Summary
Coordinating terminal differentiation with permanent exit from the cell cycle is critical for proper organogenesis, yet how the cell cycle is blocked in differentiated tissues remains unclear. Important roles for Retinoblastoma family proteins and Cyclin-dependent kinase inhibitors have been delineated, but in many cases it remains unclear what triggers cell cycle exit. This review focuses on describing recent advances in deciphering how terminal differentiation and exit from the cell cycle are coordinated.
Introduction
Terminal differentiation is usually coupled with permanent exit from the cell cycle and represents the most common cellular state in adult animals. Yet it remains unclear how cells exit the cell cycle during normal development, or maintain the non-proliferative state in adults. Upon terminal differentiation cells become refractory to proliferative signals, including those that promoted proliferation prior to differentiation. Current models for cell cycle exit invoke repression of Cyclin/Cdk activity by Cyclin dependent kinase inhibitors (CKIs), or repression of E2F-mediated transcription by retinoblastoma (Rb) family members, as the proximal mechanisms by which cell cycle progression is arrested (Fig. 1). Mutant studies in a number of organisms support this by demonstrating that the loss of various Rbs or CKIs leads to unscheduled cell proliferation in many tissues [1–4]. But several unresolved issues persist. For example, how is Rb-family and CKI activity coordinated with the process of terminal differentiation? How is cell cycle exit so robustly maintained in differentiated tissues, and do Rbs and CKIs really constitute the only essential cell cycle blockades?
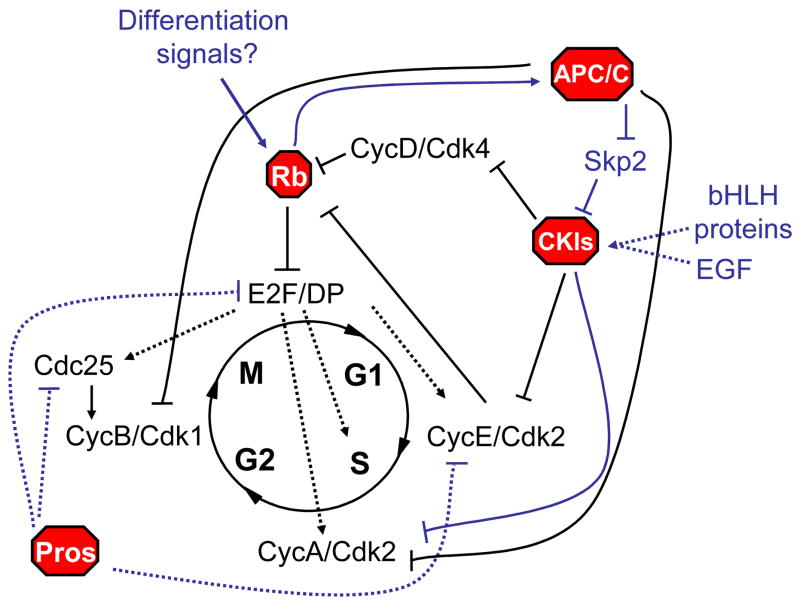
Figure 1. Molecular stop signs: Multiple mechanisms prohibit cell cycle progression upon terminal differentiation.
Negative regulators of the cell cycle act as blockades, preventing proliferation upon terminal differentiation. Recent research has identified new signals impinging on known blockades such as the retinoblastoma proteins (Rbs) and Cyclin dependent kinase inhibitors (CKIs), as well as new negative regulators such as Prospero-like homeobox transcrption factors (Pros), and a new post-mitotic role for the Anaphase Promoting Complex/Cyclosome (APC/C). Regulation can be at the level of transcription (dotted lines) or post-transcriptionally (solid lines). Recently identified pathways highlighted in blue are discussed in this review.
