Abstract
Mutations within the BRCA1 tumor suppressor gene occur frequently in familial epithelial ovarian carcinomas but they are a rare event in the much more prevalent sporadic form of the disease. However, decreased BRCA1 expression occurs frequently in sporadic tumors, and the magnitude of this decrease has been correlated with increased disease progression. The near absence of somatic mutations consequently suggests that there are alternative mechanisms that may contribute to the observed loss of BRCA1 in sporadic tumors. Indeed, both allelic loss at the BRCA1 locus and epigenetic hypermethylation of the BRCA1 promoter play an important role in BRCA1 down-regulation; yet these mechanisms alone or in combination do not always account for the reduced BRCA1 expression. Alternatively, misregulation of specific upstream factors that control BRCA1 transcription may be a crucial means by which BRCA1 is lost. Therefore, determining how regulators of BRCA1 expression may be co-opted during sporadic ovarian tumorigenesis will lead to a better understanding of ovarian cancer etiology and it may help foster the future development of novel therapeutic strategies aimed at halting ovarian tumor progression.
Introduction
Epithelial ovarian cancer is the most lethal of all gynecological malignancies [1]. The poor survival associated with ovarian carcinoma is due, at least in part, to the fact that the disease is usually asymptomatic in its early stages. As a result, detection often occurs at a late, metastatic stage when the prognosis is poor. While the etiology of ovarian carcinogenesis is poorly understood, evidence from histopathological studies and recently developed mouse models of ovarian cancer progression suggest that the majority of the tumors originate from the ovarian surface epithelium (OSE), a simple cuboidal layer that covers the surface of the ovary [2-5]. It remains unclear at this time, however, if a predictable progression of molecular events within the OSE gives rise to a well-defined neoplastic precursor that can be used to improve early detection and diagnosis. Changes in a number of genes, including p53, k-Ras, HER2/neu and c-Myc, have been implicated in ovarian carcinoma progression. However, none of these changes appear to occur in a stage-specific manner [6,7]. While global gene profiling approaches have recently identified a number of genes that are differentially expressed in epithelial ovarian cancer these alterations have not yet been be fully characterized with respect to stage, grade or functional importance [8-10]. Thus, to date, the most compelling target gene linked to the development of ovarian cancer continues to be the breast and ovarian cancer susceptibility gene 1 (BRCA1).
The protein products of the BRCA1 gene regulate, at least in part, transcriptional activation, DNA repair, cell-cycle checkpoint control, and chromosomal re-modeling [11]. Such multi-faceted contributions to essential cellular functions imply a truly fundamental role for BRCA1 in normal development but they also confound our understanding of its role in tumorigenesis [12]. This confusion was initially compounded by the finding that complete BRCA1 ablation in transgenic mice blocks embryonic proliferation [13,14]. However, the subsequent generation of a targeted knockout in the mouse mammary epithelium did result in tumor formation, which is direct experimental evidence that BRCA1 can act as a tumor suppressor in a susceptible tissue [15]. While a BRCA1 knockout has not yet been targeted to the OSE there is compelling clinical evidence that the gene is also a tumor suppressor in the ovary. Drawing on parallels with the situation in the breast, this review will focus on the possible means by which a non-mutational suppression of BRCA1 can be achieved in highly prevalent, non-familial, sporadic epithelial ovarian carcinoma.
BRCA1 in familial and sporadic tumors
BRCA1 was originally isolated using positional cloning techniques and inactivating mutations were found in families with a high incidence of breast and ovarian cancer [16]. Specifically, germline alterations in the BRCA1 gene result in a predisposed likelihood of developing early-onset breast and ovarian cancer with a dominant penetrance as high as 85% and 65% respectively [17]. Tumorigenicity only occurs in these familial BRCA1 heterozygotes if there is also a loss of the second wild type BRCA1 allele. The latter observation supports the notion that the original germline BRCA1 mutation acts recessively at the cellular level [18-20]. Although the presence of an inherited mutation in one BRCA1 allele continues to be one of the best-defined overall risk factors for the development of breast or ovarian cancer, these familial mutations, together with familial BRCA2 mutations, occur in less than 10% of all diagnosed cases [21,22]. The great majority of breast and ovarian carcinomas arise sporadically where inherited BRCA1 mutations do not occur. In addition, somatic BRCA1 mutations are virtually undetectable in sporadic breast cancers and they are extremely rare in sporadic ovarian cancers [19,23-26]. Thus, at first glance, it would not be unreasonable to conclude that BRCA1 does not play a significant role in sporadic tumor development. However, mounting evidence suggests that alternative, non-mutational, mechanisms may suppress BRCA1 expression in these tumors [27].
