Summary
Human APOBEC3G (A3G) belongs to a family of polynucleotide cytidine deaminases. This family includes APOBEC1 and AID, which edit APOB mRNA and antibody gene DNA, respectively. A3G deaminates cytidines to uridines in single-strand DNA and inhibits the replication of HIV-1, other retroviruses and retrotransposons. Although the mechanism of A3G-catalyzed DNA deamination has been investigated genetically and biochemically, atomic details are just starting to emerge. Here, we compare the DNA cytidine deaminase activities and NMR structures of two A3G catalytic domain constructs. The longer A3G191-384 protein is considerably more active than the shorter A3G198-384 variant. The longer structure has an α1 helix (residues 201–206) that was not apparent in the shorter protein and it contributes to catalytic activity through interactions with hydrophobic core structures (β1, β3, α5 and α6). Both A3G catalytic domain solution structures have a discontinuous β2 region that is clearly different than the continuous β2 strand of another family member APOBEC2. In addition, the longer A3G191-384 structure revealed part of the N-terminal pseudo-catalytic domain including the inter-domain linker and some of the last α-helix. These structured residues (191–196) enabled a novel full-length A3G model by providing physical overlap between the N-terminal pseudo-catalytic domain and the new C-terminal catalytic domain structure. Contrary to predictions, this structurally constrained model suggested that the two domains are tethered by structured residues and that the N- and C-terminal β2 regions are too distant from one another to participate in this interaction.
Keywords: APOBEC3G, DNA deamination, DNA editing, NMR structure, retrovirus restriction
Introduction
Human A3G is a prominent member of a multifunctional family of Zn2+-dependent polynucleotide cytidine deaminases [reviewed recently by1; 2; 3. Members include the founder, APOBEC1, which edits APOB mRNA C6666 to generate an early stop codon and a shorter polypeptide, and AID, which edits immunoglobulin gene DNA cytidines to trigger somatic hypermutation and class switch recombination. Physiological roles have yet to be revealed for other members such as APOBEC2 (A2) and APOBEC4. However, A3G, six other human A3s and a multitude of A3s from other mammals have elicited DNA cytidine deaminase activity and have been shown to inhibit the replication of a variety of retrotransposons and retroviruses. The most prominent human pathogen that can be restricted by A3G is the AIDS virus HIV-1.
A defining feature of DNA cytidine deaminases is a Zn2+-coordinating motif, H-x1-E-x25–31-C-x2–4-C4; 5; 6. This motif is required for catalytic activity [e.g., 7; 8; 9; 10; 11; 12; 13; 14]. Based on structures of free-base and nucleoside deaminases, the histidine, two cysteines and water are thought to directly coordinate Zn2+ and the glutamate exchanges hydrogens during catalysis 15; 16; 17. Some APOBEC3 proteins have two Zn2+-binding motifs whereas others have only one. In A3G, the C-terminal Zn2+-binding motif is catalytically active, whereas the N-terminal one is not 8; 14; 18; 19.
High-resolution structural information was reported recently for the C-terminal catalytic domain of A3G 20; 21; 22 and for residues 41–224 of the single Zn2+-domain protein A2 23, which lacks deaminase activity [e.g.,23; 24]. Both are globular proteins with mostly similar secondary structures and Zn2+-coordinating motifs. However, structural and biochemical studies of A2 have shown that it exists as a homodimer mediated by extensive anti-parallel β2-β2 contacts23. In contrast, the catalytic domain of A3G profiled as a monomer in solution and showed no obvious dimeric points of contact in the crystal20; 21; 22; 25, despite the fact that the A3G holoenzyme is capable of dimerizing and forming higher-order oligomers14; 26; 27; 28; 29. The A2 data therefore suggested that the analogous β2 regions of the N- and C-terminal domains of A3G might mediate the inter-domain connection21; 22; 23; 25; 30. However, the feasibility of such an A3G β2-β2 interaction has been debated because the A3G catalytic domain NMR structures have shown a discontinuous β2-bulge-β2′ region20; 21, whereas the A3G197-380 crystal structure was reported to have a A2-like continuous β2 strand22.
Here, we report the activity and solution structure of a longer A3G catalytic domain variant, which spans residues 191–384. These data provide several novel insights and confirm aspects of the prior NMR and crystallographic studies. First, the new structure substantiated the discontinuous β2-bulge-β2′ structure observed by NMR but not by crystallography. Second, the new structure confirmed the presence and demonstrated the importance of the α1 helix, which was apparent in the crystal but not previously in solution. Third, a comparison of the A3G191-384 and A3G198-384 NMR structures yielded a robust explanation for the increased activity of the longer construct (substantive α1 to the catalytic core structure interactions). Fourth, the new A3G191-384 solution structure revealed a region of physical overlap – one helical turn (residues 191–194) – between the predicted N-terminal pseudo-catalytic domain (A3G1-196) and the actual C-terminal catalytic domain (A3G191-384). This overlap enabled the construction of a novel full-length A3G model. Fifth, the physical constraints imposed by the new structural elements orientated the N- and C-terminal β2 regions too far away from one another to facilitate inter-domain interactions. In this report, we present the structure of the longest fragment of A3G to date and we address some of the controversies raised by previously published structures. All together, our data provide several important steps toward a robust structural understanding of full-length A3G and its central role in retroelement restriction, and they help broaden our structural and mechanistic knowledge of DNA deaminases.
Results
A longer and more active A3G catalytic domain variant
As outlined in the introduction, there are clear differences between the NMR and the crystal structures of the catalytic domain of A3G, and none of these has shed light on the A3G inter-domain connections.
To better understand the A3G catalytic domain and how it is oriented with respect to the N-terminal pseudo-catalytic domain, we revisited prior data showing that A3G175-384 was approximately 3-fold more active than A3G198-384 in an E. coli-based RifR mutation assay25. The RifR mutation assay provides a quantitative, but indirect, read-out of DNA deaminase activity, with each RifR colony reflecting an rpoB gene mutation that occurred during the outgrowth of a single cell into a saturated culture. To better define the residues responsible for this elevated mutation frequency, we examined a series of single alanine mutants and observed that residues 175–190 were dispensable (data not shown).
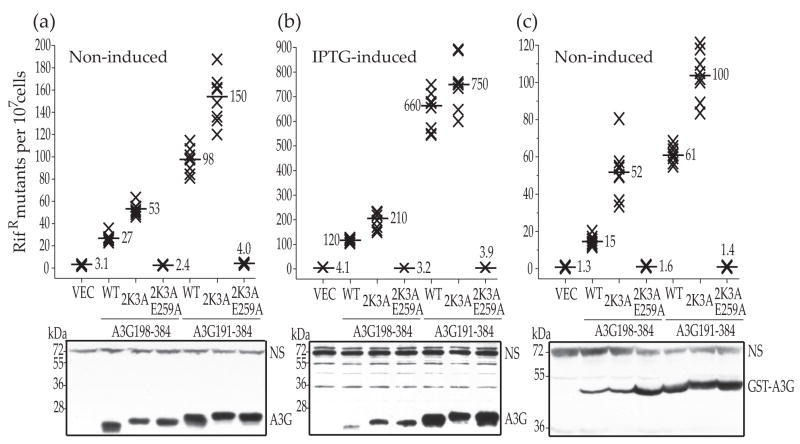
We next generated untagged and GST-tagged A3G191-384 and A3G198-384 constructs for extensive head-to-head genetic, biochemical and structural characterizations. Derivatives with five amino acid substitutions, L234K, C243A, F310K, C321A and C356A (2K3A), were also made to enable concentrated protein preparations for NMR studies21. Similar to our prior observations with A3G175-38425, A3G191-384 was 3-fold more active than A3G198-384 under non-induced, basal expression conditions (Fig. 1a). 2K3A-derivatives of these constructs had even higher RifR mutation frequency increases, but the relative activity difference between the longer and shorter constructs was still three-fold. Immunoblots showed that the A3G191-384 and A3G198-384 constructs were expressed similarly at non-induced levels, indicating that the improved activity of the longer protein is not simply due to higher expression levels or improved solubility (Fig. 1a, lower panel).
