Abstract
Ebola virus (EBOV) entry requires the surface glycoprotein, GP, to initiate attachment and fusion of viral and host membranes. Here, we report the crystal structure of EBOV GP in its trimeric, pre-fusion conformation (GP1+GP2) bound to a neutralizing antibody, KZ52, derived from a human survivor of the 1995 Kikwit outbreak. Three GP1 viral attachment subunits assemble to form a chalice, cradled by the GP2 fusion subunits, while a novel glycan cap and projected mucin-like domain restrict access to the conserved receptor-binding site sequestered in the chalice bowl. The glycocalyx surrounding GP is likely central to immune evasion and may explain why survivors have insignificant neutralizing antibody titres. KZ52 recognizes a protein epitope at the chalice base where it clamps several regions of the pre-fusion GP2 to the N terminus of GP1. This structure now provides a template for unraveling the mechanism of EBOV GP-mediated fusion and for future immunotherapeutic development.
The Ebola virus (EBOV) is an enveloped, non-segmented, negative-strand RNA virus, which together with Marburg virus, makes up the filoviridae family. The virus causes severe hemorrhagic fever associated with 50-90% human mortality1. Four species of the virus (Zaire, Sudan, Côte d’Ivoire, and Reston ebolavirus) have thus far been identified, with Zaire typically associated with the highest human lethality2. A fifth EBOV species is confirmed in a 2007 outbreak in Bundibugyo, Uganda3,4. Infection with EBOV results in uncontrolled viral replication and multiple organ failure with death occurring 6-9 days after onset of symptoms5. Fatal cases are associated with high viremia and defective immune responses, while survival is associated with early and vigorous humoral and cellular immune responses6-9. Although preliminary vaccine trials in primates have been highly successful10-13, no vaccines, specific immunotherapeutics, or post-exposure treatments are currently approved for human use. Since 1994, EBOV outbreaks have increased more than four-fold, thus necessitating the urgent development of vaccines and therapeutics for use in the event of an intentional, accidental or natural EBOV release.
The EBOV genome contains seven genes, which direct the synthesis of eight proteins. Transcriptional editing of the fourth gene (GP) results in expression of a 676-residue transmembrane-linked glycoprotein termed GP, as well as a 364-residue secreted glycoprotein termed sGP14,15. EBOV GP is the main target for the design of vaccines and entry inhibitors. GP is post-translationally cleaved by furin16 to yield disulfide-linked GP1 and GP2 subunits17. GP1 effects attachment to host cells, while GP2 mediates fusion of viral and host membranes16,18-20. EBOV is thought to enter host cells through receptor-mediated endocytosis via clathrin-coated pits and caveolae21, followed by actin and microtubule-dependent transport to the endosome21, where GP is further processed by endosomal cathepsins22-24. Essential cellular receptor(s) have not yet been identified, but DC-SIGN/L-SIGN25, hMGL26, β-integrins27, folate receptor-α28 and Tyro3 family receptors29 have all been implicated as cellular factors in entry. Here, we report the crystal structure of EBOV GP, at 3.4 Å resolution, in its trimeric, pre-fusion conformation in complex with neutralizing antibody Fab KZ52.
Ebola virus GP is in a pre-fusion conformation
In an effort to increase sample homogeneity and to promote crystal contacts, we excised the mucin-like and transmembrane domains (GP33-632Δmuc), mutated two N-linked glycosylation sites (T42V/T230V) and complexed the GP with Fab KZ52, which recognizes a conformational epitope. The resulting GP construct is fully capable of mediating virus entry and exhibits similar antibody neutralization profiles as wild-type, when expressed with a transmembrane domain on vesicular stomatitis virus (VSV) pseudovirions (Supplemental Methods and Fig. S1).
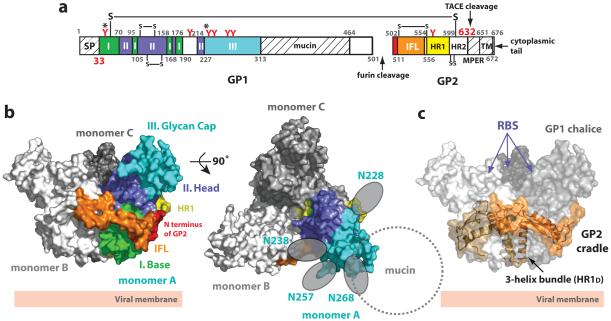
The EBOV GP trimer contains three non-covalently attached monomers (A, B and C) (Supplemental Fig. S2), which together adopt a chalice-like shape with overall dimensions of ∼95 Å × 95 Å × 70 Å (Fig. 1). Each monomer consists of two disulfide-linked subunits, GP1 and GP2. GP trimerization is mediated through multiple GP1-GP2 and GP2-GP2 contacts and occludes ∼2900 Å2 surface area on each subunit. No major contacts are observed between neighboring GP1 molecules. Three GP1 ectodomains together form a bowl-like structure encircled by helices of the three GP2 subunits (Fig. 1c).
Figure 1. Structure of Zaire-EBOV GP.