Senescence vs. quiescence vs. cell cycle exit upon terminal differentiation
Exit from the cell cycle upon terminal differentiation shares many characteristics with other quiescent states, but also appears to be distinct. Senescence, quiescence and terminal differentiation are all characterized by prolonged cell cycle arrest with a G1 DNA content, the presence of hypophosphorylated Rb proteins that mediate inhibitory E2F activity, and often, high CKI activity [5,6]. However, quiescence, a state mostly characterized in cell culture, is a more easily reversible than senescence or differentiation induced exit in vivo. In quiescent tissue culture cells, differentiation also seems to be inhibited [7] and thus may more closely resemble the quiescence of stem cells in vivo.
Senescence is a largely non-reversible state characterized by distinct cell morphology, formation of senescence-associated heterochromatin foci and expression of senescence-associated genes [8,9]. Senescence been characterized primarily in cell culture, but recent work has demonstrated that cellular senescence can and does exist in vivo, and is suggested to be induced either as a barrier against cancer [10,11], or as a result of normal aging [12]. Newer characterizations of senescence demonstrate that it involves an activated DNA damage checkpoint [13,14], and therefore seems to be a state distinct from terminal differentiation.
Terminal differentiation by contrast, has been characterized both in cell culture and in vivo, and appears to be more reversible in some cell types than others. Regeneration accompanied by de-differentiation, as observed in amphibian limbs, is a classic example of cell cycle exit reversal in vivo [15]. Yet reversal of exit in other cell types can be quite difficult [16]. It is not clear why cell cycle exit and differentiation are more or less reversible in different cell types, but continuing work examining the mechanisms of exit in different tissues will hopefully clarify this issue.
Coordinating cell cycle exit with terminal differentiation
Perhaps the most pressing question in the field of terminal differentiation is: how do the developmental signals triggering differentiation impinge on cell cycle regulators? One answer that seems to come up again and again is that differentiation signals can transcriptionally regulate CKIs to trigger cell cycle exit. One of the best-characterized examples of this involves regulation of the Cip/Kip type CKIs (p21, p27 and p57) by basic Helix Loop Helix (bHLH) transcription factors. Cip/Kip CKIs are frequently expressed in a manner spatially and temporally coordinated with terminal differentiation and initiation of exit. Examples of bHLH proteins such as MyoD or Hes1 regulating Cip/Kip expression both in vitro and in vivo exist [17–22]. In some of these cases however, the same bHLH proteins are used in earlier developmental events before terminal differentiation. So how is bHLH activity upon terminal differentiation distinguished from earlier activity in cells that continue to proliferate? A recent study in Drosophila [23] demonstrates that combinatorial regulation by bHLH activity together with EGF signaling might distinguish the proper time to induce cell cycle exit (Fig. 1). In this case both EGF signaling through the ETS transcription factor Pointed and specific bHLHs are required together at the promoter of the p21/p27 homolog, Dacapo, for proper activation in certain Drosophila neurons. Combinatorial mechanisms are not limited to bHLH-dependent regulation however. A similar mechanism has also been described for regulation of mammalian p21 by a combination of Tgfβ signaling though Smad transcription factors and developmental regulation of FoxO activity [24]. Such combinatorial control provides a satisfying mechanism to coordinate differentiation with exit, but it is important to note that this regulation may not actually be required for cell cycle exit in vivo. For example in the Drosophila eye, Dacapo acts redundantly with Rb like proteins in Drosophila (Rbfs) to promote cell cycle exit in neurons and loss of one or the other pathway can be tolerated [25,26]. How the Rb proteins are regulated upon differentiation is not known, but such redundancy has been described in a number of organisms and highlights the robustness of mechanisms that ensure cell cycle exit, a topic we will return to later in this review.