Fig. 1.
E. coli-based DNA cytidine deaminase activity of A3G catalytic domain constructs. (a) The capacity of untagged A3G198-384, A3G191-384 and the indicated mutant derivatives to trigger RifR mutations in E. coli growing under non-inducing conditions. Each X represents the mutation frequency of an independent culture and the median values are indicated. VEC and WT represent empty vector control and the indicated construct with no additional mutations, respectively. A representative anti-A3G immunoblot is shown and non-specific (NS) bands provide loading controls. 2K3A-derivates migrate slightly slower. (b) A representative RifR mutation experiment with the same constructs as in panel (a), except this was performed under IPTG-induced expression conditions. The Y-axis values are different. (c) A representative RifR mutation experiment with the indicated GST-tagged constructs performed under non-inducing expression conditions. The Y-axis values are different. The complementary IPTG-induced experiment could not be done with the GST expression constructs because induction causes cell death.
As expected, IPTG-induced expression of the A3G constructs resulted in the highest levels of RifR mutation, with the A3G191-384 and its 2K3A derivative easily triggering 100-fold increases over the vector control levels (Fig. 1b). Again, the A3G191-384 proteins were at least 3-fold more active than the A3G198-384 variants, but here activity appeared to correlate reproducibly with solubility (Fig. 1b, lower panel; see Discussion). Similar RifR mutation trends were observed for non-induced, GST-tagged versions of all of these A3G constructs (Fig. 1c). The GST tag was influential but not responsible for elevated catalytic activity as, for instance, it rendered the A3G198-384 construct almost as soluble as the A3G191-384 protein. (Fig. 1c lower panel). Overall, the E. coli activity and expression data combined to indicate that A3G191-384 is approximately 3-fold more active than the shorter A3G198-384 variant (see Discussion).
In vitro DNA cytidine deaminase activity of A3G191-384-2K3A and A3G198-384-2K3A
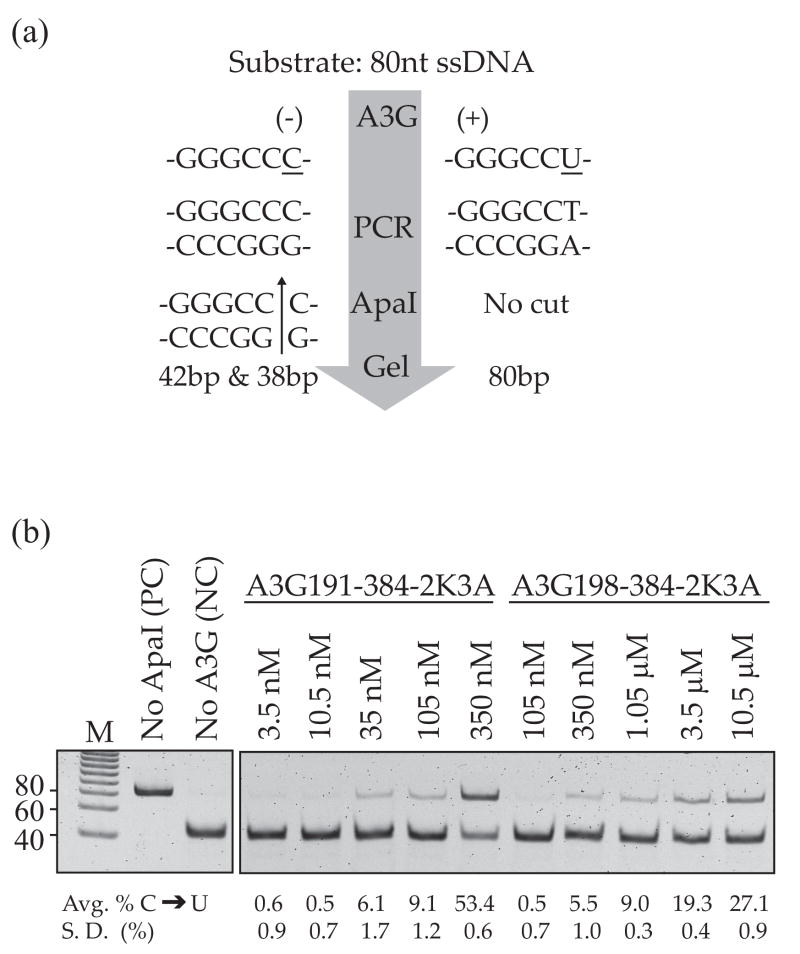
Although the E. coli-based activity assay provides a good proxy for DNA cytidine deaminase activity, it is not amenable to kinetic analyses and its activity range is limited by the spontaneous mutation rate on the lower end and cell inviability on the upper end. We therefore compared the DNA cytidine deaminase activities of purified A3G191-384-2K3A and A3G198-384-2K3A using a quantitative in vitro single-strand DNA (ssDNA) deaminase assay11 (Fig. 2a). DNA cytidine deaminase activity is measured by i) incubating A3G with an 80 nucleotide ssDNA substrate containing a 5′-GGGCCC A3G target site (the strongly preferred A3G target cytidine is underlined), ii) subjecting the deamination products to PCR, which will amplify both substrate and the 5′-GGGCCU product, and iii) digesting the products to completion with ApaI, which cleaves the deamination substrate but not the deamination product. The percentage of uncut PCR product thereby provides a quantitative measure of DNA cytidine deaminase activity.
Fig. 2.
In vitro DNA cytidine deaminase activity. (a) A schematic view of the major steps of the in vitro deamination assay11. See the text and methods for details. (b) In vitro DNA cytidine deaminase activity of A3G191-384-2K3A and A3G198-384-2K3A. The migration positions of a positive control (PC) and a negative control (NC) are indicated. The controls were processed in parallel with the experimental reactions and analyzed as part of the same gel, but non-relevant intervening lanes were removed for presentation. The percentage of deaminated substrate (uncut PCR product) from three independent experiments is shown below each lane.
The in vitro DNA cytidine deaminase activity data showed that A3G191-384-2K3A was 10-fold more active than A3G198-384-2K3A (e.g., the 105 nM product band intensity is similar to that of the 1.05 μM reaction, respectively; Fig. 2b). These data were representative of multiple experiments, and the protein concentrations were selected specifically to show the linear activity ranges. Taken together with the E. coli activity data, we concluded that A3G191-384-2K3A is intrinsically more active than A3G198-384-2K3A. The differential magnitudes of the effects in E. coli and in vitro are probably due to many factors, including the presence of a complex milieu and protein chaperones in E. coli (3-fold difference) versus a chemically defined buffer in vitro (10-fold difference) (see Discussion). It is further important to note that these in vitro studies were done with a subset of the protein preparations that were used for the following structural studies (e. g., Fig. S1)
The α1 helix in A3G191-384-2K3A contributes to active site stability
Our original NMR studies of A3G198-384-2K3A did not resolve residues 198–21021, but the subsequent A3G197-380 crystal structure showed that residues 201–206 were capable of forming an α-helix22. Since the inability to resolve this region in our original NMR experiments correlated with diminished A3G198-384-2K3A activity in E. coli and especially in vitro, we hypothesized that some additional structure in A3G191-384-2K3A (such as the α1 helix) would be directly responsible for the elevated DNA cytidine deaminase activity. We also considered an alternative hypothesis that one or more of the 2K3A mutations destabilize the α1 region.
The solution structure of A3G191-384-2K3A showed that residues 201–206 form the α1-helix of the C-terminal domain (Fig. 3a & b, Fig. S2). Since 2K3A mutations are present in both the A3G191-384 and the A3G198-384 constructs, it was clear that the 2K3A substitutions are not responsible for the lack of the α1-helix in the 198–384 NMR structure. The α1 helix is positioned anti-parallel to α5, in full agreement with the A3G197-380 crystal structure22. This positioning was supported by strong nuclear-Overhauser-enhancement signals (NOEs) between the amide protons of T201/F202/T203 at the beginning of α1 and the methyl protons of V351 at the end of α5. The NMR data also indicated that F202 has hydrophobic contacts with V351. NOEs were also detected between the amide protons of N208/E209 and Hε1 of W361 immediately proximal to α6. These data were in good agreement with the A3G197-380 crystal structure, which showed hydrophobic contacts between N208 and W361 and a hydrogen bond between Oδ1 of N208 and Hε1 of W361. These α1 interactions are crucial for DNA deamination because alanine substitutions at these positions were shown either to ablate (F202A, N208A, V351A and W361A) or compromise (T201A and T203A) catalytic activity in E. coli 20,25.