(a) Domain schematic of GP. Domains observed in the crystal structure are coloured and numbered according to the description in the text. White and hash-marked regions designate crystallographically disordered and construct-deleted regions, respectively. Abbreviations are as follows: SP, signal peptide; I, GP1 base; II, GP1 head; III, GP1 glycan cap; mucin, mucin-like domain; IFL, internal fusion loop; HR1, heptad repeat 1; HR2, heptad repeat 2; MPER, membrane-proximal external region; and TM, transmembrane domain. Red Y-shaped symbols designate the predicted N-linked glycosylation sites; those sites marked with an asterisk were mutated. The final model includes EBOV GP residues 33-189, 214-278, 299-310 and 502-599. No electron density is observed for residues 190-213, 311-312, 464-501 and 600-632. Weak or discontinuous electron density is seen in the loop containing the GP1-GP2 disulfide bridge (residues 49-56) and the outer regions of the GP1 glycan cap (residues 268-278 and 299-310); these regions are modeled as poly-alanine fragments. (b) Molecular surface of the GP trimer viewed on its side (left) and top (right), as viewed down the three-fold axis. Monomer A is coloured according to the scheme in panel (a), and monomers B and C of the trimer are shown in white and grey, respectively. Predicted N-linked oligosaccharides (N228, N238, N257 and N268) belonging to the glycan cap of monomer A are shown as grey ovals. The location of the N-linked glycan at N268 is tentative, as the sequence assignment in this region is ambiguous. (c) Molecular surface of the EBOV GP chalice and cradle. The three GP1 subunits that form the chalice are shown in various shades of grey and GP2 subunits forming the cradle are shown as ribbons, in various shades of orange, underneath the transparent molecular surface. The putative receptor-binding sites (RBS) are recessed in the inner bowl of the GP trimer.
EBOV GP is thought to assemble as a metastable trimer of heterodimers on the viral surface. Upon an as yet unidentified trigger, GP undergoes an irreversible conformational change to a lower energy state, thereby merging the viral and host cell membranes. The low energy or post-fusion conformation is characterized by a 6-helix bundle (6HB), in which the N terminus and fusion loop of GP2 are juxtaposed to its C terminus in a hairpin conformation30,31. Our EBOV GP structure likely corresponds to the metastable, pre-fusion conformation of EBOV GP, as GP2 forms a trimer intimately associated with GP1, rather than the post-fusion 6HB conformation. In addition, crystals were grown at a neutral pH and the EBOV GP presented here contains no multimerization motifs or recombinant constraints on quaternary structure that could artificially force the structure into a non-prefusion state.
Ebola virus GP1
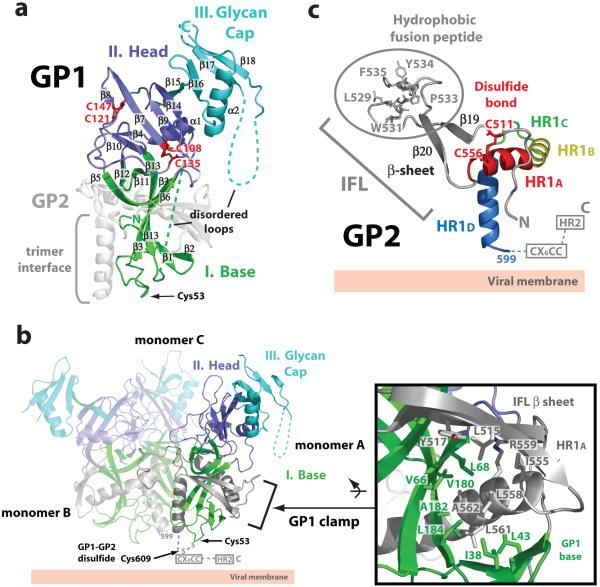
GP1 is responsible for cell surface attachment, which is probably mediated by a region including residues 54-20132. GP1 is composed of a single domain (∼65 Å × 30 Å × 30 Å), arranged in the topology shown in Fig. 2a, and can be further subdivided into the (I) base, (II) head and (III) glycan cap regions (Fig. 2a and Supplemental Fig. S3). The base (I) subdomain is composed of two sets of β sheets, forming a semi-circular surface which clamps the internal fusion loop and a helix of GP2 through hydrophobic interactions (Fig. 2b). Moreover, this subdomain contains Cys53, which is proposed to form an intermolecular disulfide bridge to Cys609 of the GP2 subunit17. Cys53 resides near GP2 in the β2-β3 loop at the viral membrane-proximal end of the base subdomain (Fig. 2a-b). Our EBOV GP contains an intact GP1-GP2 disulfide bridge, based on reducing and non-reducing SDS-PAGE analysis. However, the region containing the counterpart GP2 cysteine is disordered, which may reflect functionally important mobility in the region. The head (II) is located between the base and glycan cap regions towards the host membrane surface. Two intramolecular disulfide bonds stabilize the head subdomain and confirm the biochemically determined disulfide bridge assignments17. Cys108-Cys135 connects a surface-exposed loop (β8-β9 loop) to strand β7, while Cys121-Cys147 bridges the β8-β9 and β9-β10 loops (Fig. 2a). The glycan cap (III) contains four predicted N-linked glycans (at N228, N238, N257 and N268) in an α/β dome over the GP1 head subdomain (Fig. 1b and 2a). This subdomain does not form any monomer-monomer contacts and is fully exposed on the upper and outer surface of the chalice. The central β sheets from the head and glycan cap together form a fairly flat surface and, in the context of the GP trimer, form the three inner sides of the chalice bowl.