Another transcriptional mechanism coordinating terminal differentiation and cell cycle exit through the Drosophila homeobox transcription factor Prospero (Pros) has been elaborated recently. In neuroblasts, Pros is asymmetrically localized at the cytoplasmic membrane. Upon division, Pros is inherited by one daughter cell, the Ganglion Mother Cell (GMC), where it enters the nucleus and directs terminal differentiation. Previous studies of Pros in the Drosophila embryo had shown that Pros inhibits transcription of Cyclin E and the cdc25 homolog String, and also induces the CKI Dacapo in differentiating neurons. But it remained unclear whether this was a an indirect effect of Pros on differentiation, or a more direct effect on these cell cycle genes [27]. New work [28] investigating the genome-wide binding of Pros on chromatin confirms that Pros has a direct role in transcriptionally inhibiting several key cell cycle genes including e2f1, cyclin E and string, making it an effective blockade (Fig. 1). Importantly, Pros also has a direct role in activating a number of terminal differentiation genes in neurons, making it a dual-function signal [28].
Pros expression is highly restricted to certain cell types, and is therefore not likely to have a universal role in cell cycle exit. Pros could be a common mechanism for cell cycle exit when there is direct differentiation from asymmetric division of a multipotent progenitor without a transiently amplifying progenitor pool. Recent work in the Drosophila gut has delineated a stem cell differentiation pathway where a similar pattern of an asymmetric division followed by differentiation without intervening mitoses occurs, and Pros is specifically expressed in one of the resulting post-mitotic cell types [29,30], where it might coordinate terminal differentiation and cell cycle exit. Importantly, the mammalian Pros homolog, Prox1, regulates exit in certain cell types in the mammalian retina [31]. Other Homeobox proteins could play similar roles in other cell types. The search for such dual-function transcription factors will continue.
Destroying the destroyer: protein degradation and timely cell cycle exit
Recent work suggests new roles for the protein degradation machinery in promoting cell cycle exit. The Anaphase promoting complex/Cyclosome (APC/C) and Skp-Cullin-F-box (SCF) ubiquitin ligase complexes have been recognized as cell cycle blockades that promote quiescence. APC/C has long been known to have function in mediating exit from mitosis and entry into G1 or a quiescent state. The large APC/C complex associates with activators Cdc20 or Cdh1, which activate the ubiquitin ligase activity and specify targets such as mitotic Cyclin/Cdks for degradation [32]. In addition to limiting mitotic Cyclin/Cdk activity in proliferating cells, it had previously been shown that the APC/C also targeted Skp2, for degradation [33,34]. Binne et al. have now connected this degradation of the degradation machinery to the cell cycle exit pathway by showing that hypophosphorylated Rb associates with the APC/C specifically when activated by Cdh1 [35]. This promotes degradation of Skp2, a component of the SCFskp2 complex responsible for degradation of p27 and p21. The Rb/APC/C interaction thus results in accumulation of p27 (and possibly p21), allowing Rb to act in a second, E2F-independent manner, to inhibit the cell cycle (Fig. 1). Importantly, using a mutated Rb that cannot bind Cdh1, Binne et al. demonstrate that the Rb/APC/C interaction is required for Rb to inhibit the cell cycle in proliferating cells.
How do these observations relate to differentiation induced cell cycle exit in vivo? Although the studies of Binne et al. were performed in cells either undergoing senescence or quiescence in vitro, hypophosphorylated Rb does accumulate when cyclin/cdk activity is low in vivo, a hallmark of cell cycle exit. Furthermore, genetic studies of APC/C components in vivo suggest an important role in promoting properly timed cell cycle exit [36–39]. We must point out though, that different SCF components have also been shown to play roles in promoting timely exit in a number of organisms, through distinct SCF complexes that promote degradation of Cyclin E [40,41], thus protein degradation is important for timely cell cycle exit through at least two distinct pathways. Moreover, in all of these APC/C and SCF mutant studies in vivo, cell cycle exit is only temporarily delayed, demonstrating that while the protein degradation machinery may play a role in triggering cell cycle exit, additional mechanisms eventually compensate to ensure cell cycle exit.