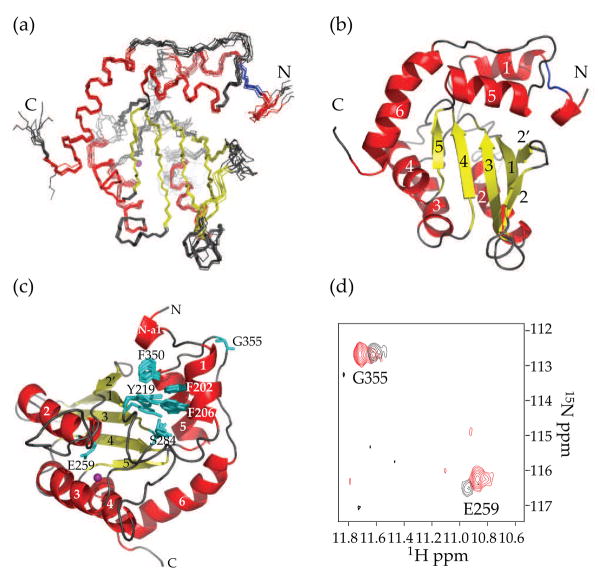
Fig. 3.
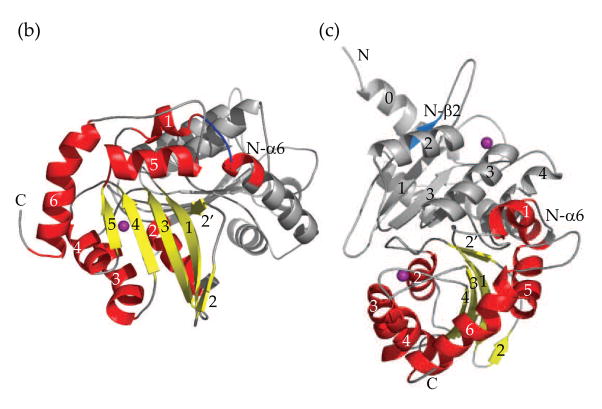
NMR structure of A3G191-384-2K3A (PDB ID code 2kem). (a) A superimposition of 10 NMR structures showing α-helices in red, β-sheets in yellow and Zn2+ in purple. Residues 195–200 are colored blue to highlight their well-defined structure. (b) A ribbon diagram of the NMR structure shown in A from the same angle. Since our prior work had detected only five out of six α-helices21, the helices were renumbered α1 (201–206), α2 (258–269), α3 (289–301), α4 (321–330), α5 (340–351) and α6 (363–380). (c) A schematic of the hydrophobic contacts involving F202, F206 and F350. The side-chains of E259 and G355 are also labeled. (d) 1H-15N correlation NMR signals of G355 and E259 of A3G191-384-2K3A in red and A3G198-384-2K3A in black.
Further direct comparison of the A3G191-384-2K3A and the A3G198-384-2K3A solution structures indicated that several residues within α1 help stabilize the catalytic core and thereby enhance DNA deaminase activity. For instance, F202 and F206 make hydrophobic contacts with Y219 located at the beginning of β1 (Fig. 3c). Additionally, F202 helps stabilize the conformation of the active site through hydrophobic contacts with F350 of α5, and the side chain of F206 contacts S284 within the conserved S284-W-S-P-C288 motif (Fig. 3c). All of these residues are conserved and required for full levels of catalytic activity21; 25. Moreover, as anticipated, we have also found that F202 and F206 are essential for activity of the full-length A3G holoenzyme (see below).
The important contributions α1 makes to catalytic site stability are evidenced by the intensity of the NMR signal of the catalytic glutamate E259, which is located within the substrate-binding pocket. The E259 signal is weak in the A3G198-384-2K3A NMR spectrum, indicating this residue is undergoing chemical exchange (black in Fig. 3d). In contrast, the E259 signal is much stronger in the NMR spectrum of A3G191-384-2K3A, indicating an increased stability (red in Fig. 3d). G355 yielded similarly intense NMR signals in each protein, indicating that G355 is unaffected by α1 stability and that similar amounts of labeled protein were used in each NMR experiment.
The β2 region of the A3G catalytic domain is discontinuous
Our original A3G198-384-2K3A solution structure revealed a loop-like bulge within the β2 region21. This unique structure is also apparent in our new A3G191-384-2K3A NMR data sets (Fig. 3a & b, Fig. 4a) and in a recently published A3G193-384 NMR structure (Fig. 4b)20. In contrast, a recent paper describing an A3G197-384 crystal structure reported that the β2 region was continuous and possibly functionally equivalent to the long continuous β2 strand of A2 that mediates homo-dimerization22.
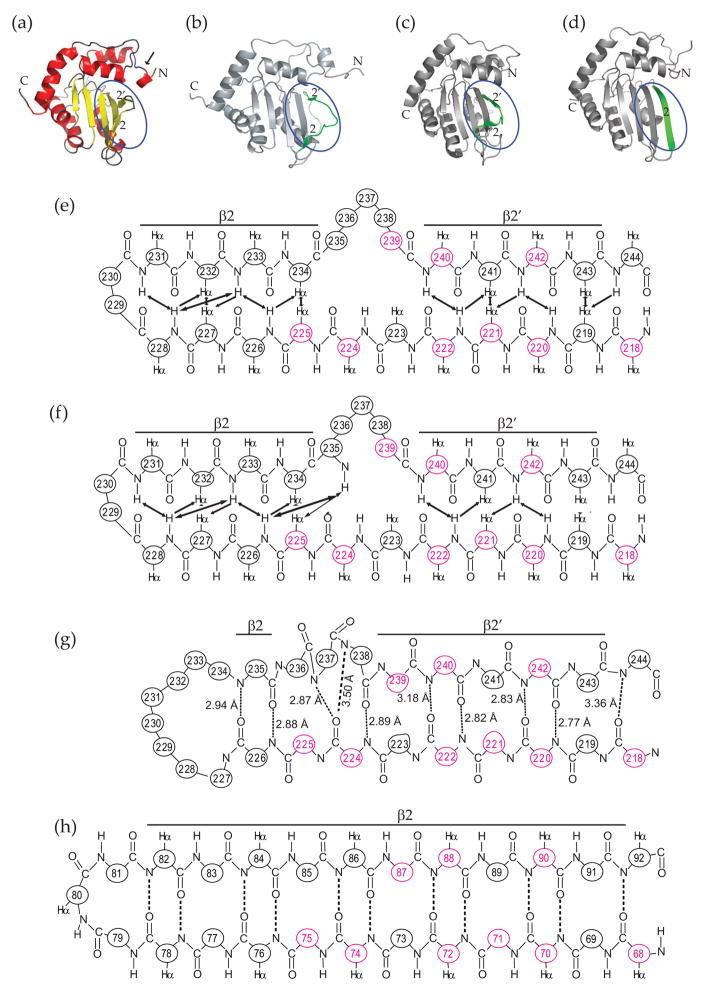
Fig. 4.
A structural comparison A3G191-384-2K3A (PDB ID code 2kem; this study), A3G193-384 [2kbo;20], A3G197-380 [3e1u;22] and APOBEC2 [2nyt;23]. (a, b, c and d)α-carbon traced ribbon schematics for the aforementioned structures. The β2 regions are highlighted by blue ovals and colored green in panels (b), (c) and (d). The inter-domain linker region in panel (a) is indicated by an arrow. (e, f, g, h) Polypeptide backbone and chemical contact schematics for the β2 and β1 regions of A3G191-384-2K3A, A3G193-384, A3G197-380 and A2, respectively. Amino acid positions are numbered and, if the residue is conserved, colored red. In panel (e) and (f), observed NOEs are indicated by arrows. In panels (g) and (h), predicted hydrogen bonds are indicated by dashed lines and labels corresponding to distances between the amide nitrogen and the carbonyl oxygen atoms.