Figure 2. EBOV GP1 and GP2.
(a) Ribbon diagram of the GP1 subunit. The base (I) subdomain (green) contains four discontinuous sections (residues 33-69, 95-104, 158-167 and 176-189), which form two mixed β sheets with strands β3 and β13 shared between the two β sheets. The head (II) subdomain (purple) is composed of residues from four discontinuous segments, 70-94, 105-157, 168-175 and 214-226, and forms a four-stranded, mixed β sheet supported by an α helix and a smaller, two-stranded, antiparallel β sheet. The glycan cap (III) region (cyan) is composed of a continuous polypeptide chain, residues 227-310, and forms an α helix packed against a four-stranded, mixed β sheet. Intramolecular disulfide bridges between Cys108-Cys135 and Cys121-Cys147 are coloured red. (b) Ribbon diagram of the Ebola virus GP trimer. Each GP1 subdomain is coloured according to panel (a) and the three GP2 subunits are shown in grey. The GP1 base subdomain forms a clamp onto the GP2 internal fusion loop and HR1A helix through interactions with hydrophobic residues (inset box). The GP1-GP2 disulfide bridge (Cys53-Cys609), CX6CC motif and HR2 region are disordered in the structure and are marked and indicated by dashed lines. (c) The pre-fusion conformation of EBOV GP2. The EBOV GP2 internal fusion loop contains a disulfide bond at its base (red), antiparallel β strand (grey) and a hydrophobic fusion peptide. The EBOV GP2 HR1 region is segmented into four parts (HR1A-HR1D). Note that the HR2 and the CX6CC motif are disordered in the pre-fusion conformation.
Ebola virus GP2
GP2 is responsible for fusion of viral and host cell membranes and contains the internal fusion loop and the heptad repeat regions, HR1 and HR2. Many viral glycoproteins have fusion peptides, located at the N terminus of their fusion subunit, which are released upon cleavage of the precursor glycoprotein. By contrast, class II and class III fusion proteins, as well as class I glycoproteins from Ebola, Marburg, Lassa and avian sarcoma leukosis viruses, contain internal fusion loops lacking a free N terminus. The crystal structure reveals that the EBOV GP internal fusion loop, which encompasses residues 511-556, utilizes an antiparallel β stranded scaffold to display a partially helical hydrophobic fusion peptide (L529, W531, I532, P533, Y534 and F535) (Fig. 2c). The side chains of these hydrophobic residues pack into a region on the GP1 head of a neighboring subunit in the trimer, reminiscent of the fusion peptide packing in the pre-fusion parainfluenza virus 5 F structure33. A disulfide bond between Cys511 at the base of β19 and Cys556 in the HR1 helix covalently links the antiparallel β sheet. This disulfide bond between the internal fusion loop and HR1 is conserved among all filoviruses, and is analogous to a pair of critical cysteines flanking the internal fusion loop in avian sarcoma leukosis virus34,35. Interestingly, the EBOV internal fusion loop has features more similar to those observed in class II and III viral glycoproteins (in particular to flaviviruses) than those previously observed for class I glycoproteins (Supplemental Fig. S4). It thus appears that regardless of viral protein class, internal fusion loops share a common architecture for their fusion function.
EBOV GP2 contains two heptad repeat regions (HR1 and HR2), connected by a 25-residue linker containing a CX6CC motif and the internal fusion loop. The crystal structures of post-fusion GP2 fragments30,31 have revealed that the two heptad repeat regions form antiparallel α helices and that a CX6CC motif forms an intrasubunit disulfide bond between Cys601 and Cys608 (Supplemental Fig. S5). In the pre-fusion EBOV GP, HR2 and the CX6CC motif are disordered. By contrast, the HR1 region is well ordered and can be divided into four segments: HR1A, HR1B, HR1C and HR1D (Fig. 2c), which together assemble the cradle encircling GP1. Similarly, heptad repeat regions in influenza and parainfluenza viruses also contain multiple segments in their pre-fusion helices that substantially rearrange in their post-fusion conformations33,36,37.
The first two segments, HR1A and HR1B (residues 554-575), together form an α helix with an ∼40° kink at T565, which delineates HR1A from HR1B. Interestingly, the bend between HR1A and HR1B contains an unusual 3-4-4-3 stutter, which may act as a conformational switch31, rather than the typical 3-4 periodicity of heptad repeats (Supplemental Fig. S6). A similar stutter has also been noted in parainfluenza virus 5 F33. The Ebola virus HR1C (residues 576-582) forms an extended coil linking HR1B to the 14-residue α helix of HR1D (residues 583-598). HR1D forms an amphipathic helix and the hydrophobic faces of each HR1D join to form a three-helix bundle at the trimer interface. Although the breakpoint maps directly to a chloride ion binding site in the post-fusion conformation of GP230,31 and at least two other viruses38,39, no chloride ion is observed here as HR1 and HR2 do not come together to form the six-helix bundle. Instead, the pre-fusion GP2 adopts a novel conformation, intimately curled around GP1 (Fig. 1c).