Maintaining cell cycle exit: Switching things up at the chromatin
Studies characterizing Rb-family/E2F complexes on the DNA have identified a number of interactions with chromatin remodeling factors that can contribute to E2F-dependent gene repression. These include Rb-family dependent recruitment of a Histone Deacetylase (HDAC) complex, association with a SWI/SNF ATP-dependent nucleosome remodeling complex, and Rb-family association with a histone methyltransferase (HMT) complex (reviewed in [42]). These associations suggest an obvious mechanism by which E2F-dependent cell cycle genes can be stably repressed by the Rb family members (Rb, p107 and p130) upon cell cycle exit. It has remained unclear, however, which associations are functionally relevant for repression of cell cycle genes upon terminal differentiation.
One hint was provided by the characterization of a repressive complex from Drosophila termed the dREAM complex, which contains Rb/E2F complexes, Myb, Myb-interacting proteins, and at least in one study, contained the HDAC Rpd3 [43,44]. A new characterization of the homologous DREAM complex in human cells shows its specific recruitment to E2F-regulated cell cycle genes in quiescent cells in culture [45]. Knockdown of DREAM components prevented repression of cell cycle genes upon serum starvation, demonstrating functional relevance in a quiescent state, although the effect was mild, ranging from a 1.5–3-fold increase in expression, suggesting other factors may compensate. Interestingly, the Drosophila dREAM complex specifically represses developmentally regulated E2F targets with no known cell cycle functions [43]. Since the studies on the human complex were performed on cultured cells in a reversible quiescent state, further work will also be needed to determine whether the human DREAM complex plays a role in maintaining cell cycle gene repression upon terminal differentiation.
Rb also associates with the HMT Suv39H, which methylates Histone H3, thereby creating a binding site for Heterochromatin Protein 1 (HP1) family members. HP1s play important roles in establishing and maintaining repressive heterochromatin (reviewed in [42]) and Rb can recruit HP1 to the Cyclin E promoter and repress Cyclin E expression in proliferating cells in culture [46]. New work extends these observations to differentiating cells by suggesting a role for Rb-family association with an HMT complex in a model of terminal differentiation [47]. Panteleeva et al. demonstrate an increase in Hp1α expression and association with the Rb-family member p130/E2F complexes at E2F responsive cell cycle gene promoters upon differentiation of neurons in the cerebellum. This association may be important for repression of cell cycle genes upon differentiation, as knockdown of Hp1α in a cell culture model of neural differentiation partially impairs cell cycle arrest and terminal differentiation.
One interesting aspect of the study by Panteleeva et al., is that they observe a developmental switch in HP1 isoform expression, from HP1γ which does not associate with p130/E2F complexes, to HP1α upon neural differentiation. This is reminiscent of another study [48] which identified a developmental switch in the composition of a neural specific SWI/SNF complex upon terminal differentiation in vivo. While this study does not demonstrate a direct effect of this subunit switch on the chromatin of cell cycle genes, they do find that the switch in subunits is essential for cell cycle exit and differentiation of neurons in vivo. This study along with the others described above suggests that developmentally regulated changes in chromatin modifying complexes are likely to play an important role in maintaining cell cycle exit upon terminal differentiation.
Cell cycle regulators with roles in terminal differentiation
Cases of dual roles in cell cycle and terminal differentiation are not limited to developmental transcription factors like Pros, but can also be found among cell cycle regulators themselves. A classic example of a cell cycle regulator with cell cycle independent functions in differentiation is Rb. Rb associates with a number of tissue-specific transcription factors either directly or indirectly, in an E2F independent manner, to potentiate their differentiation inducing activity. Some examples include Rb association with MyoD and Mef2 in muscle [49,50], association with CBFA1 and Runx2 in bone [51,52], and association with C/EBP family transcription factors in cell culture models of adipocyte and macrophage differentiation [53,54]. Roles for Rb in terminal differentiation aren’t always E2F-independent however. Recent work has demonstrated a function for Rb in directing neural migration in the mammalian brain through inhibition of E2F3, as well as induction of terminal differentiation in specific interneurons of the retina by inhibition of the E2F3a isoform, roles that appear distinct from Rb and E2F3 effects on the cell cycle [55,56]. The finding that Rb/E2F complexes can regulate differentiation targets is not surprising in light of previous studies of Rb/E2F transcriptional targets in Drosophila, where a large class of non-cell cycle developmental targets were identified, as well as a chromatin remodeling complex likely to control their expression [43,44,57].