However, a detailed atomic comparison of the β2 region of the A3G191-384-2K3A NMR structure (Fig. 4a & e), the A3G193-384 NMR structure (Fig. 4b & f), the A3G197-380 crystal structure (Fig. 4c & g) and the A2 crystal structure (Fig. 4d & h) revealed that the A3G catalytic domain crystal structure also has a discontinuous β2 region. First, for purposes of orientation, the hydrogen bonds between A3G β2′ residues 240–243 and β1 residues 219–222 are identical in both the NMR and crystal structures (Fig. 4e, f & g). Second, the structures differ at R239, which contacts β1 in the crystal but forms part of the loop in the NMR structures. Third and importantly, in the crystal structure the next N-terminal residue R238 is bulged-out such that the amide nitrogen of Q237 takes its place and bonds with the carbonyl oxygen of V224. In the NMR structure, R238 is part of the loop and it does not appear to make any chemical contacts. Fourth, in both structures, N236 is located in the central part of the loop and it does not appear to contact any β1 residues. Finally, in the crystal structure, β2-to-β1 hydrogen bonding resumes with L235 interacting with R226. This event is associated with an excess of β2 to β1 residues, with three residues in β2 (N236, Q237 and R238) juxtaposing only two in β1 (V224 and E225) (Fig. 4e). The net result of this mismatch is a bulged-out β2 strand. Altogether, the β2 strand in the A3G197-380 crystal structure is discontinuous at three positions --- residues N236, Q237 and R238. It is important to emphasize that these three residues constitute a major part of the loop-like bulge in the β2-bulge-β2′ region in the NMR structures [20; 21 and this study], in reverse-engineered protein preparations with L234 and/or C243 restored [21 and Fig. S3] and in wild-type A3G193-38420. We conclude that a loop-like bulge structure is a fundamental part of the β2 region of the A3G catalytic domain.
Nevertheless, a significant difference between the NMR spectroscopic and the crystallographic studies is the length and organization of the β2 portion of the β2-loop-like-bulge-β2′ region (compare Fig. 4e, f & g). The NMR structures showed that residues 229 and 230 form the β1-to-β2 loop and that β1 residues 225–228 form a beta-sheet with β2 residues 231–234 [regardless of whether 234 was L or K; Fig. 4e, 4f and S3 20; 21]. Similar conceptually but contrasting in some details, the crystal structure has a much larger loop from residues 227–234 and only a single β1-to-β2 interaction between R226 and L235. In total contrast, the corresponding region of A2 is clearly part of a continuous β2 strand (Fig. 4h). At present we cannot explain this difference between the A3G catalytic domain structures, but it may be due to differences in buffers and/or techniques. However, the difference may suggests that the β2 region has an intrinsic structural flexibility, which enables it to adopt multiple conformations (perhaps also influenced by nucleic acid and/or protein interactions). Indeed, it is striking that most of the A3G β2 region residues are dispensable for deaminase activity in E. coli [most single alanine substitutions within residues 232–243 were at least 50% active, with the exception of G240 which was not tested21; 25].
The NMR structure of A3G191-384-2K3A revealed a structured inter-domain linker region
Residues 191–194 resolved as a single helical turn in the A3G191-384-2K3A NMR structure (Fig. 3a & b). Amino acid sequence alignments with the C-terminal domain of A3G and A2 predicted that these residues constitute part of the last helix of the N-terminal pseudo-catalytic domain, – N-α6, which is predicted to span residues 177–194 [Fig. 5a; also see31]. The A3G191-384-2K3A NMR data also indicated that residues 195-200 form a stable loop (bridge-like-structure) that connects the final helix of the pseudo-catalytic domain to the first helix of the catalytic domain (i.e., N-α6 to C-α1). The stability of the bridge may be mediated in part by an interaction between L193 and G240/F241, as NOEs were detected between the Hδ of L193 and the amide protons of the latter residues.
Fig. 5.
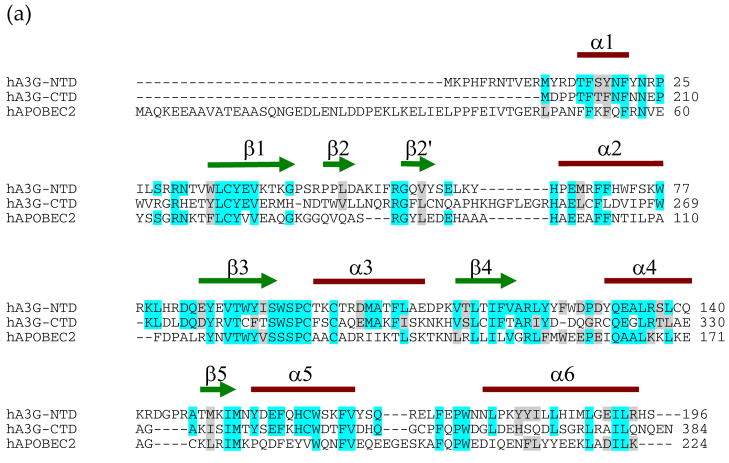
A model for full-length A3G. (a) Amino acid alignments of A3G1-196, A3G197-384 and APOBEC2. The NCBI accession numbers for these proteins are NP_068594 and NP_006780.Identity is indicated by light blue and similarity by gray shading, and the actual secondary structure elements for A3G191-384-2K3A are illustrated above the alignments. (b, c) Two different angles of a full-length A3G structural model. In (b) the catalytic domain is viewed with the same angle of Fig. 3b, which clearly shows the superposition N-α6 region. The model structure of the N-terminal pseudo-catalytic domain is colored gray. Helices and β-strands within the C-terminal catalytic domain are colored red and yellow, respectively. N-β2 and N-α6 are labeled and highlighted in blue and red, respectively. The β2 region of the pseudo-catalytic domain modeled as a continuous strand and, unlike prior A3G catalytic domain structures, the pseudo-catalytic domain has an N-terminal extension that modeled as an alpha helix.
A model for full-length A3G
The well-defined atomic coordinates of the N-terminal residues (191–196) of the A3G191-384-2K3A NMR structure enabled us to assemble a novel full-length A3G structural model consisting of the predicted pseudo-catalytic domain structure overlapped with the actual A3G191-384 catalytic domain structure (Fig. 5b & c).
Interestingly, the resulting full-length A3G model suggested that the N-terminal pseudo-active site and the C-terminal active site are situated on different faces of the holoenzyme (Fig. 5b & c). Moreover, in contrast to full-length A3G models based upon the crystal structure of the β2-β2 homo-dimeric A2 protein23; 30, the short N-α6 to C-α1 connection placed physical constraints upon our model such that it became impossible for the β2 region of the pseudo-catalytic domain (N-β2) to mediate inter-domain interactions with the β2-loop-β2′ region of the catalytic domain (colored blue and yellow, respectively, in Fig. 5c).
It should also be noted that three loops appear close to the domain interface [N-α4-loopβ5 (residues 140–149), C-α1-loop-β1 (residues 210–217) and C-β2′-loop-α2 (residues 245–256)] and that the structures of these loops have yet to be well determined [21 and this study]. For instance, the loop between N-α4 and N-β5 has a four-residue insertion relative to the A3G catalytic domain making it difficult to model precisely (Fig. 5a). Moreover, the large loops between α1 and β1 and between β2′ and α2 of the catalytic domain are flexible in solution and the structures were not resolved by NMR [20; 21 and this study]. It is likely that multiple contact points mediate the inter-domain interactions and that further structural and functional studies will be needed to precisely delineate all of the residues involved.