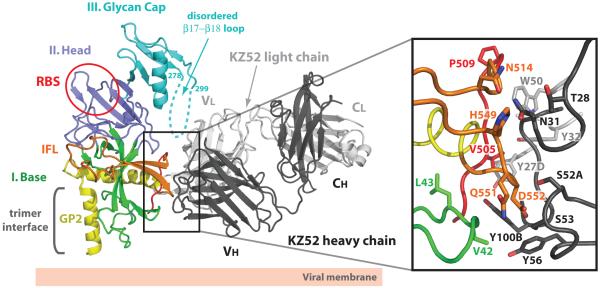
Ebola virus GP-KZ52 interface
KZ52 is an antibody isolated from a human survivor of a 1995 outbreak in Kikwit, Democratic Republic of the Congo (formerly Zaire)40. This antibody neutralizes Zaire ebolavirus in vitro40 and offers protection from lethal EBOV challenge in rodent models41, but has minimal effects on viral pathogenicity in non-human primates42. KZ52 is directed towards a vulnerable, non-glycosylated epitope at the base of the GP chalice, where it engages three discontinuous segments of EBOV GP: residues 42-43 at the N terminus of GP1, and 505-514 and 549-556 at the N terminus of GP2 (Fig. 3 and Supplemental Fig. S7). Although the majority of the GP surface buried by KZ52 belongs to GP2, the presence of both GP1 and GP2 are critical for KZ52 recognition43. It is likely that GP1 is required to maintain the proper pre-fusion conformation of GP2 for KZ52 binding. Indeed, KZ52 is the only antibody known to bridge both attachment (GP1) and fusion (GP2) subunits of any viral glycoprotein. Given that KZ52 requires a conformational epitope seen only in the GP2 pre-fusion conformation and that the KZ52 epitope is distant from the putative receptor-binding site (RBS), KZ52 likely neutralizes by preventing rearrangement of the GP2 HR1A/HR1B segments and blocking host membrane insertion of the internal fusion loop. Alternatively, IgG KZ52 may sterically hinder access to the RBS or to a separate binding site of another cellular factor, especially if multiple attachment events are required for entry.
Figure 3. EBOV GP-Fab KZ52 interactions.
KZ52 recognizes residues 505-514 (red) and 549-556 (orange) of GP2 and residues 42-43 (green) of GP1. The β17-β18 loop (residues 279-298) is disordered, but may interact with KZ52. One EBOV GP monomer is coloured and labeled according to Fig. 1b and Fab heavy and light chains are shown in black and light grey, respectively. Side-chain interactions at the GP-KZ52 interface are magnified in the inset box. The putative receptor-binding site (RBS) is outlined in a red circle on the EBOV GP ribbon diagram. Note that only selected residues from GP and KZ52 are shown for figure clarity and residue 42 is a threonine in the wild-type Zaire ebolavirus sequence.
The KZ52 epitope of GP is convex and does not have a high shape complementarity to the antibody (Sc index of 0.63), although ∼1600 Å2 of each GP monomer are occluded upon KZ52 binding. The antibody contacts a total of 15 GP residues by van der Waals interactions and 8 direct hydrogen bonds (Supplemental Fig. S7). Ten out of 15 residues in the structurally defined KZ52 epitope are unique to Zaire ebolavirus (Supplemental Fig. S6), thus explaining the Zaire specificity of KZ52.
Ebola virus GP glycosylation
We generated a fully glycosylated molecular model of EBOV GP to illustrate the native GP trimer as it exists on the viral surface (Fig. 4). The majority of N-linked glycosylation sites are concentrated in the mucin-like domain and glycan cap of GP1. Given that the mucin-like domain is ∼75 kDa in mass (protein and oligosaccharide), the volume of this domain is predicted to be similar to each GP monomer observed here. The crystal structure suggests that the mucin-like domain is linked to the side of each monomer and may further build up the walls of the chalice, forming a deeper bowl (Fig. 4). Although a mixture of complex, oligomannose and hybrid-type glycans are found on intact, mucin-containing GP144, those glycans outside the mucin-like domain are likely to be complex in nature: the mucin-deleted GP used for crystallization is sensitive to PNGaseF, but not to EndoH treatment (Supplemental Fig. S8). Modeling of complex-type oligosaccharides on the EBOV GP indicates that the majority of the GP trimer is cloaked by a thick layer of oligosaccharide, even without the mucin-like domain (Fig. 4). The ∼19 additional oligosaccharides on the full-length GP (17 on the mucin-like domain and 2 more on GP1, disordered here) further conceal the sides and top of the chalice. The KZ52 binding site and, presumably, the flexible regions of HR2 and the membrane-proximal external region (MPER) remain exposed and perhaps vulnerable to binding of antibodies and inhibitors.
