Abstract
Circular clamps tether polymerases to DNA, serving as essential processivity factors in genome replication, and function in other critical cellular processes as well. Clamp loaders catalyze clamp assembly onto DNA, and the question of how these proteins construct a topological link between a clamp and DNA remains open, especially the mechanism by which ATP is utilized for the task. Here we describe pre-steady state analysis of ATP hydrolysis, PCNA clamp opening and DNA binding by S. cerevisiae RFC, and present the first kinetic model of a eukaryotic clamp loading reaction validated by global data analysis. ATP binding to multiple RFC subunits initiates a slow conformational change in the clamp loader, enabling it to bind and open PCNA, and bind DNA as well. PCNA opening locks RFC into an active state, and the resulting RFC•ATP•PCNA(open) intermediate is ready for entry of DNA into the clamp. DNA binding commits RFC to ATP hydrolysis, which is followed by PCNA closure and PCNA•DNA release. This model enables quantitative understanding of the multi-step mechanism of a eukaryotic clamp loader, and furthermore facilitates comparative analysis of loaders from diverse organisms.
Keywords: Processive DNA replication, RFC clamp loader, PCNA clamp, ATPase kinetics
INTRODUCTION
Circular clamps are ubiquitous proteins that encircle duplex DNA, forming mobile tethers for polymerases and thereby enabling processive DNA synthesis for efficient genome replication 1,2. Clamps also influence the function of proteins involved in other critical DNA metabolic processes, including repair and recombination, as well as cell cycle regulation 3. Clamps are formed by association of two or more identical or homologous subunits into strikingly similar ring-shaped oligomers, with large enough diameters to encompass duplex DNA. They are loaded onto DNA by multi-protein complexes known as clamp loaders, in a reaction driven by ATP binding and hydrolysis 4,5. In eukaryotes, including humans, the PCNA clamp (Proliferating Cell Nuclear Antigen) is a trimer of identical subunits arranged in head-to-tail fashion 6,7. The RFC clamp loader (Replication Factor C) is a pentamer of homologous subunits arranged in a claw-like structure that binds both PCNA and primed-template DNA (ptDNA) and catalyzes formation of a topological link between the two 8,9. RFC and other clamp loaders belong to the AAA+ family of proteins, whose members possess conserved ATP binding/hydrolysis motifs within structural contexts that enable utilization of ATP for mechanical work—such as loading clamps on DNA 10,11.
Studies of clamps and clamp loaders from diverse organisms, including T4 bacteriophage gp45 clamp and gp44/62 loader 12,13, E. coli β clamp and γ complex loader 4,5, as well as P. furiosus 14, A. fulgidus 15,16, S. cerevisiae 8,17, and human 18,19 PCNA clamps and RFC loaders, have identified distinct steps in the clamp loading reaction. These include, at minimum, the clamp loader (a) binding the clamp (as an open ring, closed ring, or perhaps in disassembled/partially assembled ring form), (b) binding DNA such that it is positioned in the center of the clamp, and (c) releasing the topologically linked clamp•ptDNA product (the order of early steps in the reaction may vary). These dynamic interactions between proteins, and proteins and DNA, are driven by ATP binding, hydrolysis and product release steps of the ATPase cycle. Clamp loader proteins from the model systems noted above have the same overall structure and catalyze the same overall reaction; however, there appear to be intriguing differences in their reaction mechanisms. For example detailed kinetic analysis of E. coli γ complex (γ3δδ′χψ) supports a mechanism in which the clamp loader, which has three ATPase sites, binds β clamp with high affinity in the presence of ATP (ATP hydrolysis is not necessary for clamp opening), and then ptDNA binding leads to hydrolysis of three ATP molecules and release of β•ptDNA 5,20. In the case of bacteriophage T4 gp44/62 clamp loader, which has four ATPase sites, multiple mechanisms have been proposed, differing both in the stoichiometry of ATP and the manner in which it is utilized 13,21,22. Studies to resolve these mechanisms continue, and the possibility that gp44/62 can catalyze gp45 loading via alternate pathways has also been proposed 21. In the case of A. fulgidus RFC clamp loader, which has five ATPase sites, four ATP molecules are bound in the presence of PCNA, and according to the proposed mechanism three ATP are hydrolyzed for PCNA•ptDNA release and a fourth is hydrolyzed for catalytic turnover 16.
The S. cerevisiae RFC, which is related closely to human RFC, comprises five subunits: RFC-A (Rfc1), RFC-B (Rfc4), RFC-C (Rfc3), RFC-D (Rfc2), and RFC-E (Rfc5). Four of these subunits, A – D, have complete Walker A and B motifs, and conserved SRC or ‘arginine finger’ motifs contributed by neighboring subunits, that create ATP hydrolysis-active sites (Figure 6). RFC-E has disrupted Walker motifs and lacks input from an SRC motif, and is thus not considered to be ATPase active 9, although it may bind ATP 8. A few years ago, data from steady state analysis of S. cerevisiae RFC activities were used to propose a model in which the clamp loader binds two ATP, followed by binding of PCNA clamp and one more ATP, which leads to binding of DNA and an additional ATP and, finally, hydrolysis of an unknown number of ATP molecules to release PCNA•ptDNA 9,23. A more recent steady state analysis of RFC clamp loaders containing mutated ATPase sites led to the proposal that hydrolysis of one ATP molecule is associated with PCNA closure and hydrolysis of the rest leads to release of PCNA•ptDNA complex 24.
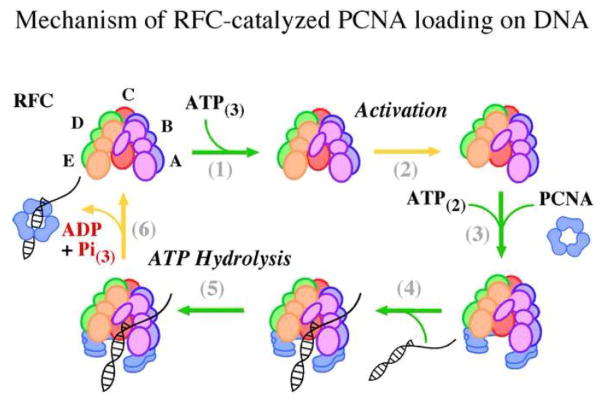
Figure 6.
Mechanism of RFC-catalyzed PCNA loading on ptDNA. Schematic depicting key steps in the clamp loading reaction determined by this study (proposed ATP stoichiometry is shown in subscript), (1) ATP binding to RFC initiates (2) slow activation of the clamp loader, (3) PCNA induces further ATP binding, fixes RFC in an active conformation and is opened in the process, (4) ptDNA binding acts as a switch to induce (5) rapid ATP hydrolysis, which allows (6) PCNA closure around ptDNA and release of the reaction products.
Thus far, kinetic analysis at a level of detail comparable to the prokaryotic systems has not been reported for a eukaryotic clamp loader. The order of events in the clamp loading reaction, the nature of the changing conformations and interactions, and the manner in which they are driven by ATP binding and hydrolysis catalyzed by the clamp loader subunits remains in question. We measured the ATPase, DNA binding, and PCNA opening/closing activities of S. cerevisiae RFC under pre-steady state conditions to observe progression of the first clamp loading cycle and thereby gain insights into the reaction mechanism. The data revealed key events, including ATP-, PCNA-, and DNA-mediated changes in RFC conformation, that occur in particular order as the reaction advances. Based on information from the current and previous studies, we designed a computational model that captures our understanding of the RFC mechanism, and used global data analysis to determine the parameters of the model and show that it is consistent with the observed ATPase kinetics. The RFC model sets the stage for greater understanding of the mechanism of action of eukaryotic clamp loader proteins. It also facilitates detailed comparisons of the clamp loading mechanism among diverse organisms, enabling discovery of common principles that characterize evolution of this essential reaction in DNA metabolism.