Holoenzyme studies to evaluate the role of N-α6 and the inter-domain region
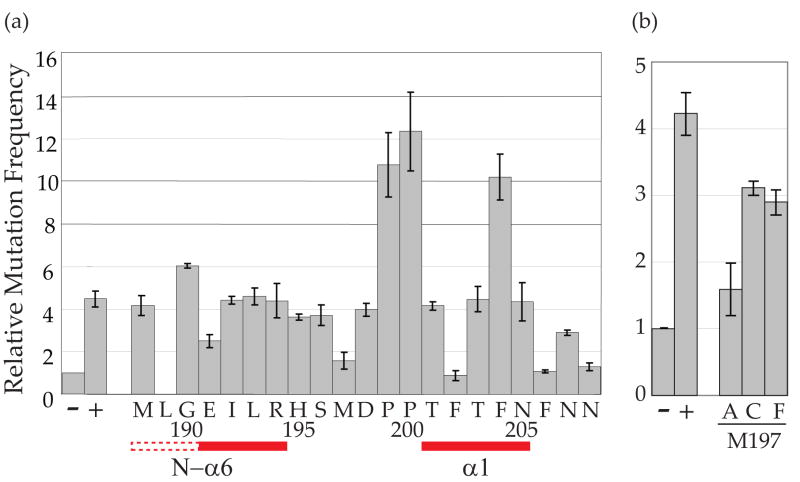
RifR mutation assays were used to test whether N-α6 residues and inter-domain loop residues are required for full-length holoenzyme catalytic activity. Interestingly, although most mutants showed near wild-type levels of activity, several had phenotypes. First, A3G-E191A showed a 50% reduction in the E. coli-based assay (Fig. 6a). A reasonable structural explanation for this observation is the likelihood that E191 is required to form the portion of the N-α6 helix (191–194) observed here using NMR and that this helix contributes to the integrity of the catalytic domain. Specifically, since four residues are required to form one stable helical turn, the CO of E191 is probably hydrogen bonding with the HN of R194. The importance of this N-α6 helix for catalytic domain stability in solution is apparent by comparing the A3G191-384-2K3Astructure described here and the A3G193-384 structure reported recently – only the former shows the α1 helix buttressing core structural elements [compare Figs. 4a and b 20; 32]. We conclude that at least part of the N-α6 helix (191–194) and the inter-domain region (195–200) are important for the structural integrity and activity of the C terminal catalytic domain. It is also worth noting that the catalytic domain α1 helix is similarly positioned in our 191–384 NMR structure and in the 197–380 crystal structure, suggesting that crystal packing chemical contacts may also be able to promote the proper orientation of the α1 helix [compare Fig. 4a & b and22].
Fig. 6.
E. coli-based DNA cytidine deaminase activity of full-length A3G amino acid substitution mutants. (a) Mutator phenotype of untagged A3G1-384 alanine substitution mutants. The histogram bars show the relative RifR mutation frequencies of cells expressing the vector control (−), A3G1-384 (+) or the indicated alanine substitution derivatives. Each histogram bar reports the average and SEM of the median mutation frequency from multiple independent experiments (each with 6–8 cultures per condition). Residue 189 was not examined due to construction difficulties. The primary amino acid sequence is indicated under the histogram. The positioning of N-α6 and C-α1 is indicated by solid red lines and the predicted part of N-α6 by a dashed line. (b) Mutator phenotype of three M197 variants. The experimental conditions were identical to those described in (a), but the Y-axis was adjusted to represent lower maximal mutation values.
Second, we observed that A3G-M197A triggered levels of mutation that were almost as low as the vector-control background (Fig. 6a). At first glance, this result appeared at odds with our other data, because the entire N-terminal half of A3G (including M197) is dispensable for catalytic activity in E. coli [25 and Fig. 1]. However, subsequent analyses revealed that A3G-M197C or A3G-M197F constructs were still catalytically active, indicating that M197 itself is non-essential and that an alanine substitution at this position somehow compromises the activity of the full-length protein (Fig. 6b). The precise reason for this was not pursued further (also because this residue is not conserved with other APOBEC3 family members).
Finally, as reported previously for A3G198-384 constructs, alanine substitutions at F202, F206 or N208 resulted in a total loss of catalytic activity [compare Fig. 6a and 21; 25]. These data further confirm the importance of the α1 region for DNA cytidine deaminase activity.
HIV restriction activity of full-length A3G-2K3A
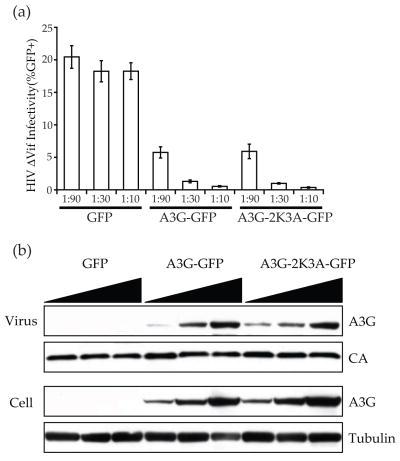
To dispel concerns that the 2K3A substitutions that facilitate long-term solubility might compromise the function of the full-length A3G holoenzyme, we compared the HIV-1 restriction activities of wild-type A3G and a full-length 2K3A derivative. Both proteins inhibited the infectivity of a Vif-deficient HIV-1 reporter virus similarly (Fig. 7a). In agreement with these infectivity data, the amount of each protein detected in cells and in viral particles was also indistinguishable (Fig. 7b). Thus, these studies demonstrate that the five amino acid substitutions that were used here to render A3G191-384 amenable to solution studies do not have a significant impact on the HIV restriction activity, cellular expression level or encapsidation ability of the A3G holoenzyme. These data thereby counter suggestions20; 22 that these five substitutions may have compromised the structure and function of the A3G catalytic domain, and they lend additional support to our conclusions (above) (i) that the α1 helix is an essential structural feature of the A3G catalytic domain (and likely all DNA cytidine deaminase family members), (ii) that the loop-like bulge in the β2 region is a bona fide feature of the protein, and (iii) that the structured inter-domain region positions the N- and C-terminal β2 regions too far from one another to facilitate inter-domain interactions (akin to dimerization of single domain family members).
Fig. 7.
HIV restriction activity of A3G-2K3A. (a) A histogram reporting the infectivity of Vif-deficient HIV-1 produced in the presence of a GFP control expression vector, wild-type A3G-GFP or full-length A3G-2K3A-GFP. (b) Representative immunoblots of virus particles (unti-A3G, anti-p24) and cellular lysates (anti-A3G and anti-tubulin).
Discussion
A direct comparison of minimal and slightly longer A3G catalytic domain proteins revealed striking differences in activity and structure. The longer A3G191-384-2K3A was approximately 10-fold more active in vitro than the protein that was 7 residues shorter. The main structural explanation for this difference was the presence of an α-helix spanning residues 201–206 in the longer protein. Residues within this α1 region buttress several core structural elements and indirectly help stabilize the enzyme’s catalytic center, in full agreement with the recently published A3G197-380 crystal structure22. Several residues within the α1 region are conserved and required for activity indicating that this structural feature will be an integral part of all DNA cytidine deaminase family members.
We previously showed that the entire α1 region and most of its individual residues are required for DNA cytidine deaminase activity21; 25. These data are further supported here by results demonstrating that F202, F206 and N208 are required for holoenzyme activity in E. coli (Fig. 6). Given these observations and our new α1 structural insights, it was surprising that the shorter, α1-deficient A3G198-384-2K3A protein shows any catalytic activity (e.g., Figs. 1 & 2). A possible explanation is that a proportion of this protein is folded properly in E. coli and, following purification, a significantly smaller fraction maintains α1 integrity and catalytic activity. This fraction would not be large enough to yield structural data, but it could still elicit activity. Such an explanation also agrees with the bigger activity differential in vitro than in E. coli and the fact that a longer A3G catalytic domain construct was required to resolve the α1 region by NMR. This explanation is further supported by the fact that another slightly shorter A3G193-384 construct failed to resolve the α1 helix and reveal its critical interactions with several core structures20.