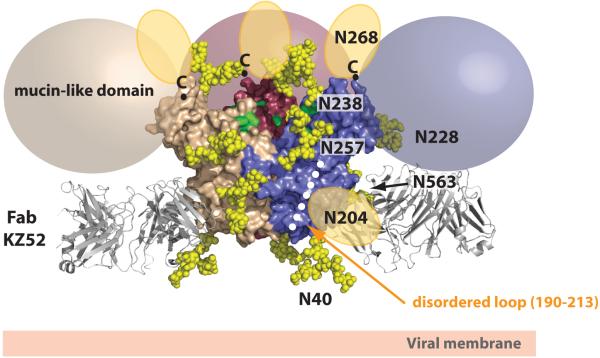
Figure 4. Model of the fully glycosylated GP.
N-linked bi-antennary complex-type glycans (Gal2Man3GlcNAc4) were modeled onto the GP1 glycan cap subdomain. Oligosaccharides are shown as yellow space-filling spheres and for figure clarity, only those glycans belonging to the purple monomer are labeled. Note that the glycans on N228 and N563 reside on the back of the purple monomer and are partially obscured. The glycans at N204 and N268 are found in regions that are poorly ordered in the structure and as a result, their tentative locations are shown as orange ovals. The C termini of the last ordered residues of GP1, to which mucin-like domains are linked, are marked with ’C’ (top of the chalice) and coloured spheres (beige, pink and purple) outline the predicted positions of the mucin-like domains attached in each of these regions. Surface residues previously identified to be critical for viral entry, recessed in the chalice bowl and RBS, are coloured green. Fab KZ52 (coloured grey) recognizes a non-glycosylated, predominantly GP2-containing epitope at the base of the chalice.
The development of neutralizing antibodies is limited in natural Ebola virus infection. Many survivors have low or insignificant titres1,7, and those antibodies that are elicited preferentially recognize a secreted version of the viral glycoprotein that features an alternate quaternary structure and lacks the mucin-like domain43. The glycocalyx surrounding EBOV GP likely forms a shield that protects it from humoral immune responses and/or confers stability inside or outside a host. The mucin-like domain and glycan cap sit together as an external domain to the viral attachment and fusion subunits, reminiscent of the glycan shields of HIV-1 gp12045,46 and Epstein-Barr virus gp35047, perhaps pointing to a common theme for immune evasion. Alignment of filoviral sequences indicate that regions involved in immune evasion have a low degree of sequence conservation [i.e. GP1 glycan cap (∼5%) and mucin-like domain (0%)], but the N-glycosylation sites in the glycan cap are mostly conserved among all EBOV subtypes (Supplemental Fig. S6), indicating the functional importance of these post-translational modifications.
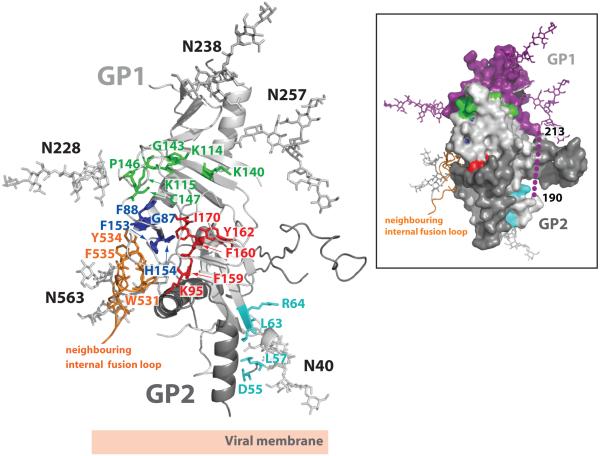
Sites of receptor binding and cathepsin cleavage
Although a definitive receptor for EBOV remains to be identified, previous studies32,48,49 have determined that residues 54-201, which map to the base and head subdomains of GP1, form a putative receptor-binding site (RBS) for attachment to host cells. Additional experimental studies have identified at least 19 GP1 residues, assigned into four groups based on the location in the structure, that are critical for viral entry48-50 (Fig. 5). Many of these residues are apolar or aromatic and are involved in maintaining the structural integrity of GP1 for receptor binding or fusion. However, six residues (K114, K115, K140, G143, P146 and C147) cluster within a ∼20 × 15 Å surface in the inner bowl of the chalice and may thus represent important receptor contact sites. All residues in the putative RBS are highly conserved among Ebola virus species (Supplemental Fig. S6).
Figure 5. Sites of receptor binding and cathepsin cleavage.
Residues coloured in cyan, green, royal blue and red were previously identified by mutagenesis to be important for viral entry48-50. Residues coloured in cyan (D55, L57, L63 and R64) reside at the base of the chalice, near the GP1-GP2 disulfide bond and HR1D, and are likely important for fusion-mediated conformational changes rather than receptor binding. Residues coloured in red (F159, F160, Y162 and I170) are primarily buried hydrophobic amino acids that help to maintain the structural stability of GP1. Residues coloured in royal blue (G87, F88, F153 and H154) are in proximity to the putative receptor-binding site (RBS) and pack against the hydrophobic residues from a neighbouring internal fusion loop (coloured orange). Mutations to these residues may affect viral entry by altering the structural integrity of the RBS and/or by affecting packing of the fusion loop. Residues coloured in green (K114, K115, K140, G143, P146 and C147) reside on or near the GP surface and may contribute to receptor binding. A molecular surface representation of a GP monomer, coloured and oriented according to the ribbon diagram, is presented in the inset box. Indicated in this view: cleavage at a site on the loop (shown as purple dots) between residues 190-213 would remove the entire glycan cap (coloured in purple) and the mucin-like domain (not shown), leaving GP2 and an ∼18 kDa fragment of GP1.