RESULTS
PCNA and DNA enhance ATP binding and hydrolysis by RFC
RFC was purified from E. coli cells and shown to catalyze PCNA loading on primed-template DNA 25,26. To determine how many RFC subunits bind ATP and how PCNA and primed-template DNA (ptDNA) modulate RFC interactions with ATP, nitrocellulose membrane filtration experiments were performed using ATPγS (adenosine 5′-3-O-(thio)-triphosphate), which is not hydrolyzed by RFC. ATPγS binding saturates at 3 molecules per RFC (alone), and when PCNA or ptDNA (40/65 nt primer-template) is present in the reaction, ATPγS binding saturates at 5 molecules per RFC (Figure 1A). In addition, PCNA increases the affinity of RFC for ATPγS, and the tightest binding is detected in the presence of both PCNA and ptDNA (Figure 1A; KD = 0.3 μM). These data indicate that interaction with either PCNA or DNA promotes changes in the conformational state of RFC such that ATP can occupy all five subunits with high affinity.
Figure 1.
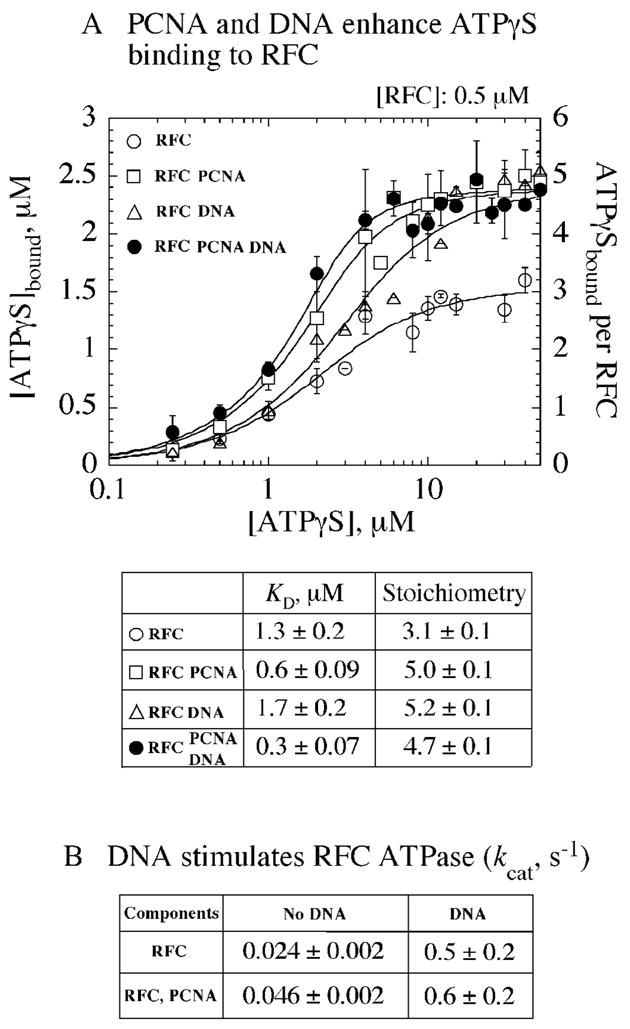
Interaction of RFC with PCNA, ptDNA stimulates ATP binding and hydrolysis. (A) RFC (0.5 μM) alone (○) binds 3 ATPγS, whereas in the presence of PCNA (□) or ptDNA (△), or both (●), it binds 5 ATPγS molecules; PCNA also helps increase the binding affinity of RFC for ATPγS. (B) Steady state ATPase assays show that ptDNA increases RFC kcat by ~ 20-fold, while PCNA increases it by only ~ 2-fold (calculated as Vmax/(4*[RFC]), assuming 4 ATPase active sites in RFC).
Complementary to the ATPγS binding data, steady state ATPase experiments following the conversion of [α32P]ATP to [α32P]ADP reveal that ptDNA and PCNA also stimulate RFC ATPase activity (Figure 1B). While PCNA alone has a small but reproducible effect, increasing the kcat by 2-fold from about 0.024 s−1 to 0.046 s−1, ptDNA has a more striking effect, increasing kcat by 20-fold to 0.5 s−1. Further addition of PCNA does not change the kcat ignificantly (kcat was calculated as Vmax/4*[RFC], assuming 4 ATPase active sites in RFC). These data suggest that PCNA has greater influence on the ATP binding phase, and ptDNA has greater influence on the ATP hydrolysis phase of the RFC-catalyzed reaction. The steady state ATPase experiments, however, do not reveal individual steps; therefore, we examined the reaction under pre-steady state conditions to observe the transactions RFC, ATP, PCNA, and ptDNA within the first catalytic turnover.
DNA triggers ATP hydrolysis by RFC
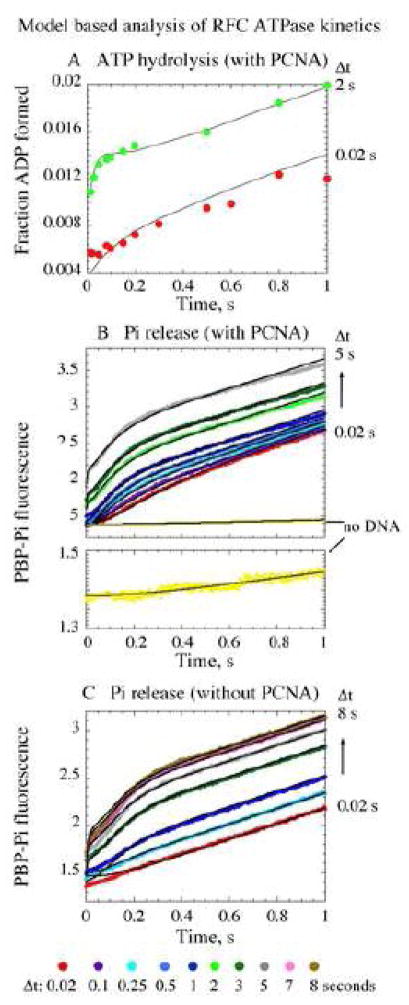
The pre-steady state kinetics of ATP hydrolysis were measured initially by following the production of phosphate (Pi) in real time. Pi release from RFC was detected by monitoring the increase in MDCC-PBP reporter fluorescence on binding free Pi 27 (Phosphate Binding Protein labeled with 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin); MDCC-PBP binds Pi rapidly and with high affinity under our experimental conditions (Table I, kPBP-Pi_On = 1.3 ± 0.1 × 107 M−1 s−1). Systematic series of experiments were performed to determine the specific effects of PCNA and ptDNA on the ATPase activity of RFC. First, two-syringe, single-mixing experiments were performed in which RFC from one syringe of the stopped-flow instrument was mixed rapidly with ATP from the other syringe, and the reaction observed for up to 1 second (Figure 2). PCNA was added to the reaction along with either RFC or ATP, and ptDNA was added along with ATP; mixing ptDNA with RFC in the absence of nucleotides causes protein aggregation, therefore this reaction configuration could not be tested.
Table I.