Cell cycle genes moonlighting as terminal differentiation inducers go beyond Rb and E2F. Studies in the last year have also described additional roles for the p27Kip CKI and the APC/C in differentiation. Xenopus p27 has a domain separate from the Cyclin/Cdk inhibitory domain that promotes differentiation of neurons and muscle [58,59], and mammalian p27 can promote neural differentiation through stabilization of Ngn2, even when its binding to Cyclin/Cdks is inhibited. This binding deficient mutant (p27ck-) also plays a role in neural migration, through regulation of Rho [60]. The APC/CCdh1 complex, the same shown to interact with Rb, has also been suggested to play a role in terminal differentiation of neurons by regulating the stability of Inhibitor of Differentiation or ID proteins, which can regulate axon growth in culture [61].
Differentiation and cell cycle exit, independent events or not?
The dual roles of cell cycle regulators in terminal differentiation have complicated genetic studies and led to confusion about whether cell cycle exit and terminal differentiation are separable events. Although many studies have demonstrated that cells can exit the cell cycle without differentiating, it has remained controversial whether cells can terminally differentiate without exiting the cell cycle. The answer to this question is likely to be tissue specific, since the roles for cell cycle regulators in terminal differentiation are cell-type specific. Some apparent examples of separability exist [25,26,62] (Fig. 2), but difficulty arises in how to determine whether cells are fully terminally differentiated or not. Current definitions of terminal differentiation often rely on molecular markers, and the discovery of new, more specific markers can change the criteria for assaying terminal differentiation. In general, specific cellular morphologies (such as the formation of multinucleated myotubes in the case of muscle), or functions (such as firing in the case of neurons) is held to be the gold standard, and in this case the hair cells of the mammalian inner ear provide an excellent in vivo example in which exit and terminal differentiation can indeed be separated [62]. Loss of Rb in hair cells leads to continued entry into S-phase and proliferation, even after hair cells exhibit functional mechanotransduction (Fig. 2) [62]. Thus cell cycle exit and terminal differentiation are indeed separable, but may not appear so by genetic studies, in cases where cell cycle regulators also play essential functions in terminal differentiation.
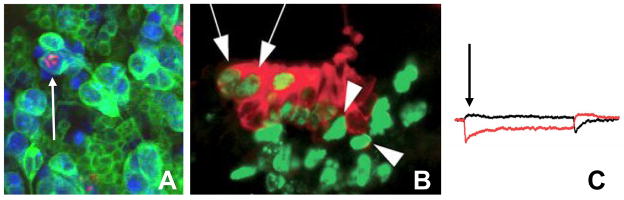
Figure 2. Terminal differentiation and cell cycle exit are separable.
Although terminal differentiation and cell cycle exit are coordinated, in some contexts they are separable. A. For example a neuron in the late Drosophila retina can continue cycling and undergo mitosis (indicated by phosphohistone H3 in red) while maintaining characteristics of terminal differentiation, such as expression of Elav (in blue) and projection of an axon (cell membrane in green, indicated by an arrow) if both E2F and Cyclin/Cdk activities are kept high [25]. B. Hair cells of the mouse inner ear lacking Rb expression also continue cycling upon terminal differentiation, providing another example where differentiation and cell cycle exit are separable. Myo7A expression in red labels hair cells of the cochlea, while BrdU in green labels cells in S-phase. Co-labeled cells differentiate but continue into S-phase (arrow) C. Recorded transduction currents in Rb−/− hair cells demonstrate mechanosensitivity [B and C adapted from 62].