Importantly, this putative RBS is recessed beneath the glycan cap and perhaps further masked by the mucin-like domain (Fig. 4), suggesting that additional conformational change or removal of the mucin-like domain could reveal additional surfaces required for receptor or cofactor binding. It has been demonstrated that endosomal proteolysis of EBOV GP by cathepsin L and/or B removes the mucin-like domain to produce a stable ∼18 kDa GP1 intermediate which has enhanced viral binding and infectivity22-24. The precise site of cathepsin cleavage is unknown and the role of cathepsins in natural infection is as yet unclear. However, formation of an ∼18 kDa GP1 fragment implies that cathepsin may cleave near the GP1 β13-β14 loop (residues 190-213). Indeed, this loop is unresolved in the pre-fusion structure, suggesting enhanced mobility and accessibility to enzymatic cleavage. Cleavage within this loop would remove the entire mucin-like domain and glycan cap region (Fig. 5). As a result, β7 to β9 strands and their associated loops would become exposed. These regions of GP are in proximity to the previously identified residues critical for viral entry. The fold, location and physicochemical properties of this site should now provide new leads in the search for the elusive filoviral receptor(s).
A summary of the Ebola virus mechanism of infection, including the events of cathepsin cleavage and conformational changes to GP2 during fusion, is presented in Supplemental Materials and Fig. S9.
METHODS SUMMARY
T42V/T230V GP33-632Δmuc was transiently expressed in HEK293T cells with an N-terminal haemaglutinin tag for purification via affinity chromatography. GP was natively deglycosylated using PNGaseF and complexed with a SeMet-containing KZ52 antibody, and the resulting trimeric GP-Fab complex was purified by size exclusion chromatography. Crystals were grown in 8.5% (w/v) PEG 10,000, 0.1 M Tris-HCl pH 8.5, 0.4 M sodium acetate and 10% (v/v) PEG 200 and diffracted to 3.4 Å resolution. Phases were obtained by combination of the Fab selenium anomalous signal and molecular replacement using an independently determined structure of the uncomplexed KZ52 Fab as a search model. Interpretable electron density maps with clear secondary structural elements and solvent boundaries were obtained after NCS-averaged density modification. The structure was refined to final Rwork and Rfree values of 26.2% and 30.2%, respectively. Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession code 3CSY.
Supplementary Material
Acknowledgements
We thank Christopher Kimberlin (TSRI) and the staff at the Advanced Light Source (ALS) beamlines 8.3.1, 5.0.2, 4.2.2, 8.2.2 and 12.3.1 and Stanford Synchrotron Radiation Laboratory (SSRL) beamlines 11-1 and 9-2 for data collection support, and Dafna Abelson and members of the Ollmann Saphire Lab for assistance, comments and suggestions. The ALS and SSRL are national user facilities operated on behalf of the U.S. Department of Energy. We thank Dr. Arthur Olson and Jon Huntoon for generation of tangible molecular models for analysis of receptor binding surfaces. E.O.S. and D.R.B. are funded by the U.S. National Institutes of Health and E.O.S. and J.E.L. are supported by a Career Award from the Burroughs Wellcome Fund and a fellowship from the Canadian Institutes of Health Research, respectively. This is manuscript #19375 from The Scripps Research Institute.
Appendix
METHODS
Expression of Ebola virus GP and antibody
Zaire ebolavirus (Mayinga strain) glycoprotein DNA was codon-optimized, whole-gene synthesized (Blue Heron Biotech) and cloned into the pDISPLAY vector (Invitrogen, Carlsbad, CA) with a stop codon introduced before the internal transmembrane segment of the vector. GP is composed of >50% oligosaccharides by weight, primarily due to a heavily glycosylated mucin-like domain that is non-essential for cellular attachment or entry51. In an effort to increase sample homogeneity and solubility, deletion of the mucin-like and transmembrane domains (residues 312-462 and 633-676) and mutations of two N-linked glycosylation sites (T42V and T230V) were created by overlap PCR and the QuikChange II site-directed mutagenesis kit, respectively. Large-scale expression of T42V/T230V GP33-632Δmuc was performed using HEK293T cells transfected by standard calcium phosphate precipitation in 10-layer CellStacks (Corning). The DNA-calcium phosphate mixture was added to 70% confluent cells grown in DMEM plus 1X Pen/Strep and 5% (v/v) FBS. The supernatant was harvested four days post-transfection, concentrated using a Centramate tangential flow system, purified by anti-HA immunoaffinity chromatography and natively deglycosylated with PNGaseF in 1X PBS supplemented with 1.5 M urea at 37°C.