Best Fit Parameters for RFC Mechanism (±PCNA)
| Parameters | PCNA | No PCNA | ||
|---|---|---|---|---|
| kR-ATP_On | ATP binding | (μM−1 s−1) | 100 | 100 |
| kR-ATP_Off | ATP release | (s−1) | 100 | 100 |
| kR_Activation | RFC activation | (s−1) | 4.64 ± 0.02 | 1.55 ± 0.01 |
| kR_ATPase | RFC ATPase | (s−1) | 0.046 ± 0.002 | 0.017 ± 0.0004 |
| kR(A)_ATPase | RFC(A) ATPase | (s−1) | 0.072 ± 0.0006 | 0.12 ± 0.001 |
| kR(A)-DNA_On | RFC(A)-DNA binding | (μM−1 s−1) | 50 | 50 |
| kR(A)-DNA_Off | RFC(A)-DNA release | (s−1) | 0.5 | 0.5 |
| kR(A)DNA_ATPase | RFC(A)-DNA ATPase | (s−1) | 53.9 ± 0.04 | 11.1 ± 0.1 |
| kR-Pi_Off | Pi release | (s−1) | 6.41 ± 0.003 | 13.53 ± 0.01 |
| kR-DNA_Off | DNA(PCNA) release | (s−1) | 1.7 ± 0.003 | 25.63 ± 2.7 |
| NATP | ATPase sites | 3.11 ± 0.001 | 2.12 ± 0.0004 | |
| kPBP-Pi_On | PBP-Pi binding | (μM−1 s−1) | 14 | 12 |
| kPBP-Pi_Off | PBP-Pi release | (s−1) | 1.2 | 1.2 |
Parameter values shown without confidence intervals were fixed during global data fitting.
Figure 2.
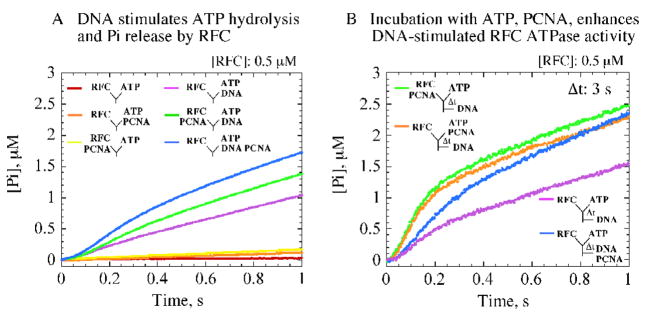
Pre-incubation of RFC with ATP, PCNA enables ptDNA-dependent rapid ATP hydrolysis. (A) Single-mixing stopped-flow experiments measuring pre-steady state Pi release kinetics show that ptDNA is required for rapid ATP hydrolysis and Pi release by RFC; highest activity is detected with PCNA plus ptDNA (blue trace).(B) Sequential-mixing stopped-flow experiments show that pre-incubation of RFC with ATP (Δt = 3 s shown) enables ptDNA to trigger a burst of ATP hydrolysis and Pi release (purple trace). The burst kinetics are maximal when PCNA is included during pre-incubation (orange and green traces versus blue trace). Final reagent concentrations: 0.5 μM RFC, 500 μM ATP, 2.5 μM PCNA, 2.5 μM ptDNA, 10 μM MDCC-PBP.
Pi is released at a rate of 0.025 s−1 by RFC alone and 0.043 s−1 by RFC with PCNA (Figure 2A, red and orange traces, respectively; Vmax/4x[RFC]). These Pi release rates are similar to the kcat values determined earlier under steady state conditions (Figure 1B), and direct measurement of [α32P]ATP hydrolysis confirms this finding (Figure 3A). In the presence of ptDNA, the Pi release rate increases (Figure 2A, purple trace), and a linear fit yields a rate of 0.5 s−1, indicating that RFC ATPase activity is stimulated by ptDNA. This rate is similar to the kcat (Figure 1B), suggesting that a step before ATP hydrolysis and Pi release limits the reaction under these conditions. In the presence of both PCNA and ptDNA, however, the kinetics of Pi release display three phases. A burst of Pi is detectable before the linear steady state phase (Figure 2A, blue and green traces), indicating that RFC hydrolyzes ATP rapidly under these conditions and that a subsequent slow step limits the catalytic turnover. A lag phase (~ 100 ms) precedes the burst, indicating a slow step occurs before ATP hydrolysis as well. These pre-steady state data exposed changes in RFC activity induced by ptDNA and PCNA at particular stages in the ATPase reaction, and we probed them further in our exploration of the clamp loading mechanism.
Figure 3.
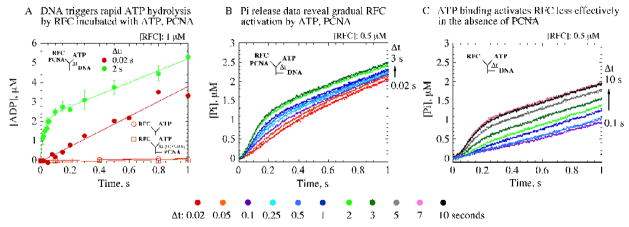
Activation of RFC on binding ATP and PCNA. (A) Sequential-mixing rapid-quench experiments, measuring [α-32P]ADP production, show that ptDNA triggers ATP hydrolysis; compare reactions with (● ) and without ptDNA (○ no PCNA, □ with PCNA). The data also verify slow step(s) during RFC, PCNA, ATP pre-incubation (Δt), before ptDNA-induced ATP hydrolysis; compare Δt = 2 s (green ●; kBurst = 35 ± 3 s−1) versus 0.02 s (red ●); final concentrations: 1 μM RFC, 500 μM ATP, 2.5 μM PCNA, 2.5 μM ptDNA. (B, C) Sequential-mixing stopped-flow experiments show gradual increase in ATP hydrolysis and Pi release with Δt (0.02 – 10 s), and maximal activity is detectable at Δt = 2 s with PCNA (B, light green trace) and at Δt = 7 s without PCNA (C, pink trace); final concentrations: same as above except for 0.5 μM RFC, 10 μM MDCC-PBP. kcat is 0.6 – 0.8 s−1 in all cases in the presence of ptDNA.
First, we sought to discover rate-limiting steps that govern the early stages of the reaction. As noted above, the initial lag phase (Figure 2A, blue and green traces) implied at least one slow event in the reaction before ATP hydrolysis. In order to parse this phase, we performed three-syringe, sequential-mixing experiments by varying the mixing order of reactants. Among all the conditions tested, the highest activity was detected on pre-incubation of RFC with ATP for a short period (Δt) prior to addition of ptDNA and observation of the reaction up to 1 second. As shown in Figure 2B for Δt = 3 s, a rapid burst of Pi release is observed after ptDNA is added to the reaction (note: RFC hydrolyzes only a small, sub-stoichiometric fraction of ATP during the pre-incubation period in the absence of ptDNA; Figure 2A). The rate and amplitude of the burst phase are maximal when PCNA is added with either RFC or ATP (green and orange traces), compared with reactions in which PCNA is added with ptDNA (e.g., blue trace) or is absent (purple trace). These results suggest an important role for the PCNA clamp during the RFC, ATP pre-incubation period.
Comparison of data from single-mixing and sequential-mixing experiments clearly reveals that the slow step early in the reaction is associated with ATP binding to RFC. For example, the green traces in Figures 2A and 2B are from reactions in which RFC and PCNA were mixed with ATP and ptDNA. In the former case, there is a long lag phase (~ 100 ms) and minimal burst of ATP hydrolysis, whereas in the latter case, prior incubation of RFC and PCNA with ATP results in a significantly smaller lag phase (~ 20 ms) and greater burst of ATP hydrolysis on addition of ptDNA. Thus, completion of the slow step during the pre-incubation period exposes subsequent fast ATP hydrolysis and Pi release triggered by ptDNA.