The first group to conclude that BRCA1 may be important in sporadic tumor development observed a significant decrease in BRCA1 mRNA in high grade, invasive breast tumors [28]. The same investigators also demonstrated that a suppression of BRCA1 activity has functional consequences in cultured breast carcinoma cells [28]. Since then, a number of studies have confirmed the association between decreased BRCA1 mRNA and sporadic tumorigenesis in both the breast and the ovary [29-31]. Furthermore, immunohistochemical analyses of BRCA1 expression in sporadic breast and ovarian cancers revealed a significant reduction in BRCA1 protein [32,33]. This suppression of BRCA1 expression appears to be achieved through multiple means. For example, loss of heterozygosity (LOH) at the BRCA1 locus occurs in a significant proportion of sporadic of ovarian tumors [19,34]. Additionally, a comprehensive study by Russell et al. [35] found that 44% of the tumors had BRCA1 allelic loss, yet, strikingly, this event did not account for the loss of BRCA1 protein expression. Finally, 18% of the tumors exhibited a complete loss of BRCA1 protein in the absence of both LOH and allelic loss [35]. Taken together, these data indicate that epigenetic misregulation also contributes to the reduction of BRCA1 expression in sporadic tumors.
Epigenetic regulation of BRCA1
Hypermethylation of the BRCA1 promoter
Promoter hypermethylation is used during normal development to epigenetically downregulate gene expression in a tissue-specific manner. Methylation of the DNA occurs most frequently on the 5' cytosine residues within 5'-CpG-3' di-nucleotides, which often cluster together in CpG islands that can stretch for several kilobases [36]. In actively transcribed genes, CpG islands within regulatory regions are often unmethylated. In contrast, methylation at these sites represses transcription by altering chromatin structure such that the transcriptional machinery does not have proper access to functionally important regions of the promoter [37]. The proximal BRCA1 promoter lies within such a methylation-sensitive island and a developmentally inappropriate hypermethylation of the promoter does occur in some sporadic breast and ovarian tumors [38,39]. This hypermethylation may be functionally significant as it correlates with decreased BRCA1 mRNA [40]. An important question, yet to be answered, is whether such an abnormal promoter hypermethylation is the cause or consequence of an initial transcriptional repression [41]. Regardless, promoter hypermethylation has only been found in tumors where BRCA1 LOH has also occurred [40]. Thus, hypermethylation may serve as an epigenetic 'second hit' that inactivates the remaining wild type BRCA1 allele after LOH has occurred. While inappropriate promoter hypermethylation is very likely a powerful repressor of BRCA1 expression it is important to point out that it only occurs in a small subset of sporadic tumors [42,43].
Transcriptional regulation of BRCA1
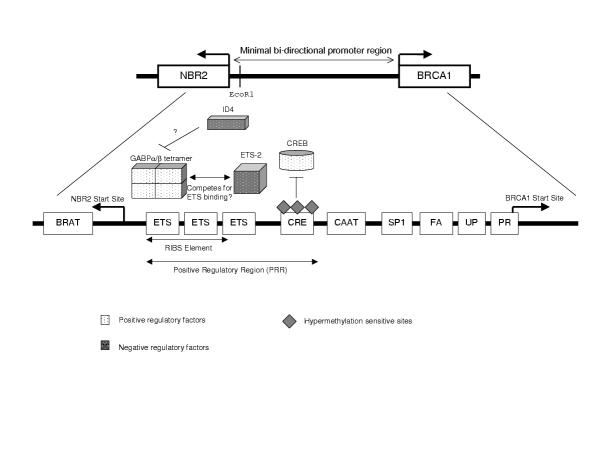
The primary proximal BRCA1 promoter, which consists of less than 300 base pairs (bp), lies immediately upstream of the major breast-specific transcription start site located within the gene's first exon [44,45]. The regulation of this promoter is complex and a number of candidate regulatory sites have been identified and partially characterized (Fig 1). One of these elements, the positive regulatory region (PRR) at the 5' end of the promoter is both necessary and sufficient to maximally activate BRCA1 transcription [46]. Of particular interest, one putative regulatory site located within the PRR is a cyclic-AMP response element, which is capable of specifically binding to CREB, the cyclic-AMP response element binding protein [47]. Importantly, this element is a site of frequent hypermethylation in breast and ovarian tumors [48] and experimental methylation of this site decreases BRCA1 promoter activity in vitro [49]. Mutation of the CREB-binding consensus sequence within the context of an intact BRCA1 promoter [49] also causes a significant reduction in promoter activation in non-tumorigenic human OSE cells in culture [50]. Taken together, these findings indicate that the cAMP-response element is very likely an important positive regulator of BRCA1 expression in both the normal OSE and in tumor tissue. As an initial transcriptional repression is capable of contributing to the later hypermethylation of sensitive sites [41] misregulation of the transcriptional complex that binds to the cAMP-response element in the BRCA1 promoter could contribute to sporadic ovarian carcinoma development. Therefore, a careful examination of the signal transduction pathways that influence the activity and/or binding of transcriptional complexes to the cAMP-response element in the BRCA1 promoter may identify potentially important oncogenic events in sporadic ovarian carcinoma development.