It is also important to emphasize all high-resolution structural studies to date have focused on fundamental details of the A3G catalytic domain [20; 21; 22& this study]. The GST tagged full-length A3G holoenzyme is 40-fold more active than GST-A3G191-384-2K3A in our in vitro ssDNA deamination assay [R.N. & M.K., unpublished data &11]. Similarly, GST-tagged full-length protein showed 25-fold more activity than GST-A3G197-38022. This activity differential between the holoenzyme and its C-terminal domain is likely due to a combination of several factors such as specific activity, substrate affinity, processivity and stoichiometry [e.g.,7; 8; 11; 22; 26; 29]. For instance, the ssDNA dissociation constant for full-length A3G was 76 ± 21 nM8 or 50 ± 7 nM7, in contrast to approximately 400 μM for A3G198-384-2K3A or A3G191-384-2K3A [21 and data not shown]. A full A3G holoenzyme structure in the presence and absence of a single-strand DNA substrate will help address many of these remaining questions.
The β2 region has been a subject of controversy since the A2 crystal showed a continuous β-strand at the dimer interface23 and our original A3G catalytic domain NMR structure showed a discontinuous loop-like bulge structure21. Similar to A2, initial analyses of the A3G197-380 crystal structure suggested the existence of a continuous β2 (residues 235–243)22. However, our examination of the atomic coordinates of the crystal structure also revealed evidence for discontinuous structure in the β2 region (Fig. 4). A similar loop-like bulge was also apparent between the β2 and β2′ regions in our new A3G191-384-2K3A solution structure (Figs. 3–5) and in a wild-type A3G193-384 NMR structure20. These data strongly indicated that, in solution, the β2 region of the A3G catalytic domain is not continuous like it is in A2, with the functional implication being that it is unlikely to participate in a similar β2-β2 dimerization event. our studies do not exclude the possibility that this region may participate in other intra- or inter-molecular interactions.
A major difference between the A3G191-384-2K3A structure and all prior structures is a well-resolved N-terminal end, which extends into the predicted final α-helix of the A3G pseudo-catalytic domain (residues 1–196). The significant overlap between pseudo-catalytic domain residues and our new catalytic domain structure enabled the construction of a full-length model (Fig. 5). The structural restrictions imposed by the well-resolved N- to C-terminal domain connecting region place the β2 regions of the N- and the C-terminal halves of A3G too far away from one another to forge the inter-domain connection [i.e., unlike the A2 β2-β2 dimer23; 30]. Future studies will be directed toward defining the potentially extensive inter-domain loop-loop contacts suggested by the model and addressing their functional importance. Our full-length A3G model may also prove useful for understanding other features of this enzyme, such as intersegmental transfer, oligomerization and ribonucleoprotein complex formation [e.g.,11; 26; 27; 29].
Materials and Methods
Plasmids
pTrc99A was used to express untagged A3G derivatives24. pTrc99A-A3G191-384 (or 198–384) was constructed by sub-cloning an NcoI/SalI-digested PCR fragment, which was generated using primers 5′-NCC-ATG-GAA-CCA-TGG-AGA-TTC-TCA-GAC-ACT-CG or 5′-NCC-ATG-GAT-CCA-CCC-ACA-TTC-ACT-TTC and 5′-NGT-CGA-CTA-GTT-TTC-CTG-ATT-CTG-GAG-AAT-GGC, respectively. pTrc99A-A3G was constructed similarly with 5′-NCC-ATG-GCG-CCT-CAC-TTC-AGA-AAC-AC and the same downstream primer. pGEX-6P2-A3G198-384 and its mutant derivatives were described previously21. PCR with primers 5′-GGA-ATT-CGA-GCT-CGG-TAC-CAC-CGA-GAT-TCT-CAG-ACA-CTC-GAT-GGA-TC and 5′-ATC-CAT-CGA-GTG-TCT-GAG-AAT-CTC-GGT-GGT-ACC-GAG-CTC-GAA-TTC-C was used to create pGEX6P1-A3G191-384 from a full-length A3G parental plasmid25. The mammalian cell expression vector pEGFP-N3 (Clontech) and the full-length A3G-expressing derivative were used previously33. A3G-2K3A-GFP was constructed by replacing the BamH1/SalI fragment of the parental construct with a similarly cut 2K3A-encoding PCR amplicon made using GST-A3G191-384 2K3A plasmid template and primers 5′-GAG-CTC-AGG-TAC-CAC-CAT-GGA-TCCA-CCC-ACA-TTC-ACT-TTC and 5′-GTC-GAC-TCC-GTT-TTC-CTG-ATT-CTG-GAG-AAT. Mutant derivatives of the parental constructs were constructed by site-directed mutagenesis (Stratagene) and a full list of primers is available upon request.
E. coli-based deaminase activity assays
The intrinsic DNA cytidine deaminase activity of A3G and its variants was assayed by expressing these proteins in ung-deficient E. coli BW310 and quantifying the frequency of rifampicin-resistance (RifR)-conferring RNA polymerase B (rpoB) mutations [e.g.,21; 24; 25]. A broad number of single base mutations in rpoB cause active site amino acid substitutions that confer RifR. For each condition, 8 single colonies were grown overnight at 37°C in LB medium containing 100 μg/mL ampicillin. Under inducing conditions, 1 mM IPTG was added to the medium. Appropriate volumes of cells were then spread to plates containing 100 μg/ml rifampicin to select for RifR mutants and to plates containing 100 μg/ml ampicillin to determine the number of viable cells. Mutation frequencies were calculated as the number of RifR mutants per 107 viable cells.
E. coli immunoblots
A3G constructs were expressed in E. coli strain BW310. Proteins were produced by overnight expression at 37°C in LB medium containing 100 μg/mL ampicillin. To induce expression, cells were diluted 1:10 in LB medium containing 100 μg/mL ampicillin and 1 mM IPTG and grown for 1 hour at 37°C. Cells were pelleted and resuspended in SDS gel loading buffer [50mM Tris-Cl (pH 6.8), 100mM β-mercaptoethanol, 2% SDS (w/v), 0.1% (w/v) bromophenol blue, 10% (v/v) glycerol]. Lysates were heated at 95°C for 5 min and fractionated by SDS-PAGE. Proteins were transferred to a PVDF membrane (Millipore) and probed with a rabbit anti-A3G polyclonal serum. The primary antibody was detected by incubation with HRP-coupled anti-rabbit IgG (Bio-Rad) followed by chemiluminescent imaging (Roche).
HIV infectivity studies and immunoblots
HIV-GFP reporter viruses were produced by Fugene-mediated transfection (Roche) of 293T cells with a five plasmid cocktail34. The HIV-GFP proviral plasmid CS-CG, the Gag-Pol expression plasmid, the Rev expression plasmid and the VSV-G envelope expression plasmid constituted 0.9 μg of the cocktail, and the vector control or the A3G expression plasmid another 0.01, 0.03 or 0.09 μg. The total amount of transfected DNA was adjusted to 1 μg by adding pcDNA3.1 (Invitrogen). Virus-containing supernatants were harvested 48 hrs post-transfection and purified from cell debris by filtration (0.22 μm PVDF, Millipore). Viral supernatants (1 mL) were further purified by centrifugation through a 20% sucrose cushion (2hr, 20,000 g). The resulting viral pellet was resuspended directly in SDS gel loading buffer (above), fractionated by SDS-PAGE, transferred to a PVDF membrane (Millipore) and probed with an anti-GFP antibody JL-8 (Invitrogen) to detect GFP, A3G-GFP or A3G-2K3A-GFP. An anti-p24 monoclonal antibody35 provided by M. Malim through the NIH AIDS Research and Reference Reagent Program was used as a loading control. Both monoclonal antibodies were detected using an HRP-conjugated goat anti-mouse IgG serum (BioRad) followed by chemiluminescent imaging (Roche). After harvesting viral supernatants, A3G levels in virus producing cells were monitored by extracting soluble proteins with RIPA buffer (1hr, 4°C, gentle rotation), removing particulate by centrifugation (10 min, 20,000 g) and immunoblotting as described above. An anti-tubulin monoclonal antibody (Covance) was used for a cellular lysate loading control.