Recombinant IgG1 KZ52 was expressed in Chinese hamster ovary (CHO-K1) cells in glutamine-free custom-formulated Glasgow minimum essential medium (GMEM) (MediaTech Cellgro). For large-scale tissue culture, media was supplemented with 3.5% (v/v) Ultra Low Bovine IgG Fetal Bovine Serum (FBS) (Invitrogen) and grown in 10-layer CellStacks (Corning). The supernatants were centrifuged, sterile-filtered, purified over Protein A affinity matrix (GE Healthcare) and eluted in 0.1 M citric acid, pH 3.0. The pH of the eluted antibody was immediately brought to neutrality by the addition of 2 M Tris pH 9.0 and subsequently dialyzed against phosphate-buffered saline (PBS, pH 7.4). Antibody concentration was determined by absorbance at 280 nm. Purity (>98%) and concentrations were confirmed by SDS-PAGE. Selenomethionine-incorporated KZ52 was produced similarly except cysteine- and methionine-free RPMI (supplemented with 60 mg/mL L-selenomethionine and 60 mg/L L-cysteine) media was used for growth in T-225 cm2 tissue culture flasks (Corning). Supernatant was harvested every four days over two weeks and concentrated using the Centramate system prior to Protein A purification. IgG1 KZ52 was cleaved by standard papain digestion, the released Fc and uncleaved IgG were removed via Protein A chromatography and the resulting Fab fragments were further purified by MonoS ion exchange chromatography.
Preparation and crystallization of the GP-antibody complex
To promote the formation of crystal contacts, purified GP was mixed with excess Fab KZ52 and separated on a Superdex-200 GL 10/300 equilibrated with 10 mM Tris-HCl pH 7.5 and 150 mM NaCl. Fractions containing trimeric GP-Fab KZ52 were concentrated to ∼8 mg/mL and crystallized by hanging drop vapour diffusion at 22°C in 8.5% (w/v) PEG 10,000, 0.1 M Tris-HCl pH 8.5, 0.4 M sodium acetate and 10% (v/v) PEG 200. These crystals were gently cross-linked with glutaraldehyde and cryoprotected with sequential soaks of 10%, 20%, 30% and 40% (v/v) glycerol plus mother liquor prior to flash cooling in liquid nitrogen. Although >800 crystals were screened at seven synchrotron beamlines, only three crystals diffracted to 4.0 Å resolution or better. Two complete datasets were collected at the Advanced Light Source (ALS, Berkeley, CA) beamline 5.0.2 from SeMet-derivatized Fab KZ52-GP crystals. All data were indexed, integrated and scaled using d*TREK52 to 3.4 Å and 4.0 Å resolution, respectively (Supplemental Table S1). The T42V/T230V GP33-632Δmuc-KZ52 crystals belong to space group H32, with one and one-third trimers in the asymmetric unit.
Structure determination
Using an independently determined, uncomplexed Fab KZ52 structure as a search model, Phaser53 identified one clear Fab solution residing on a three-fold crystallographic axis (z-score=9.3). A trimeric arrangement of KZ52 Fab was generated by applying crystallographic symmetry and the trimer of antibodies was subsequently used in a molecular replacement search while keeping the original Fab KZ52 solution fixed. Phaser identified a clear unique hit for the trimer (z-score=10.4). A difference Fourier anomalous electron density map was calculated and selenium peaks for all 20 SeMet residues were picked manually using Xfit54. Phase calculation combining Fab KZ52 model phases with Se-SAD anomalous data from two crystals was performed in SHARP55. Initial phases from SHARP55 were improved by solvent flipping, histogram matching and 1500 cycles of non-crystallographic symmetry (NCS) averaging using DM56. Strong electron density was seen for the central regions of GP1 and GP2 in all four monomers, whereas weaker density was associated with the outer regions of GP1. Using the DM-improved phases, Resolve57 was used to initially trace poly-alanine fragments in the map. These starting fragments were extended manually using Coot (v. 0.3.3)58. All β strands and α helices were real-space refined with torsional secondary structural restraints. Calculation of B-value sharpened electron density maps improved the resolution of side-chain electron density and were used to help build the model. Torsion-angle simulated-annealing refinement, starting at 5000 K59 with a maximum-likelihood amplitude target, NCS restraints and no σ-cutoffs, was employed in CNS-SOLVE (v. 1.2)60. A final round of refinement using riding hydrogens, grouped atomic displacement and TLS parameters was implemented using Phenix.refine61. Identification of proper sequence registry was confirmed by secondary structural predictions and by anchoring N-linked glycosylation sequons to associated electron density. This strategy proved particularly useful as the N-linked glycan at N257 allowed initial assignment of the residues in the glycan cap (residues 214-267). Moreover, alternate sequence registers were considered in several portions of the GP1 and GP2 subunits, but these models could be eliminated based on inspection of σA-weighted 2Fo-Fc and Fo-Fc electron density maps. Procheck62, MolProbity63, and composite omit electron density maps (Supplemental Fig. S10) were used to validate the quality of the model. Although the protein used for crystallization was treated with protein N-glycosidase F (PNGaseF), two glycan chains at N257 on GP1 and N563 on GP2, resistant to deglycosylation, were retained. Electron density is visible for the chitobiose core and up to three mannose residues of these glycans (Supplemental Fig. S10). The pdb-care64,65 and carp66 servers were used to detect discrepancies in glycan connectivity, nomenclature and glycosidic torsion angles. The final model contains EBOV GP residues 33-189, 214-278, 299-310 and 502-599 and Fab KZ52 residues 1-228 (heavy chain) and 1-211 (light chain). Electron density is missing for GP residues 190-213, 311-312, 464-501 and 600-632. Weak or discontinuous electron density is observed in the outer regions of the glycan cap (residues 268-278 and 299-310) and the loop containing the GP1-GP2 disulfide bridge (residues 49-56); these regions are tentatively assigned as poly-alanine fragments. Interestingly, a small chain-like piece of electron density is visible near N512 in GP2, but is not sufficiently resolved for identification. Final refinement statistics and analysis are presented in Supplemental Table S1. Molecular surface and ribbon diagrams were generated using MacPyMOL67. For Figure 4, N-linked complex-type glycans were modeled using the GlyProt server68. Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession code 3CSY.