ATP binding-induced activation of RFC is enhanced by PCNA
The preliminary analysis above suggested that ATP induces, and PCNA enhances, changes in RFC that must occur before DNA can induce rapid ATP hydrolysis. This hypothesis predicts that the extent of ATP hydrolysis is dependent on the rate of these prior events in the reaction. In order to address the hypothesis, a series of sequential-mixing experiments was performed in which RFC was pre-incubated with ATP and PCNA for varying periods before addition of ptDNA to the reaction (similar data were obtained whether PCNA was added to the reaction along with RFC or with ATP). First, direct measurement of [α32P]ADP production confirmed that a larger burst of ATP hydrolysis correlates with longer RFC, ATP, PCNA pre-incubation time (Figure 3A; Δt = 0.02s and 2s; Δt 2s data fit to a burst equation yield 35 s−1 hydrolysis rate); in the absence of ptDNA, the ATPase activity remains minimal even at the longest Δt tested (Figure 3A). A more comprehensive analysis of the RFC, ATP, PCNA pre-incubation period using Pi release assays shows that the burst phase increases gradually with Δt and concomitantly the lag phase becomes shorter (Figure 3B; Δt = 0.02 – 3 s). In the absence of PCNA, a similar trend is observed; however, longer Δt is required to achieve the maximal burst, which nevertheless remains lower than that in the presence of PCNA (Figure 3C; Δt = 0.02 – 10 s).
The gradual increase in RFC ATPase activity with increasing Δt is not due to slow ATP binding, as these reactions contain 750 μM ATP during the pre-incubation period, and the kinetics are independent of ATP concentrations over 200 μM. Another possibility is that ATP binding is fast, but the slow step is an ATP-induced change in RFC conformation that ‘activates’ the clamp loader for subsequent steps in the reaction. Data from reactions both with and without PCNA support this hypothesis, since prolonged pre-incubation with ATP enables ptDNA-dependent ATP hydrolysis by RFC in both cases. As noted above, maximal ATPase activity is higher and detectable at significantly shorter Δt in the presence of PCNA (Figure 3B; ~ 2 s), relative to that in the absence of PCNA (Figure 3C; ~ 7 s). Thus, ATP binding to RFC alone appears sufficient to activate the clamp loader, at least partially, for ATP hydrolysis on addition of ptDNA; however, inclusion of PCNA in the complex results in faster, more effective activation of RFC, which is likely an important feature of the ATPase-coupled clamp loading mechanism.
Global analysis of RFC-catalyzed ATPase kinetics
The observed ATPase kinetics suggest that ATP-bound RFC has to undergo a slow ‘activation’ step, enhanced by PCNA, before it can bind DNA and hydrolyze ATP efficiently. Details of the reaction mechanism, such as the rate of the activation step and the number of ATP molecules hydrolyzed by RFC in a single catalytic turnover can be extracted by model based global analysis of all the data. Furthermore, such quantitative analysis can generate specific hypotheses that can be experimentally tested and verified. To this end, we developed a computational model of the RFC-catalyzed reaction and globally fit the available data with it (Scheme I, Figure 4). RFC has four ATPase sites that could bind and hydrolyze ATP. Enumerating all possible ATP-bound/hydrolyzed RFC states produced a model with many unknown rate constants, which could not be estimated with confidence based on available experimental data. We therefore modeled RFC with a single ATPase site with floating stoichiometry (Scheme I). The mechanism was further simplified by excluding explicit RFC-PCNA interactions, assuming that RFC•ATP•PCNA complex is saturated and that, upon hydrolysis, PCNA dissociates from RFC simultaneously with ptDNA (these transactions are being measured separately). Raw data from 31 separate [α32P]ATP hydrolysis and Pi release experiments performed with PCNA (including datasets shown in Figure 3A and 3B) were fit simultaneously to the kinetic model using gfit, a program designed for global analysis of data from different types of experiments (details of model development and analysis are in Supplementary Material) 28. The fits for a representative dataset each from [α32P]ATP hydrolysis and Pi release experiments performed with PCNA are presented in Figure 4A and 4B, respectively (as black lines overlaying the corresponding experimental data). The best fit kinetic parameters obtained from the analysis are listed in Table I. Raw data from 18 Pi release experiments performed without PCNA (including the dataset in Figure 3C), were fit similarly and are presented in Figure 4C, and the best fit kinetic parameters are listed in Table I.
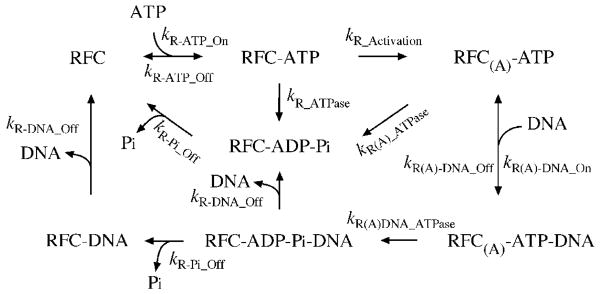
Scheme I.
Minimal kinetic mechanism of RFC
Figure 4.
Global analysis of ATPase kinetics supports a model for RFC-catalyzed PCNA loading on ptDNA. Raw data from sequential-mixing experiments measuring [α-32P]ATP hydrolysis (A, with PCNA) and Pi release (B, with PCNA; C, without PCNA) were fit using gfit to a kinetic model for RFC-catalyzed ATP binding and hydrolysis coupled with DNA binding and release (± PCNA), shown in Scheme I. The experimental traces are shown in colors corresponding to Δt. The black lines represent simulations by the model using the parameters listed in Table I.
The reaction starts with RFC binding ATP to form RFC•ATP complex (Scheme I) at a fast bimolecular rate constant of 100 μM−1 s−1 (kR-ATP_On), with a dissociation rate of 100 s−1 (kR-ATP_Off). These rates were set to be consistent with the affinity of RFC for ATPγS (Figure 1; KD ~ 1 μM), and with the fact that the ATPase rates are independent of ATP concentration in our experiments, indicating rapid binding. ATP binding activates RFC at a relatively slow rate of 4.6 s−1 in the presence of PCNA, and an even slower rate of 1.5 s−1 in the absence of PCNA (Table I, kR_Activation). The activated RFC binds ptDNA at fast bimolecular rate constant of 50 μM−1 s−1 (kR(A)-DNA_On), with a dissociation rate of 0.5 s−1 (kR(A)-DNA_Off). These rates are estimated from data shown in Figure 5, and reflect the high affinity of RFC for ptDNA (KD~ 10 nM) 26. In the absence of ptDNA the ATPase rates of all RFC species in the reaction are slow (Figure 4B lower panel; Table I, kR_ATPase = 0.02 – 0.05 s−1, kR(A)_ATPase = 0.07 – 0.1 s−1). In contrast, ptDNA binding to RFC triggers rapid ATP hydrolysis, and the fit yields a rate constant of 54 s−1 in the presence of PCNA (Table I, kR(A)DNA_ATPase, similar to the measured constant of 35 s−1 in Figure 3A). In the absence of PCNA, the ptDNA-dependent ATP hydrolysis rate is far slower at 11 s−1. The last steps include dissociation of ptDNA, PCNA, and the ATP hydrolysis products, ADP and Pi. The experimental data are not sufficient to resolve the preferred order of dissociation of RFC•ADP•Pi•ptDNA(±PCNA) complex; therefore, we assumed that either ADP and Pi (kR-Pi_Off) or ptDNA and PCNA (kR-DNA_Off) leave the complex first. The assumption that ptDNA and PCNA are released simultaneously from RFC is being tested by direct kinetic measurements of these interactions, and similarly for ADP and Pi, by direct measurements of ADP dissociation. The fits yield only slightly different rate constants for Pi release in the presence or absence of PCNA (kR-Pi_Off= 6.4 s−1 and 13.5 s−1, respectively). In contrast, release of PCNA•ptDNA is very slow compared with release of ptDNA alone (kR-DNA_Off = 1.7 s−1 and 25.6 s−1, respectively). Also shown in Table I are the measured rate constants for Pi binding by reporter protein MDCC-PBP, which were used in data fitting (note: trace amounts of Pi formed during pre-incubation are bound rapidly by MDCC-PBP, and detected as a small initial burst within the first ~ 5 milliseconds of the reaction, especially at longer Δt; Figure 4).