In vitro DNA cytidine deamination assays
A3G deamination reactions were performed as described11 in a 10 μl reaction volume containing 25 mM Tris (pH 7.0), 0.1 mg/ml BSA and 10 fmol ssDNA substrate 5′-GGA-TTG-GTT-GGT-TAT-TTG-TTT-AAG-GAA-GGT-GGA-TTA-AGG-GCC-CAA-TAA-GGT-GAT-GGA-AGT-TAT-GTT-TGG-TAG-ATT-GAT-GG. Reactions were incubated for 8 min at 37°C and then terminated for 5 min at 95°C. One-tenth of the reaction mix was used as a PCR template for amplification by the target-flanking primers (underlined above) in 20 μl buffer S (Larova Inc.; 1 denaturation cycle at 95°C for 3 min followed by 14 rounds of annealing at 61°C for 30 sec and denaturation at 94°C for 30 sec). One-fourth of each PCR reaction was incubated with 5 units ApaI (Fermentas) for 1 hr at 30°C (the cleavage site is indicated above). The resulting restriction products were fractionated by 14% PAGE, stained with SYBR gold (Molecular Probes) diluted 1:10,000 in 1xTris-Borate-EDTA buffer (pH 7.8), excited by UV light (302nm), imaged by an Olympus C-5050 CCD camera and quantified using TINA2.0 densitometry software (Raytest).
Protein expression and purification
GST-A3G191-384-2K3A and GST-A3G198-384-2K3A were expressed in E. coli strain BL21-DE3(RIL) (Stratagene). Unlabeled proteins were produced by overnight expression at 17°C in LB medium containing 1 mM IPTG and 100 μg/mL ampicillin. Isotope-labeled proteins were produced by overnight expression at 17°C in M9 supplemented with 15NH4Cl, 13C-labeled D-glucose and 2H water21. Proteins were purified by sonicating cell pellets in lysis buffer [100 mM NaCl, 50 mM Na2HPO4/NaH2PO4 (pH7.0), protease inhibitor (Roche)], separating the soluble (supernatant) and insoluble (pellet) fractions by centrifugation (12,110g, 20 min, 4°C), binding to glutathione sepharose (GE Healthcare), washing with lysis buffer, eluting with PreScission protease (GE Healthcare) in 1 mM DTT, 50 mM Na2HPO4/NaH2PO4 [pH7.4] and, finally, concentrating with Centricon filters (Millipore). Representative protein preparations were also used for in vitro DNA cytidine deaminase assays.
NMR spectroscopy and structure determination
NMR assignments of A3G198-384-2K3A were used to help assign signals for A3G191-384-2K3A. For backbone assignments, we used HNCA 36; 37; 38 and HNCACB 39 spectra of the uniformly 13C and 15N labeled and 90% perdeuterated protein. The side chain assignments were completed using 15N-, 13C- edited NOESY-HSQC40. NOE-derived distance restraints were obtained from 15N- or 13C- edited NOESY-HSQC and 2D NOESY spectra of protonated, 90% perdeuterated and 50% perdeuterated proteins using mixing times between 80 and 200 ms. To collect NOEs between amide proton and methyl proton, methyl proton and methyl proton, δCH3 protons of Leu and Ile and γCH3 protons of Val were selectively protonated in an otherwise fully deuterated protein41. All spectra were taken with Varian 800 MHz, Varian 600 MHz, Bruker 900 MHz, Bruker 800 MHz or Bruker 700 MHz spectrometers. NMR spectra were processed with NMRPipe42 and analyzed with CARA43. 300 torsion angle restraints were taken from TALOS prediction44. 145 hydrogen bonds were set for residues consistent with the chemical shifts deviations and NOE pattern defined secondary structure. Newly found NOEs for residues 191–209 were added to NOEs found in A3G198-384-2K3A construct to calculate initial structure of A3G191-384-2K3A using CNS45. This initial structure was used for further Atnos 46/Candid 47 structure dependent cycles with spectra of A3G191-384-2K3A construct. A total of 2224 NOEs were assigned. The final calculations employed 259 intra-residue, 640 sequential, 613 medium-range and 756 long-range NOE-derived constraints. 100 structures were calculated with CNS torsion angle molecular dynamics. Ten of the calculated structures were chosen based on energy to represent the ensemble. The NMR constraints and structure statistics of A3G191-384-2K3A are provided in Table 1.
TABLE 1.
NMR refinement statistics.
| Distance restraints | Average RMSD | ||
|---|---|---|---|
| NOEs | From distance restraints | 0.0321 ± 0.0010 | |
| All | 2224 | From dihedral restraints | 0.5424 ± 0.0371 |
| Intra-residue | 259 | From idealized geometry: | |
| Inter-residue | 1772 | Bonds (Å) | 0.0030 ± 0.0001 |
| Sequential (|i−j|=1) | 640 | Angles (°) | 0.4912 ± 0.0096 |
| Medium | 613 | Impropers (°) | 0.3933 ± 0.0091 |
| Long | 756 | ||
| Hydrogen bonds | 145 | Average pairwise between 10 structures: | |
| Dihedral angle restraints: | Backbone (Å)a | 0.86 ± 0.18 | |
| φ | 150 | All heavy atoms (Å)a | 1.84 ± 0.26 |
| ψ | 150 | Backbone (Å)b | 0.54 ± 0.13 |
| All heavy atoms (Å)b | 1.26 ± 0.15 | ||
| Ramachandran plot appearance: | |||
| Most favored regions (%) | 64 | ||
| Additional allowed (%) | 27 | ||
all residues are included except loops between β2 and β2′ and between β2′ and α2.
all secondary structure elements are included.
A3G1-194 and full-length structural models
The full-length A3G model was constructed in three steps. First step, a model for the A3G pseudo-catalytic domain residues 1–194 was generated based on amino acid sequence homology between the A3G pseudo-catalytic domain, the A3G catalytic domain and A2 48 (Fig. 5a). Sequence alignment was generated by pair-wising A3G1-196 with A3G197-384, then with A2, using the NCBI algorithm BLAST 2 SEQUENCES set to the similarity matrix BLOSUM62. This similarity matrix uses amino acid side chain and genetic code similarities, and it calculated 35% identity and 52% similarity for the A3G1-196 and A3G197-384 alignment and 29% identity and 46% similarity for the A3G1-196 and A2 alignment. The actual A3G191-384 secondary structural elements from this study were superimposed on the alignment to delineate core residues (Fig. 5a). Considering only these residues within the secondary structural elements, the percentages of identity and similarity were recalculated to yield 49% identity and 62% similarity between A3G1-196 and A3G197-384 alignment and 39% identity and 49% similarity for the A3G1-196 and A2 alignment. These high levels of identity strongly indicated that the A3G1-196 model structure would closely approximate the pseudo-catalytic domain of A3G, because homology models with 30–50% identity typically position greater than 90% of the backbone atoms within 1.5 Angstroms RMSD to the real structure49. The A3G1-194 model was generated using a program called YASARA48. This program used multiple parameters to systematically generate an A3G1-194 model structure. First, each of the three most related structures, A3G191-384-2K3A (PDB ID code 2kem; this study), A3G197-380 [3e1u;22] and A2 [2nyt;23], was aligned with the A3G1-194 primary and predicted secondary amino acid structures (Fig. 5a). Second, YASARA accommodated insertions by searching the PDB for loops with start and end points that superimposed well with anchor points in the model and then loops were further optimized to adopt the lowest energy conformation. Third, amino acid side chain geometry was refined using electrostatic and knowledge based interactions. Fourth, an unrestrained high-resolution refinement was performed and one model was built for each related structure. Individual models were ranked based on a residue-for-residue score. Finally, the top model was taken and lower-scoring regions were replaced iteratively with higher-scoring regions of the other two models. Hence, the resulting model resembled both the A3G catalytic domain (most regions) and A2 (β2 region). The pseudo-catalytic domain β2 region modeled as a continuous short strand because it shared little homology with the catalytic domain β2 region (Fig. 5b & c).