Excessive redundancy
Do Rbs and CKIs constitute the only essential downstream cell cycle blockades for cell cycle exit upon terminal differentiation? Mammals have 3 Rb family members and 7 Cip/Kip or Ink type CKIs. The sheer number of paralogs has made answering this question difficult in mammals. Studies knocking out all three Rb family members have been able to delineate their roles in mouse embryonic fibroblasts [63,64]. Although these triple knockout or “TKO” fibroblasts fail to senesce or undergo quiescence upon contact inhibition, they can undergo cell cycle arrest in response to serum starvation [65].
Drosophila have a simpler system with only two Rb family members, a single Cip/Kip type CKI and no INK homologs. Investigations into this question in Drosophila suggest that Rb/E2F repressive activity is not required for cell cycle exit in vivo [66–68] and that Rb/E2F repression and CKI activity act redundantly to ensure exit in neurons and non-neural cells in the eye [25,26]. Other cell types however, such as the epithelial cells of the wing, still exit from the cell cycle upon differentiation even when E2F activity is high and the CKI Dacapo is absent [25], indicating that additional cell cycle blocking mechanisms must act in these cells. Currently we do not know what these additional mechanisms are, but since Cyclin/Cdk activity seems to be inhibited even when Cyclin and Cdk expression levels are quite high, a novel mechanism repressing CDK activity may be turned on at the onset of terminal differentiation in certain epithelial cells.
We have described some examples of how CKIs may be regulated by differentiation signals and their downstream transcription factors, but how are Rb proteins temporally regulated? In general Rb regulation is thought to be at the level of phosphorylation rather than transcription. The current model is that CKIs induced upon terminal differentiation trigger cell cycle exit by inhibiting CDK phosphorylation of Rbs. This leads to accumulation of hypophosphorylated Rbs which associate with E2F and recruit repressive chromatin modifying complexes to E2F bound promoters, thereby inhibiting cell cycle gene expression. So how does hypophosphorylated Rb accumulate when CKIs are absent, or in aberrant situations when Cyclin/CDK activity remains high? Previous investigations have searched for phosphatases that act on Rbs, and identified Protein Phosphatase 1 (PP1) as a potential Rb phosphatase [69–71]. However recent studies in Drosophila have demonstrated that PP1 is not required for regulation of Rbs [72]. How Rb phosphatases could be regulated by differentiation signals is also not clear. Thus, an essential connection between differentiation signals and Rb-family members still needs to be made.
Conclusions
The extent of redundancy in mechanisms ensuring cell cycle exit upon terminal differentiation is beginning to be fully appreciated. Given the double and even triple redundancy in exit mechanisms; will we ever be able to delineate a common cell cycle exit mechanism in all tissues? While no particular pathway appears to be universally dedicated to specifically direct cell cycle exit, the types of mechanisms used may be categorized by cell type or lineage. Hopefully, this will allow us to make some sense, or even predictions, of which exit mechanisms are used in different developmental contexts.
Acknowledgments
L.A.B. thanks the members of the Edgar Lab for their help in her work on cell cycle exit, and in particular Dr. J. Bandura for helpful comments on this manuscript. Work on cell cycle exit in Dr. Edgar’s lab is supported by NIH grant GM070887. L.A.B. was supported by a Damon Runyon postdoctoral fellowship (DRG#1838-04).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
•• Recent papers of interest on cell cycle exit and terminal differentiation.