Figure 5.
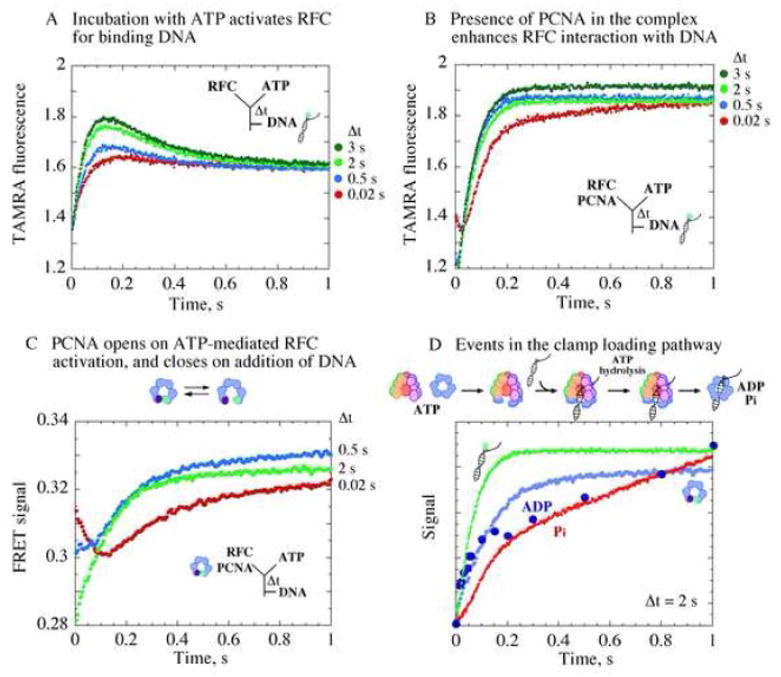
Formation of an active RFC•ATP•PCNA(open) complex that binds ptDNA for clamp loading. (A) Changes in fluorescence intensity of TAMRA-labeled ptDNA on binding RFC show increasing interaction correlated with increasing RFC, ATP pre-incubation time (Δt). (B) Presence of PCNA during pre-incubation enhances the interaction. (C) Kinetics of PCNA opening/closing measured by FRET between Tryptophan and AEDANS at PCNA(WCAEDANS) clamp interfaces. At shortΔt (0.02 s, red trace), PCNA opening is detected at first after ptDNA addition (FRET decrease) followed by PCNA closing (FRET increase). At longer Δt, PCNA is completely opened already (2 s, green trace), and only its closure is detected after ptDNA addition (kPCNA_Close ~ 5 s−1). (D) Overlay of data from PCNA opening/closing, DNA binding/release, and ATPase kinetics shows rapid ptDNA binding to RFC•ATP•PCNA(open) complex formed during pre-incubation (Δt = 2 s), triggering ATP hydrolysis, followed by PCNA clamp closure, release of Pi and other products, and catalytic turnover.
An important parameter assessed by global fitting of the data is the number of ATP molecules hydrolyzed by RFC in a single catalytic turnover. ATP hydrolysis occurs in one step in the model and, as noted above, we introduced a variable to account for the stoichiometry of [α32P]ATP or Pi generated by RFC per turnover (NATP). In this case also, the fits indicate significant differences in the presence or absence of PCNA, yielding a stoichiometry of 3.1 ATP per RFC for the reaction with PCNA and 2.1 ATP per RFC without PCNA (Table I). These values may reflect the actual number of ATP molecules hydrolyzed by RFC in the rapid burst phase, or varying fractions of ‘active’ RFC in the reaction, as will be considered further in Discussion. All parameters that vary markedly with the status of PCNA in the reaction are noted in bold text in Table I. All these parameters have generated specific hypotheses about the reaction mechanism that we have begun to address, as demonstrated below.
Key events in the clamp loading pathway
One interesting hypothesis is that ATP binding to RFC activates the clamp loader for rapid interaction with ptDNA, which in turn triggers rapid ATP hydrolysis. We tested this hypothesis by directly measuring RFC binding to DNA, as reported by increase in fluorescence of 40/65 nt ptDNA labeled with 5-(6)-carboxytetramethylrhodamine at the 3′ primer end (TAMRA). As predicted by the model, the rate and amplitude of DNA binding increases with longer RFC and ATP pre-incubation time (Figure 5A, 0 – 0.15 s interval). Addition of PCNA during pre-incubation results in greater increase in TAMRA fluorescence, and the ptDNA dissociation phase observed in the absence of PCNA (Figure 5A, 0.15 – 0.6 s interval) becomes undetectable (Figure 5B). These differences in DNA binding kinetics may arise because PCNA facilitates RFC activation and thus shifts the equilibrium in favor of bound complex, or RFC•ATP•PCNA•ptDNA complex dissociates more slowly than RFC•ATP•ptDNA, or because PCNA remains at the ptDNA junction and affects TAMRA fluorescence even after dissociation from RFC 29 (it is also possible that TAMRA fluorescence differs when PCNA is in complex with RFC and ptDNA). Slower dissociation of RFC•ATP•PCNA•ptDNA (Figure 5B) compared with RFC•ATP•ptDNA (Figure 5A) is consistent with the rate constants for PCNA•ptDNA versus ptDNA release from RFC obtained from global fitting of the ATPase data (Table I, kR-DNA_Off), and its implication for the reaction mechanism will be considered further in Discussion.
The data above confirm that RFC activation by ATP is influenced by PCNA. We next investigated what happens to PCNA during the activation step. We measured PCNA opening/closing kinetics using a PCNA construct with a FRET pair across the subunit interfaces (Tryptophan and 5-[2(acetyl)aminoethyl-]aminonaphthalene-1-sulfonate (AEDANS)-labeled Cysteine) 17. Initial single mixing experiments performed by mixing RFC and PCNA(WCAEDANS) with ATP showed that the clamp opens at a rate of ~ 2 s−1 (FRET decrease) prior to ATP hydrolysis (Supplementary Material Figure S1). In sequential mixing experiments, incubation of RFC, ATP, and PCNA(WCAEDANS) for a short time (Δt = 0.02 s) before addition of DNA revealed a fast PCNA(WCAEDANS) opening phase (Figure 5C, FRET decrease) followed by a closing phase (FRET increase). As Δt was increased (0.5 s, 2 s), we noted that more PCNA(WCAEDANS) was opened during the pre-incubation period, and we detected primarily its closure during the reaction (a single exponential fit of the green Δt 2 s trace yields kPCNA_Close ~ 5 s−1). An overlay of the DNA binding, ATP hydrolysis, Pi release, PCNA opening/closing kinetics following Δt of 2 s (Figure 5D), provides visual confirmation that following ATP binding, ATP-induced activation of RFC, coincident with PCNA opening, leads to an activated RFC•ATP•PCNA(open) complex that binds ptDNA and hydrolyzes ATP rapidly, resulting in PCNA closure, Pi release and catalytic turnover.