Methods describing the assays for EBOV GP infectivity and KZ52 recognition at low pH are described in the Supplementary Methods section.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Sanchez A, et al. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott, Williams, and Wilkins; Philadelphia: 2001. pp. 1279–1304. [Google Scholar]
- 2.Johnson KM, Lange JV, Webb PA, Murphy FA. Isolation and characterization of a new virus causing acute hemorrhagic fever in Zaire. Lancet. 1977;1:569–571. doi: 10.1016/s0140-6736(77)92000-1. [DOI] [PubMed] [Google Scholar]
- 3.Alsop Z. Ebola outbreak in Uganda “atypical”, say experts. Lancet. 2007;370:2085. doi: 10.1016/s0140-6736(07)61880-7. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . Ebola haemorrhagic fever in Uganda. 2007. [Google Scholar]
- 5.Peters CJ, Sanchez A, Rollin PE, Ksiazek TG, Murphy GA. In: Fields Virology. Fields BN, Knipe DM, Howley PM, editors. Vol. 1. Lippincott-Raven Press, Inc.; Philadelphia, PA: 1996. pp. 1161–1176. [Google Scholar]
- 6.Baize S, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 7.Ksiazek TG, et al. Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 1999;179(Suppl 1):S177–187. doi: 10.1086/514321. [DOI] [PubMed] [Google Scholar]
- 8.Leroy EM, et al. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. doi: 10.1016/s0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez A, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J. Virol. 2004;78:10370–10377. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampton T. Vaccines against Ebola and Marburg viruses show promise in primate studies. JAMA. 2005;294:163–164. doi: 10.1001/jama.294.2.163. [DOI] [PubMed] [Google Scholar]
- 11.Jones SM, et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005;11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 12.Kobinger GP, et al. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology. 2006;346:394–401. doi: 10.1016/j.virol.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan NJ, et al. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med. 2006;3:e177. doi: 10.1371/journal.pmed.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez A, et al. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the Ebola virus glycoprotein. J. Virol. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldmann H, Klenk HD, Sanchez A. Molecular biology and evolution of filoviruses. Arch. Virol. Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 19.Takada A, et al. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wool-Lewis RJ, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez A. Analysis of filovirus entry into Vero E6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007;196(Suppl 2):S251–258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- 22.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez CP, et al. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takada A, et al. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takada A, et al. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- 28.Chan SY, et al. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106:117–126. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- 29.Shimojima M, et al. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 2006;80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn JH, et al. Conserved receptor-binding domains of Lake Victoria Marburgvirus and Zaire Ebolavirus bind a common receptor. J. Biol. Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 33.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delos SE, et al. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J. Virol. 2008;82:3131–3134. doi: 10.1128/JVI.02266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delos SE, White JM. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol. 2000;74:9738–9741. doi: 10.1128/jvi.74.20.9738-9741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 37.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 38.Fass D, Harrison SC, Kim PS. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 39.Rosenthal PB, et al. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama T, et al. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parren PWHI, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and post-exposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J. Virol. 2002;76:6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oswald WB, et al. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3:62–66. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama T, et al. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 1999;179(Suppl 1):S235–239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie GE. D.Phil. thesis. University of Oxford; 2005. [Google Scholar]
- 45.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 47.Szakonyi G, et al. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat. Struct. Mol. Biol. 2006;13:996–1001. doi: 10.1038/nsmb1161. [DOI] [PubMed] [Google Scholar]
- 48.Brindley MA, et al. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 2007;81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006;121:205–214. doi: 10.1016/j.virusres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Yang ZY, et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 52.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McRee DM. Practical Protein Crystallography. Academic Press; San Diego: 1993. [Google Scholar]
- 55.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Meth. Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 56.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 57.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Brünger AT, Krukowski A, Erickson JW. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. 1990;A47:195–204. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 60.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 62.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 63.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lutteke T, Frank M, von der Lieth CW. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr Res. 2004;339:1015–1020. doi: 10.1016/j.carres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 65.Lutteke T, von der Lieth CW. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics. 2004;5:69. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lutteke T, Frank M, von der Lieth CW. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeLano WL. The PyMol Molecular Graphics System. 2002. [Google Scholar]
- 68.Bohne-Lang A, von der Lieth CW. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 2005;33:W214–219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.