Second step, residues 191–194 occurred in both the A3G1-194 model and the A3G191-384-2K3A NMR structure and, importantly, these residues form one helical turn in both structures. Therefore, these helical turns were simply superimposed to connect the N- and C-terminal domains and render a full-length A3G model.
Third step, three loops were located close to the opposing domains, including 140–149 of the A3G1-194 model and 210–217 and 245–256 of the A3G191-384-2K3A NMR structure. Since these loops were not well determined in NMR solution structures or the A3G1-194 model, they were regenerated in the full-length A3G model using YASARA48 to find lowest energy conformation.
All structural schematics were made with PyMOL (DeLano Scientific; www.pymol.org) and labeled manually.
Data bank accession codes
The atomic coordinates and NMR restraints for A3G191-384-2K3A have been deposited in the Protein Data Bank (www.pdb.org) with RCSB ID code rcsb101025 and PDB ID code 2kem.
Supplementary Material
Fig. S1. A3G191-384-2K3A purification. a, SDS-PAGE gel of the purified A3G191-384 2K3A. b, UV spectrum of A3G191-384 2K3A. The strong absorbance at 230 nm is due to DTT in the buffer. c, a 15N-HSQC spectrum of A3G191-384-2K3A recorded on Varian INOVA 800MHz equipped with a cryogenically cooled probe.
Fig. S2. Superimposed 15N-HSQC spectra of the A3G191-384-2K3A (red) and the A3G198-384-2K3A (black). N-terminal residues 192–209 and residues 219, 284 and 350 that are interacting with α1 helix are labeled. Signals of residues 198–205 did not appear in the 198–384 spectrum. Residues 206–209 as well as residues 219, 284 and 350 were moved in the 191–384 spectrum (shifts are shown by arrows).
Fig. S3. NOESY spectra showing that neither K234 nor A243 affect the β2-loop like bulge-β2′ secondary structure. a, a schematic diagram of the interactions detected between β1 and β2/β2′. Red arrows represent NOE interactions observed in NMR spectra, and they correspond to the boxed signals in (b) and (c). Black, dashed arrows represent observed NOE interactions not shown in (b) and (c). b, representative strips of the 15N-edited 3D NOESY spectrum of A3G-2K3A showing NOE signals between β1 and β2/β2′. c, representative strips of the 15N-edited 3D NOESY spectrum of A3G-1K2A (both K234 and A243 were reverse engineered to L234 and C243, respectively) showing NOE signals nearly identical to (b).
Acknowledgments
We thank S. Harjes for assistance with YASARA modeling, J. Albin, M. Stenglein and K. Walters for feedback, S. Gad for technical assistance and laboratory members for helpful discussions. The University of Minnesota Supercomputing Institute and NMR Core (NSF BIR-961477) and the UCSF and UC Berkeley QB3 NMR Cores provided key instrumentation. This work was done in part in the Krueger Laboratory with the support of N. & L. Glick and P. & M. Weiss. This work was supported by grants from the National Institutes of Health (AI073167 to H.M., AI064046 to R.S.H. and GM082250 to the HARC center at UCSF and Berkeley), the Medica Foundation Minnesota Partnership for Biotechnology and Medical Genomics (to H.M. and R.S.H.) and the United States-Israel Binational Science Foundation (to M.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 2.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–77. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 5.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 6.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–16. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 7.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 8.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 10.Miyagi E, Schwartzkopff F, Plishka R, Buckler-White A, Clouse KA, Strebel K. APOBEC3G-independent reduction in virion infectivity during long-term HIV-1 replication in terminally differentiated macrophages. Virology. 2008;379:266–74. doi: 10.1016/j.virol.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol. 2008;15:1059–66. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 12.Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–60. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278:44412–6. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 14.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–86. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994;235:635–56. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 16.Carter CW., Jr The nucleoside deaminases for cytidine and adenosine: structure, transition state stabilization, mechanism, and evolution. Biochimie. 1995;77:92–8. doi: 10.1016/0300-9084(96)88110-7. [DOI] [PubMed] [Google Scholar]
- 17.Xiang S, Short SA, Wolfenden R, Carter CW., Jr Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase. Biochemistry. 1995;34:4516–23. doi: 10.1021/bi00014a003. [DOI] [PubMed] [Google Scholar]
- 18.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–4. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 19.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–70. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. Embo J. 2009;28:440–51. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–9. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 22.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008 doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–51. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 24.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett RP, Salter JD, Liu X, Wedekind JE, Smith HC. APOBEC3G subunits self-associate via the C-terminal deaminase domain. J Biol Chem. 2008 doi: 10.1074/jbc.M803726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–14. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 28.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–96. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 29.Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, Raina J, Smith HC. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J Biol Chem. 2006;281:38122–6. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang KL, Mangeat B, Ortiz M, Zoete V, Trono D, Telenti A, Michielin O. Model structure of human APOBEC3G. PLoS ONE. 2007;2:e378. doi: 10.1371/journal.pone.0000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008 doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furukawa A, Nagata T, Habu Y, Sugiyama R, Hayashi F, Yokoyama S, Takaku H, Katahira M. NMR assignments and the identification of the secondary structure of the anti-retroviral cytidine deaminase. Nucleic Acids Symp Ser (Oxf) 2008:183–4. doi: 10.1093/nass/nrn093. [DOI] [PubMed] [Google Scholar]
- 33.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–41. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 34.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–91. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 35.Simon JH, Fouchier RA, Southerling TE, Guerra CB, Grant CK, Malim MH. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–67. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–67. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 37.Kay L, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990;89:496. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Matsuo H, Kupce E, Li H, Wagner G. Increased sensitivity in HNCA and HN(CO)CA experiments by selective C beta decoupling. J Magn Reson B. 1996;113:91–6. doi: 10.1006/jmrb.1996.0161. [DOI] [PubMed] [Google Scholar]
- 39.Wittekind M, Mueller J. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta- carbon resonances in proteins. J Magn Reson B. 1993;101:201–205. [Google Scholar]
- 40.Zhang O, Kay LE, Olivier JP, Forman-Kay J. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 41.Medek A, Olejniczak ET, Meadows RP, Fesik SW. An approach for high-throughput structure determination of proteins by NMR spectroscopy. J Biomol NMR. 2000;18:229–38. doi: 10.1023/a:1026544801001. [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Keller RLJ. The computer aided resonance assignment tutorial. Cantina Verlag; Swizerland: 2004. pp. 1–81. [Google Scholar]
- 44.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallog sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–89. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 47.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–27. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 48.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 49.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–6. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A3G191-384-2K3A purification. a, SDS-PAGE gel of the purified A3G191-384 2K3A. b, UV spectrum of A3G191-384 2K3A. The strong absorbance at 230 nm is due to DTT in the buffer. c, a 15N-HSQC spectrum of A3G191-384-2K3A recorded on Varian INOVA 800MHz equipped with a cryogenically cooled probe.
Fig. S2. Superimposed 15N-HSQC spectra of the A3G191-384-2K3A (red) and the A3G198-384-2K3A (black). N-terminal residues 192–209 and residues 219, 284 and 350 that are interacting with α1 helix are labeled. Signals of residues 198–205 did not appear in the 198–384 spectrum. Residues 206–209 as well as residues 219, 284 and 350 were moved in the 191–384 spectrum (shifts are shown by arrows).
Fig. S3. NOESY spectra showing that neither K234 nor A243 affect the β2-loop like bulge-β2′ secondary structure. a, a schematic diagram of the interactions detected between β1 and β2/β2′. Red arrows represent NOE interactions observed in NMR spectra, and they correspond to the boxed signals in (b) and (c). Black, dashed arrows represent observed NOE interactions not shown in (b) and (c). b, representative strips of the 15N-edited 3D NOESY spectrum of A3G-2K3A showing NOE signals between β1 and β2/β2′. c, representative strips of the 15N-edited 3D NOESY spectrum of A3G-1K2A (both K234 and A243 were reverse engineered to L234 and C243, respectively) showing NOE signals nearly identical to (b).