DISCUSSION
Clamp loaders perform the essential task of loading circular clamps onto DNA where they are utilized in DNA replication and repair, as well as several other cellular processes. This task requires the clamp loader to direct transient conformational changes and interactions among the reaction components towards formation of a topologically linked clamp•DNA product. For example, early in the reaction the clamp loader, clamp, and DNA must develop high affinity for each other, adopting conformations that enable positioning of DNA within the clamp, and later their interactions must change to allow the release of clamp•DNA. ATP binding and hydrolysis catalyzed by the clamp loader promote and coordinate events in this intricate reaction mechanism. Past investigation of the eukaryotic S. cerevisiae RFC clamp loader has revealed that it binds both PCNA clamp and primed-template DNA in the presence of non-hydrolyzable ATPγS 23,24,26. ATPγS-bound RFC is also capable of opening PCNA, possibly enough to allow entry of duplex DNA into the center of the ring 17,24. In this study, we aimed to elucidate the mechanism of RFC-catalyzed PCNA loading by measuring the kinetics of several transactions, including ATP binding/hydrolysis, DNA binding, PCNA opening/closing and product release that occur during the reaction.
The ATPase reaction promotes two distinct phases, one that involves assembly of an RFC•ATP•PCNA•ptDNA complex, and another that involves disassembly of the complex. Our kinetic data show directly that complex assembly is coupled to ATP binding and disassembly is coupled to ATP hydrolysis (Figure 6). RFC has five nucleotide binding sites, three of which are occupied by ATPγS in the absence of PCNA and ptDNA substrates, while the remaining two are occupied when RFC binds PCNA or ptDNA or both (Figure 1; schematic depiction in Figure 6). A previous report suggested very different stoichiometry, with only four ATPγS molecules binding per RFC—two to free RFC, one more to RFC•ATP•PCNA, and fourth one to RFC•ATP•PCNA•ptDNA complex 23. However, the clamp loader examined in that study contained an N-terminal deletion of 269 amino acids from the RFC-A (Rfc1) subunit. Our analysis of a similar N-terminal truncated clamp loader, missing 282 amino acids from RFC-A, also indicates that this mutant (ΔN-RFC) binds fewer ATPγS molecules than full length RFC (data not shown) and, significantly, the pre-steady state burst ATPase activity catalyzed by ΔN-RFC is also lower, indicating that the activity of this mutant deviates from that of the wild type clamp loader (Supplementary Material, Figure S2). The crystal structure of RFC•ATPγS•PCNA complex shows nucleotides bound at all five sites 8.
Pre-steady state ATPase data reveal important aspects of the assembly phase of the reaction. First, comparison of the ATPase kinetics in the absence and presence of primed-template DNA shows an absolute requirement for ptDNA binding in order for RFC to catalyze rapid ATP hydrolysis (Figures 2, 3A, 4B). Thus, ATP-bound RFC remains in a practically ‘ATPase-inactive’ conformation until ptDNA swiftly switches it to a conformation that is competent for hydrolysis. This DNA-dependent switch occurs whether or not RFC is in complex with PCNA; however, the ATPase burst is substantially larger in the presence of PCNA than in its absence (Figure 3B versus Figure 3C). Specifically, global analysis of the data suggests that RFC with PCNA hydrolyzes more ATP and at a faster rate than without PCNA (Table I, NATP = 3 versus 2 and kR(A)DNA_ATPase = 54 s−1 versus 11 s−1). These differences in ATPase stoichiometry and kinetics could reflect distinct properties of the RFC•ATP•PCNA and RFC•ATP complexes. They could also arise from a larger fraction of RFC•ATP•PCNA achieving ‘ATPase-active’ conformation compared with RFC•ATP following ptDNA binding, or from other steps in the reactions not measured explicitly yet (e.g., differential changes in RFC conformation in these two complexes). Alternately, these complexes may hydrolyze additional ATP molecules at relatively slow rates that are not detected in the ATPase burst phase. In any case, the data reveal a step that blocks RFC ATPase activity in the first phase of the reaction, presumably to allow assembly of RFC, ATP and PCNA into a complex with minimal futile ATP hydrolysis, until ptDNA binding triggers the second, disassembly phase of the reaction.
We also discovered a slow RFC activation step that occurs after ATP binding (Figure 6). This step occurs at a faster rate of 4.6 s−1 in the presence of PCNA compared with 1.5 s−1 in the absence of PCNA (Scheme I, Table I; kR_Activation). During the activation period RFC binds and opens PCNA, generating an RFC•ATP•PCNA(open) intermediate complex that can allow entry of ptDNA into the clamp (Figure 5C). ATP binding-induced RFC activation appears absolutely necessary for its interaction with ptDNA leading to rapid ATP hydrolysis (Figures 3 and 5). Crystal structures of S. cerevisiae RFC and other clamp loaders suggest that ATP binding drives the clamp loader subunits into a spiral arrangement such that their DNA binding sites can track the duplex portion of ptDNA when it enters the complex 4,8,15. PCNA docks under RFC and is predicted to open in a spiral conformation complementary to that adopted by the clamp loader subunits, which could explain how it stimulates activation of RFC 14,17,30. That RFC activation occurs at a faster rate when the clamp loader is in complex with PCNA increases the likelihood that during DNA replication in vivo, with high concentrations of PCNA and limited primed-template sites, ptDNA will bind a pre-assembled RFC•ATP•PCNA(open) complex rather than RFC•ATP complex, resulting in a productive PCNA loading reaction cycle.
Once the ptDNA is bound, RFC conformation changes to enable ATP hydrolysis and initiate the second, disassembly phase of the reaction. In our minimal RFC mechanism, we sorted dissociation of the four reaction products, ADP, Pi, PCNA, and ptDNA into two distinct steps, assuming that either ADP and Pi or ptDNA and PCNA leave the complex first (Scheme I). Global analysis of the ATPase data indicates that dissociation of PCNA•ptDNA from RFC occurs at a rate of 1.7 s−1, which is slow compared to dissociation of ptDNA alone at 25 s−1 (Scheme I, Table I; kR-DNA_Off). Dissociation rates for each of these products have to be measured and incorporated into the model before we can determine explicitly which step(s) controls the catalytic turnover rate of RFC. Based on the parameters obtained from ATPase data fitting, we can estimate a steady state velocity of 3.5 μM s−1 for the reaction containing PCNA and ptDNA (kcat = 1.2 s−1 for 3 sites or 0.9 s−1 for 4 ATPase sites per RFC). There are several slow steps following rapid ATP hydrolysis (54 s−1), including PCNA closing (~ 5 s−1, Figure 5C), Pi release (6.4 s−1), PCNA•ptDNA release (1.7 s−1), and the next round of RFC activation (4.6 s−1), which likely limit the steady state clamp loading rate (Figure 6).