Figure 1.
Schematic of the proximal BRCA1 promoter and the 8 functional sites that have been well characterized in breast cells. The RIBS and CREB sites have recently been partially characterized in ovarian surface epithelial and in ovarian carcinoma cells (see text for details).
It is conceivable that a number of the other transcriptional regulators of the BRCA1 promoter may also play a role in sporadic ovarian and breast carcinogenesis [27]. One such example is the GA-binding protein α/β (GABPα/β) which is a member of the ETS family of transcription factors [51]. GABPα/β specifically binds to three consecutive ETS factor-binding domains (described as the RIBS element) located immediately upstream of the CRE element in the positive response region (Fig 1) [52]. Transient overexpression of GABPα/β in breast carcinoma cells is able to stimulate BRCA1 promoter transactivation, thus demonstrating that it is potentially a direct positive regulator of BRCA1 expression [52]. Thus, loss of GABPα/β or lost responsiveness to GABPα/β-containing transcription factor complexes could result in decreased BRCA1 expression in sporadic tumors. Indeed, the promoter binding ability and transcriptional induction by GABPα/β is significantly reduced in mammary carcinoma cells. Comparatively, in the ovary the GABPα/β-binding RIBS element is highly active in normal OSE cells but not in ovarian carcinoma cells [50]. This intriguing observation suggests that the RIBS element, and perhaps aberrant function of the factors responsible for its regulation, may be specifically important in the repression of BRCA1 in sporadic ovarian tumors.
Other DNA-binding proteins that may act on the RIBS element in the BRCA1 promoter are ID4 and ETS-2. ID4 is a recognized repressor of BRCA1 expression in both breast and ovarian carcinoma cell lines [53] that negatively regulates transcription by forming heterodimers with transcription factors through their helix-loop-helix domain [54]. ETS-2 is a member of the ETS family of transcription factors related to GABPα/β that also directly binds to the BRCA1 promoter in the RIBS domain [55]. Unlike GABPα/β, ETS-2 overexpression represses BRCA1 transcription which raises the possibility that multiple ETS factors may compete for binding within the RIBS domain such that upstream signaling pathways which differentially modulate the activation of these transcriptional complexes may be the critical regulators of BRCA1 expression.
Conclusion
Transcriptional suppression of the BRCA1 promoter occurs in sporadic ovarian carcinoma. Clearly, hypermethylation of the promoter plays an important role in this process. In addition, alterations in the transcription factor complexes that bind to the promoter also have a role to play, either upstream of hypermethylation or independent of it. Therefore, the future identification of critical regulators of these complexes will point the way to functionally important signaling pathways that are co-opted during sporadic ovarian tumor formation and progression.
Acknowledgments
Acknowledgements
Work in the authors' laboratories is supported by grants from the Canadian Institutes of Health Research (C.D.R.) and the US Army Breast Cancer Research Program (C.R.M.; #DAMD17-01-1-0381). M.L.M. is a recipient of the Evelyn Martin Pre-doctoral Research Scholarship.
Contributor Information
Marcia L McCoy, Email: mmccoy@interchange.ubc.ca.
Christopher R Mueller, Email: mueller@post.queensu.ca.
Calvin D Roskelley, Email: roskelly@interchange.ubc.ca.
