1990.tb01769.x. [DOI] [PubMed] [Google Scholar]
- Redline S. Genetic and perinatal risk factors for the development of chronic obstructive pulmonary disease. New York: Marcel Dekker; 1990. [Google Scholar]
- Redline S, Tishler PV, et al. Assessment of genetic and nongenetic influences on pulmonary function. A twin study. Am Rev Respir Dis. 1987;135:217–22. doi: 10.1164/arrd.1987.135.1.217. [DOI] [PubMed] [Google Scholar]
- Redline S, Tishler PV, Lewitter FI, et al. Assessment of genetic and nongenetic influences on pulmonary function: atwin study. American Review of Respiratory Diseases. 1987;135:217–22. doi: 10.1164/arrd.1987.135.1.217. [DOI] [PubMed] [Google Scholar]
- Robertson B, Kobayashi T, et al. Experimental neonatal respiratory failure induced by a monoclonal antibody to the hydrophobic surfactant-associated protein SP-B. Pediatr Res. 1991;30:239–43. doi: 10.1203/00006450-199109000-00007. [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Beaty TH, et al. Major genetic mechanisms in pulmonary function. J Clin Epidemiol. 1990;43:667–75. doi: 10.1016/0895-4356(90)90037-p. [DOI] [PubMed] [Google Scholar]
- Sakao S, Tatsumi K, et al. Association of tumor necrosis factor alpha gene promoter polymorphism with the presence of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163:420–2. doi: 10.1164/ajrccm.163.2.2006031. [DOI] [PubMed] [Google Scholar]
- Sakao S, Tatsumi K, et al. Association of tumor necrosis factor-alpha gene promoter polymorphism with low attenuation areas on high-resolution CT in patients with COPD. Chest. 2002;122:416–20. doi: 10.1378/chest.122.2.416. [DOI] [PubMed] [Google Scholar]
- Sandford AJ, Chagani T, et al. Susceptibility genes for rapid decline of lung function in the lung health study. Am J Respir Crit Care Med. 2001;163:469–73. doi: 10.1164/ajrccm.163.2.2006158. [DOI] [PubMed] [Google Scholar]
- Sandford AJ, Weir TD, et al. Z and S mutations of the alpha 1-antitrypsin gene and the risk of chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 1999;20:287–91. doi: 10.1165/ajrcmb.20.2.3177. [DOI] [PubMed] [Google Scholar]
- Schellenberg D, Pare PD, et al. Vitamin D binding protein variants and the risk of COPD. Am J Respir Crit Care Med. 1998;157:957–61. doi: 10.1164/ajrccm.157.3.9706106. [DOI] [PubMed] [Google Scholar]
- Schurch S, Green FH, et al. Formation and structure of surface films: captive bubble surfactometry. Biochim Biophys Acta. 1998;1408:180–202. doi: 10.1016/s0925-4439(98)00067-2. [DOI] [PubMed] [Google Scholar]
- Seifart C, Dempfle A, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens. 2005;65:93–100. doi: 10.1111/j.1399-0039.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- Seifart C, Plagens A, et al. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers. 2002;18:129–36. doi: 10.1155/2002/194075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenoaks MJ, Stockley RA. Chronic obstructive pulmonary disease, inflammation and co-morbidity – a common inflammatory phenotype? Respir Res. 2006;7:70. doi: 10.1186/1465-9921-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SD, Senior RM. Matrix metalloproteinases. Matrix degradation and more. Am J Respir Cell Mol Biol. 1999;20:1100–2. doi: 10.1165/ajrcmb.20.6.f151. [DOI] [PubMed] [Google Scholar]
- Sherrill D, Lebowitz M, Burrows B. Epidemiology of cigarette smoking and ist impact on chronic obstructive pulmonary disease. Clin Chest Med. 1990;11:375–87. [PubMed] [Google Scholar]
- Silverman EK, Mosley JD, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet. 2002;11:623–32. doi: 10.1093/hmg/11.6.623. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Palmer LJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet. 2002;70:1229–39. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EK, Province MA, et al. Family study of alpha 1-antitrypsin deficiency: effects of cigarette smoking, measured genotype, and their interaction on pulmonary function and biochemical traits. Genet Epidemiol. 1992;9:317–31. doi: 10.1002/gepi.1370090504. [DOI] [PubMed] [Google Scholar]
- Smith CA, Harrison DJ. Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet. 1997;350:630–3. doi: 10.1016/S0140-6736(96)08061-0. [DOI] [PubMed] [Google Scholar]
- Spira A, Beane J, et al. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–10. doi: 10.1165/rcmb.2004-0273OC. [DOI] [PubMed] [Google Scholar]
- Steer S, Abkevich V, et al. Genomic DNA pooling for whole genome accosiation scans in complex diseases: epirical demonstration of efficacy in rheumatoid arthritis. Genes Immun. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- Stockley RA. Proteases and antiproteases. Novartis Found Symp. 2001;234:189–99. doi: 10.1002/0470868678.ch12. discussion 199–204. [DOI] [PubMed] [Google Scholar]
- Suthanthiran M, Li B, et al. Transforming growth factor-beta 1 hyperexpression in African-American hypertensives: A novel mediator of hypertension and/or target organ damage. Proc Natl Acad Sci USA. 2000;97:3479–84. doi: 10.1073/pnas.050420897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tager IB, Rosner B, et al. Household aggregation of pulmonary function and chronic bronchitis. Am Rev Respir Dis. 1976;114:485–92. doi: 10.1164/arrd.1976.114.3.485. [DOI] [PubMed] [Google Scholar]
- Tokieda K, Ikegami M, et al. Surfactant protein B corrects oxygen-induced pulmonary dysfunction in heterozygous surfactant protein B-deficient mice. Pediatr Res. 1999;46:708–14. doi: 10.1203/00006450-199912000-00014. [DOI] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, Kucukaycan M, et al. Chronic obstructive pulmonary disease is associated with the −1055 IL-13 promoter polymorphism. Genes Immun. 2002;3:436–9. doi: 10.1038/sj.gene.6363896. [DOI] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, van Veen A, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–5. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, et al. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wilk JB, DeStefano AL, et al. A genome-wide scan of pulmonary function measures in the National Heart, Lung, and Blood Institute Family Heart Study. Am J Respir Crit Care Med. 2003;167:1528–33. doi: 10.1164/rccm.200207-755OC. [DOI] [PubMed] [Google Scholar]
- Wilson AG, de Vries N, Pociot F, et al. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. J Exp Med. 1993;177:557–60. doi: 10.1084/jem.177.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AG, Symons JA, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Stockley RA. The genetics of chronic obstructive pulmonary disease. Respir Res. 2006;7:130. doi: 10.1186/1465-9921-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim JJ, Park GY, et al. Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax. 2000;55:121–5. doi: 10.1136/thorax.55.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Hiyama K, et al. Microsomal epoxide hydrolase genotypes and chronic obstructive pulmonary disease in Japanese. Int J Mol Med. 2000;5:49–53. doi: 10.3892/ijmm.5.1.49. [DOI] [PubMed] [Google Scholar]
- Young RP, Hopkins R, et al. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61:394–9. doi: 10.1136/thx.2005.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Zhu Z, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106:1081–93. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Huang SG, et al. Genetic polymorphism in matrix metalloproteinase-9 and the susceptibility to chronic obstructive pulmonary disease in Han population of south China. Chin Med J (Engl) 2004;117:1481–4. [PubMed] [Google Scholar]