Our results indicate that RFC can load and release PCNA onto a primed-template site on DNA at a rate of about 1 s−1, which is compatible with the estimated rate of Okazaki fragment synthesis in vivo (0.5 – 1 s−1). Thus, PCNA clamps can be loaded in time for DNA polymerase to cycle efficiently from completing an Okazaki fragment to beginning processive synthesis of a new one. It is also noteworthy that in the absence of PCNA, RFC activation limits the start of the reaction, and ptDNA bound to RFC under these conditions undergoes rapid dissociation at the end of the reaction. These kinetics reflect a preferred order of events in the clamp loading reaction, with RFC slow to bind ptDNA without PCNA; however, if it does happen to bind ptDNA ahead of PCNA, ATP hydrolysis and ptDNA release quickly reset the clamp loader to the beginning of the cycle.
This study reveals some common features between the reaction mechanisms of S. cerevisiae RFC and E. coli γ complex (γ3δδ′χψ) clamp loaders. The ATPase kinetics of γ complex also indicate a slow change in conformation following ATP binding and prior to ATP hydrolysis, which activates the clamp loader for interaction with DNA 5,20. The presence of β initially suppresses ATP hydrolysis by γ complex and then enhances ATP hydrolysis following addition of ptDNA, consistent with the proposition that early rate limiting steps in the ATPase reaction favor a productive clamp loading reaction. Binding of ptDNA to γ complex•ATP•β triggers rapid ATP hydrolysis at the 3 γ subunits and release of β•ptDNA to complete the reaction. Intriguingly, analysis of the RFC ATPase data based on the minimal mechanism in Scheme I also indicates that 3 ATP molecules are hydrolyzed rapidly by RFC in one turnover (Table I; NATP = 3.1). One can therefore consider the possibility that hydrolysis of 3 ATP may be sufficient for ATPase-coupled loading of clamps onto DNA. The E. coli γ complex contains only 3 ATPase-active γ subunits, with the homologous δ′ and δ subunits having lost their ability to bind and hydrolyze ATP 4. In the case of RFC, Walker ATP binding and hydrolysis motifs are disrupted in RFC-E subunit (Rfc5), whose position in RFC is analogous to that of δ′ in γcomplex, rendering it unable to hydrolyze ATP (although it is still capable of binding ATP; Figure 1) 8. The RFC-A subunit (Rfc1), whose position in RFC is analogous to that of δ in γcomplex, has an apparently functional ATPase site, but according to an in vivo study its ATPase activity does not appear to be essential for PCNA loading 31; the same study found that the ATPase activities of RFC-B, RFC-C and RFC-D subunits (Rfc4, Rfc3, Rfc2) are essential for PCNA loading.
It remains possible that a sub-stoichiometric fraction of RFC is active (or activated) for ATP hydrolysis in our experiments, despite being fully active for ATPγS binding. Alternately, hydrolysis of a fourth ATP by RFC may occur at a different rate, possibly associated with another step in the reaction. The latter scenario is analogous to that proposed for archaeal A. fulgidus RFC, which also has 4 ATPase-active subunits, and in this case the activity of 3 subunits appears necessary up to PCNA•DNA release and that of a fourth subunit affects the turnover rate 16. The bacteriophage T4 gp44/62 clamp loader also has 4 ATPase-active subunits and the reaction kinetics indicate similarities with γ complex and RFC mechanisms, such as the requirement of ATP hydrolysis for gp45 clamp and ptDNA release from gp44/62, but also some key differences 13,22. For example, gp44/62 is reported to rapidly hydrolyze 2 ATP molecules in the presence of gp45 alone, 1 ATP in the presence of ptDNA alone, and all 4 ATP molecules in the presence of both gp45 and ptDNA 21,22. It has been proposed that the T4 clamp loader utilizes alternate pathways for clamp assembly on DNA (perhaps the open gp45 clamp affords more mechanistic flexibility than the closed β and PCNA clamps), and that the stoichiometry and pattern of ATP hydrolysis varies with the pathway 12,21. In the case of RFC, there is no specific evidence for multiple clamp assembly pathways, but additional kinetic analysis, for example of RFC complexes containing individual ATPase-inactive subunits, can further resolve the question of how ATP is utilized in the clamp loading reaction.
In summary, our findings have provided direct evidence for an ordered series of ATP binding and hydrolysis-coupled events that drive clamp assembly on ptDNA by a eukaryotic clamp loader. Important questions remain regarding the mechanism of RFC and related eukaryotic clamp loaders, especially pertaining to the transactions of PCNA and ptDNA in the reaction and the nature of the conformational changes in RFC. Intriguing questions arise also from studies of more distant clamp loaders in the superfamily, such as the possible existence of multiple clamp assembly pathways. The discovery and quantitative description of key steps in the S. cerevisiae RFC-catalyzed reaction provide useful leads for focused inquiry into the workings of this essential DNA metabolic protein in multiple organisms.
MATERIALS AND METHODS
Proteins, DNAs, other reagents
RFC, ΔN-RFC (282 aa RFC-A N-terminal deletion) 25 and PCNA (His tag or no tag) 26,32 were purified from E. coli as described. PCNA(WCAEDANS) (F185W/K107C/C22S/C30S/C62S/C81S) clone was a gift from Dr. Stephen J. Benkovic (Pennsylvania State University, University Park, PA), and the protein was purified and labeled with N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine (1,5-IAEDANS) as described 17. E. coli PBP was purified and labeled with MDCC as described 27. DNAs were purchased from Integrated DNA Technologies: 40 nt primer, 5′-ATT TCC TTC AGC AGA TAG GAA CCA TAC TGA TTC ACA TGG C-3′ and 65 nt template, 5′-TAG TTA GAA CCT AAG CAT ATT AGT AGC CAT GTG AAT CAG TAT GGT TCC TAT CTG CTG AAG GAA AT-3′. Both strands were purified and annealed to form 40/65 nt ptDNA; 40 nt primer was also modified with 3′ amino linker and labeled with TAMRA as described 33. ATP and ATPγS were purchased from Sigma Aldrich, [α32P]ATP and [35S]ATPγS from Perkin Elmer Life Sciences, and 1,5-IAEDANS, MDCC, and TAMRA dyes from Invitrogen. Unless noted otherwise, all experiments were performed in buffer A (30 mM Hepes-NaOH, pH 7.5, 10 mM Mg(OAc)2 or MgCl2, 100 mM NaCl, 2 mM DTT) at 25 °C.
ATPγS binding, ATP hydrolysis, Pi release assays
ATPγS binding to RFC was measured by nitrocellulose membrane assays 34. Briefly, in 15 μl reactions, 0.5 μM RFC, ± 2 μM PCNA, ± 2 μM ptDNA, and 0 – 50 μM [35S]ATPγS were incubated for 15 min at 4 °C, and filtered through membranes. Molar amount of bound ATPγS was quantified (PhosphorImager; GE Healthcare), and the data fit to a quadratic equation. Steady state ATPase kinetics were measured by incubating 500 μM [α32P]ATP over time with 0.5 μM RFC, ± 1 μM PCNA, ± 1 μM ptDNA. Hydrolysis products were resolved by PEI-cellulose TLC (0.5 M formic acid, 0.5 M LiCl) and quantified. ATPase kcat was determined from the slope of the time course (slope/4x[RFC]). Pre-steady state ATPase kinetics were measured using a three-syringe quench-flow instrument (KinTek Corp, Austin TX). RFC was in syringe A; [α32P]ATP was in syringe B; PCNA was either in syringe A, B, or C; ptDNA was in syringe C. Contents of syringes A and B were mixed, pre-incubated for varying times (Δt = 0.02 – 2 s), mixed with contents of syringe C, and incubated for varying times again. The final concentrations were 1 μM RFC, 500 μM ATP, 2.5 μM PCNA, and 2.5 μM ptDNA. The reaction was quenched with 4M formic acid, the products were resolved by TLC, and the data were fit to the burst equation: D = A(1 − exp(−kBurstt)) + kSt, where A is burst amplitude, kBurst is the burst rate, and kS is the slope of the linear phase and yields kcat.