- 1.Vidal A, Koff A. Cell-cycle inhibitors: three families united by a common cause. Gene. 2000;247:1–15. doi: 10.1016/s0378-1119(00)00092-5. [DOI] [PubMed] [Google Scholar]
- 2.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 3.Koreth J, van den Heuvel S. Cell-cycle control in Caenorhabditis elegans: how the worm moves from G1 to S. Oncogene. 2005;24:2756–2764. doi: 10.1038/sj.onc.1208607. [DOI] [PubMed] [Google Scholar]
- 4.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 6.Pajalunga D, Mazzola A, Puggioni E, Crescenzi M. Non-proliferation as an active state: conceptual and practical implications. Cell Cycle. 2007;6:1415–1418. [PubMed] [Google Scholar]
- 7••.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. Identifies a transcriptional program induced upon quiescence in culture and demonstrates that quiescence protects against MyoD induced terminal differentiation in a fibroblast cell culture model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Narita M, Lowe SW. Senescence comes of age. Nat Med. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- 10••.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. Along with [10, 12 and 13] demonstrates the importance of senescence in tumor suppression and describes senescence as a state of active DNA damage checkpoints. [DOI] [PubMed] [Google Scholar]
- 11••.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. See note for [9] [DOI] [PubMed] [Google Scholar]
- 12.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 13••.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. See note for [9] [DOI] [PubMed] [Google Scholar]
- 14••.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. See note for [9] [DOI] [PubMed] [Google Scholar]
- 15.Tsonis PA. Regeneration in vertebrates. Dev Biol. 2000;221:273–284. doi: 10.1006/dbio.2000.9667. [DOI] [PubMed] [Google Scholar]
- 16.Camarda G, Siepi F, Pajalunga D, Bernardini C, Rossi R, Montecucco A, Meccia E, Crescenzi M. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J Cell Biol. 2004;167:417–423. doi: 10.1083/jcb.200408164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo K, Wang J, Andres V, Smith RC, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 20.Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Encinas M, Comella JX, Aldea M, Gallego C. Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004;24:2662–2672. doi: 10.1128/MCB.24.7.2662-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 23••.Sukhanova MJ, Deb DK, Gordon GM, Matakatsu MT, Du W. Proneural basic helix-loop-helix proteins and epidermal growth factor receptor signaling coordinately regulate cell type specification and cdk inhibitor expression during development. Mol Cell Biol. 2007;27:2987–2996. doi: 10.1128/MCB.01685-06. Identifies a combinatorial mechanism of p27Dacapo transcriptional regulation by bHLH proteins and transcription factors downstream of EGF signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 25••.Buttitta LA, Katzaroff AJ, Perez CL, de la Cruz A, Edgar BA. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev Cell. 2007;12:631–643. doi: 10.1016/j.devcel.2007.02.020. Along with [24] demonstrates redundancy of Rb-like and CKI homologs upon cell cycle exit in particular Drosophila tissues, and shows that cell cycle exit and differentiation are separable in neurons. [DOI] [PubMed] [Google Scholar]
- 26••.Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. See note for [23] [DOI] [PubMed] [Google Scholar]
- 27.Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- 28••.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. Demonstrates direct binding of Pros to both cell cycle and differentiation genes to regulate gene expression upon terminal differentiation of neurons in Drosophila. [DOI] [PubMed] [Google Scholar]
- 29.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 30.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 31.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 32.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 33.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 35••.Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr, Naar AM, Dyson NJ. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. Identifies a new interaction between Rb and APC/C components, important for Rb induced quiescence, that targets Skp2 for destruction. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka-Matakatsu M, Thomas BJ, Du W. Mutation of the Apc1 homologue shattered disrupts normal eye development by disrupting G1 cell cycle arrest and progression through mitosis. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehman AM, Staub W, Baier H. The anaphase-promoting complex is required in both dividing and quiescent cells during zebrafish development. Dev Biol. 2007;303:144–156. doi: 10.1016/j.ydbio.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 38.Wirth KG, Ricci R, Gimenez-Abian JF, Taghybeeglu S, Kudo NR, Jochum W, Vasseur-Cognet M, Nasmyth K. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 2004;18:88–98. doi: 10.1101/gad.285404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimentel AC, Venkatesh TR. rap gene encodes Fizzy-related protein (Fzr) and regulates cell proliferation and pattern formation in the developing Drosophila eye-antennal disc. Dev Biol. 2005;285:436–446. doi: 10.1016/j.ydbio.