Pre-steady state measurements of Pi release were performed on stopped-flow instruments (KinTek Corp, Austin, TX and Applied Photophysics, Surrey, UK) in single- and double-mixing modes (Δt = 0.02 – 10 s). RFC was in syringe A; ATP was in syringe B; PCNA was either in syringe A, B, or C; ptDNA and MDCC-PBP were in syringe B or C. Final concentrations were 0.5 μM RFC, 500 μM ATP, 2.5 μM PCNA, 2.5 μM ptDNA, and 10 μM MDCC-PBP. Change in MDCC-PBP fluorescence on binding Pi was measured over time (λex = 425 nm, λem > 450 nm), as described previously 33. Specific increase in fluorescence from MDCC-PBP binding to Pi and binding rates were determined in separate calibration experiments. Five or more individual kinetic traces were averaged, and for Figures 2 and 3, Pi concentration was determined and plotted versus time, after subtracting [Pi] formed during Δt in the absence of DNA.
DNA binding kinetics
In stopped-flow experiments, RFC and ATP (± PCNA) were pre-incubated for various times (Δt = 0.02 – 10 s) in buffer A with 0.1 mg/ml BSA and mixed with TAMRA-labeled ptDNA to final concentrations of 0.4 μM RFC, 500 μM ATP, 2 μM PCNA, 0.04 μM TAMRA-ptDNA, and the fluorescence change measured over time (λex = 535 nm, λem > 550 nm). Five or more individual kinetic traces were averaged, and the signal was corrected for background fluorescence and plotted versus time.
PCNA opening/closing kinetics
In stopped-flow experiments, RFC, ATP, and PCNA-WCAEDANS and were pre-incubated for various times (Δt = 0.02 – 5 sec) and mixed with ptDNA to final concentrations 0.6 μM RFC, 500 μM ATP, 0.25 μM PCNA, 0.25 μM ptDNA. FRET changes due to PCNA opening/closing were measured over time (λex = 290 nm, λem > 450 nm). Five or more individual kinetic traces were averaged, and the signal plotted versus time.
Data analysis
Mathematical model
A simplified model of the RFC-catalyzed reaction, consisting of a system of ordinary differential equations (ODE) corresponding to the mass-action reactions shown in Scheme I, was prepared. Interactions between RFC and PCNA were not included in the model; therefore, measurements without PCNA or with saturating PCNA were analyzed separately.
Simulation of experimental results
The model was programmed in MATLAB (The MathWorks, Inc., Natic, MA). The reaction kinetics were simulated using rsys library. Manipulations involved in the experiments, such as pre-incubation of reagents, were replicated by the model.
Estimation of model parameters
To find a set of parameters that make the model consistent with observations, the results of a total of 49 stopped-flow and quench-flow experiments were fit globally using gfit software. Seven kinetic rates and a stoichiometry constant were optimized. An extensive search of parameter space was performed by coupling local optimization with random restart and simulated annealing. Further details of the analysis are provided in Supplementary Material; rsys and gfit software, as well as the RFC model are available at http://gfit.sourceforge.net.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grant GM64514-01 to M.M.H. We thank Stephen J. Benkovic and Michael O’Donnell for clones, Smita S. Patel for helpful discussions, and Carol Teschke for use of the Applied Photophysics stopped-flow instrument.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Hingorani MM, O’Donnell M. Sliding clamps: a (tail)ored fit. Curr Biol. 2000;10:R25–9. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 3.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–60. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Bloom LB. Dynamics of loading the Escherichia coli DNA polymerase processivity clamp. Crit Rev Biochem Mol Biol. 2006;41:179–208. doi: 10.1080/10409230600648751. [DOI] [PubMed] [Google Scholar]
- 6.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–43. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 7.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 8.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 9.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–60. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 10.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Neuwald AF. Evolutionary clues to eukaryotic DNA clamp-loading mechanisms: analysis of the functional constraints imposed on replication factor C AAA+ ATPases. Nucleic Acids Res. 2005;33:3614–28. doi: 10.1093/nar/gki674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smiley RD, Zhuang Z, Benkovic SJ, Hammes GG. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: multiple pathways of holoenzyme formation. Biochemistry. 2006;45:7990–7. doi: 10.1021/bi0603322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietroni P, von Hippel PH. Multiple ATP binding is required to stabilize the “activated” (clamp open) clamp loader of the T4 DNA replication complex. J Biol Chem. 2008;283:28338–53. doi: 10.1074/jbc.M804371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seybert A, Singleton MR, Cook N, Hall DR, Wigley DB. Communication between subunits within an archaeal clamp-loader complex. EMBO J. 2006;25:2209–18. doi: 10.1038/sj.emboj.7601093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seybert A, Wigley DB. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. Embo J. 2004;23:1360–71. doi: 10.1038/sj.emboj.7600130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci U S A. 2006;103:2546–51. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O’Donnell M, Hurwitz J. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci U S A. 1998;95:11607–12. doi: 10.1073/pnas.95.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Gibbs E, Kelman Z, O’Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 1999;96:1869–74. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams CR, Snyder AK, Kuzmic P, O’Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the gamma complex. J Biol Chem. 2004;279:4376–85. doi: 10.1074/jbc.M310429200. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang Z, Berdis AJ, Benkovic SJ. An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex. Biochemistry. 2006;45:7976–89. doi: 10.1021/bi0601205. [DOI] [PubMed] [Google Scholar]
- 22.Trakselis MA, Berdis AJ, Benkovic SJ. Examination of the role of the clamp-loader and ATP hydrolysis in the formation of the bacteriophage T4 polymerase holoenzyme. J Mol Biol. 2003;326:435–51. doi: 10.1016/s0022-2836(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 23.Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–83. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 24.Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281:35531–43. doi: 10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- 25.Finkelstein J, Antony E, Hingorani MM, O’Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 26.Hingorani MM, Coman MM. On the specificity of interaction between the Saccharomyces cerevisiae clamp loader replication factor C and primed DNA templates during DNA replication. J Biol Chem. 2002;277:47213–24. doi: 10.1074/jbc.M206764200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, Webb MR. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–80. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 28.Levin MK, Hingorani MH, Holmes RM, Patel SS, Carson JH. Model-based global analysis of heterogeneous experimental data using gfit. In: Maly IV, editor. Methods in Molecular Biology/Systems Biology. Humana Press, Inc.; 2008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazmirski SL, Zhao Y, Bowman GD, O’Donnell M, Kuriyan J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2005;102:13801–6. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt SL, Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem. 2001;276:34784–91. doi: 10.1074/jbc.M011633200. [DOI] [PubMed] [Google Scholar]
- 32.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–9. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Coman MM, Sakato M, O’Donnell M, Hingorani MM. Conserved residues in the delta subunit help the E. coli clamp loader, gamma complex, target primer-template DNA for clamp assembly. Nucleic Acids Res. 2008;36:3274–86. doi: 10.1093/nar/gkn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–93. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.