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 41.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 42.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 43.Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 44.Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. Describes the identification of the human DREAM complex, and shows its interaction with cell cycle genes upon quiescence in culture. Provides evidence that components of the DREAM complex are required for full inhibition of cell cycle gene expression in quiescence. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 47••.Panteleeva I, Boutillier S, See V, Spiller DG, Rouaux C, Almouzni G, Bailly D, Maison C, Lai HC, Loeffler JP, et al. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. Embo J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. Identifies a switch in HP1 isoform expression associated with neural terminal differentiation in the mouse cerebellum. Demonstrates association of HP1α with Rb-family proteins and cell cycle genes in differentiating neurons and provides evidence of this association inhibiting cell cycle gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. Describes a developmental switch in SWI/SNF subunit expression required for proper cell cycle exit and terminal differentiation of neurons in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novitch BG, Spicer DB, Kim PS, Cheung WL, Lassar AB. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 51.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 52.Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen PL, Riley DJ, Chen-Kiang S, Lee WH. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 55••.Chen D, Opavsky R, Pacal M, Tanimoto N, Wenzel P, Seeliger MW, Leone G, Bremner R. Rb-Mediated Neuronal Differentiation through Cell-Cycle-Independent Regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. Identifies a role for Rb/E2F3a complexes in terminal differentiation of specific neural cell types in the retina, independent of effects on the cell cycle and apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. Identifies a role for Rb/E2F3 complexes in neural migration in the mouse brain, independent of effects on the cell cycle and apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vernon AE, Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development. 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- 59.Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 60••.Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. Finds a role for p27 in neural terminal differentiation and migration in the brain, even in the absence of Cyclin/Cdk binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. Demonstrates a role for the APC/C in regulating axon growth in culture, through degradation of Id proteins. [DOI] [PubMed] [Google Scholar]
- 62••.Sage C, Huang M, Karimi K, Gutierrez G, Vollrath MA, Zhang DS, Garcia-Anoveros J, Hinds PW, Corwin JT, Corey DP, et al. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science. 2005;307:1114–1118. doi: 10.1126/science.1106642. Describes that inner ear hair cells lacking Rb continue to proliferate upon terminal differentiation and demonstrates terminal differentiation and cell cycle exit are separable in this cell type. Also shows that acute loss of Rb can lead to cell cycle re-entry in post-mitotic hair cells. [DOI] [PubMed] [Google Scholar]
- 63.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Foijer F, Wolthuis RM, Doodeman V, Medema RH, te Riele H. Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell. 2005;8:455–466. doi: 10.1016/j.ccr.2005.10.021. Identifies a context in which fibroblasts lacking all 3 Rb-family members will arrest. [DOI] [PubMed] [Google Scholar]
- 66.Weng L, Zhu C, Xu J, Du W. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. Embo J. 2003;22:3865–3875. doi: 10.1093/emboj/cdg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frolov MV, Moon NS, Dyson NJ. dDP is needed for normal cell proliferation. Mol Cell Biol. 2005;25:3027–3039. doi: 10.1128/MCB.25.8.3027-3039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Genes Dev. 2001;15:2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson DA, Krucher NA, Ludlow JW. High molecular weight protein phosphatase type 1 dephosphorylates the retinoblastoma protein. J Biol Chem. 1997;272:4528–4535. doi: 10.1074/jbc.272.7.4528. [DOI] [PubMed] [Google Scholar]
- 70.Vietri M, Bianchi M, Ludlow JW, Mittnacht S, Villa-Moruzzi E. Direct interaction between the catalytic subunit of Protein Phosphatase 1 and pRb. Cancer Cell Int. 2006;6:3. doi: 10.1186/1475-2867-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubin E, Mittnacht S, Villa-Moruzzi E, Ludlow JW. Site-specific and temporally-regulated retinoblastoma protein dephosphorylation by protein phosphatase type 1. Oncogene. 2001;20:3776–3785. doi: 10.1038/sj.onc.1204518. [DOI] [PubMed] [Google Scholar]
- 72.Swanhart LM, Sanders AN, Duronio RJ. Normal regulation of Rbf1/E2f1 target genes in Drosophila type 1 protein phosphatase mutants. Dev Dyn. 2007;236:2567–2577. doi: 10.1002/dvdy.21265. [DOI] [PubMed] [Google Scholar]