References
- Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- Scully RE. Pathology of ovarian cancer precursors. J Cell Biochem. 1995;23:208–218. doi: 10.1002/jcb.240590928. [DOI] [PubMed] [Google Scholar]
- Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38:87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/S1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, Harris SS, Vanderhyden BC, Hamilton TC. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–1397. [PubMed] [Google Scholar]
- Aunoble B, Sanches R, Didier E, Bignon YJ. Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer. Int J Oncol. 2000;16:567–576. doi: 10.3892/ijo.16.3.567. [DOI] [PubMed] [Google Scholar]
- Wenham RM, Lancaster JM, Berchuck A. Molecular aspects of ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2002;16:483–497. doi: 10.1053/beog.2002.0298. [DOI] [PubMed] [Google Scholar]
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up-regulated genes in ovarian cancer. Cancer Res. 2001;61:3869–3876. [PubMed] [Google Scholar]
- Sawiris GP, Sherman-Baust CA, Becker KG, Cheadle C, Teichberg D, Morin PJ. Development of a highly specialized cDNA array for the study and diagnosis of epithelial ovarian cancer. Cancer Res. 2002;62:2923–2928. [PubMed] [Google Scholar]
- Kerr P, Ashworth A. New complexities for BRCA1 and BRCA2. Curr Biol. 2001;11:R668–676. doi: 10.1016/S0960-9822(01)00389-X. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Hakem R, de la Pompa JL, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, Firpo E, Hui CC, Roberts J, Rossant J, Mak TW. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of BRCA1 early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of BRCA1 in mammary epithelial cells results in blunted ductal morphogenesis and tumor formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- Berchuck A, Heron KA, Carney ME, Lancaster JM, Fraser EG, Vinson VL, Deffenbaugh AM, Miron A, Marks JR, Futreal PA, Frank TS. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998;4:2433–2437. [PubMed] [Google Scholar]
- Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, Jack E, Vesprini DJ, Kuperstein G, Abrahamson JL, Fan I, Wong B, Narod SA. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003;97:2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- Hosking L, Trowsdale J, Nicolai H, Solomon E, Foulkes W, Stamp G, Signer E, Jeffreys A. A somatic BRCA1 mutation in an ovarian tumor. Nat Genet. 1995;9:343–344. doi: 10.1038/ng0495-343. [DOI] [PubMed] [Google Scholar]
- Merajver SD, Pham TM, Caduff RF, Chen M, Poy EL, Cooney KA, Weber BL, Collins FS, Johnston C, Frank TS. Somatic mutations in the BRCA1 gene in sporadic ovarian tumors. Nat Genet. 1995;9:439–443. doi: 10.1038/ng0495-439. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Behbakht K, McGovern PE, Chiu HC, Couch FJ, Weber BL, Friedman LS, King MC, Furusato M, LiVolsi VA, et al. Mutation analysis of the BRCA1 gene in ovarian cancers. Cancer Res. 1995;55:2998–3002. [PubMed] [Google Scholar]
- Khoo US, Ozcelik H, Cheung AN, Chow LW, Ngan HY, Done SJ, Liang AC, Chan VW, Au GK, Ng WF, Poon CS, Leung YF, Loong F, Ip P, Chan GS, Andrulis IL, Lu J, Ho FC. Somatic mutations in the BRCA1 gene in Chinese sporadic breast and ovarian cancer. Oncogene. 1999;18:4643–4646. doi: 10.1038/sj.onc.1202847. [DOI] [PubMed] [Google Scholar]
- Mueller CR, Roskelley CD. Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res. 2002;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- Taylor J, Lymboura M, Pace PE, A'hern RP, Desai AJ, Shousha S, Coombes RC, Ali S. An important role for BRCA1 in breast cancer progression is indicated by its loss in a large proportion of non-familial breast cancers. Int J Cancer. 1998;79:334–342. doi: 10.1002/(SICI)1097-0215(19980821)79:4<334::AID-IJC5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Honda K, Inamoto T, Shinohara H, Yamauchi A, Suga K, Okuyama T, Shimada T, Kodama H, Noguchi S, Gazdar AF, Yamaoka Y, Takahashi R. Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss in BRCA1-associated cases. Clin Cancer Res. 1999;5:1249–1261. [PubMed] [Google Scholar]
- Seery LT, Knowlden JM, Gee JM, Robertson JF, Kenny FS, Ellis IO, Nicholson RI. BRCA1 expression levels predict distant metastasis of sporadic breast cancers. Int J Cancer. 1999;84:258–262. doi: 10.1002/(SICI)1097-0215(19990621)84:3<258::AID-IJC10>3.3.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D, Zuch RH, Kanter MH, Cohen S, Calzone FJ, Slamon DJ. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- Zheng W, Luo F, Lu JJ, Baltayan A, Press MF, Zhang ZF, Pike MC. Reduction of BRCA1 expression in sporadic ovarian cancer. Gynecol Oncol. 2000;76:294–300. doi: 10.1006/gyno.1999.5664. [DOI] [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Russell PA, Pharoah PD, De Foy K, Ramus SJ, Symmonds I, Wilson A, Scott I, Ponder BA, Gayther SA. Frequent loss of BRCA1 mRNA and protein expression in sporadic ovarian cancers. Int J Cancer. 2000;87:317–321. doi: 10.1002/1097-0215(20000801)87:3<317::AID-IJC2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/S0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- Bianco T, Chenevix-Trench G, Walsh DC, Cooper JE, Dobrovic A. Tumor-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21:147–51. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- Catteau A, Morris JR. BRCA1 methylation: a significant role in tumor development? Semin Cancer Biol. 2002;12:359–371. doi: 10.1016/S1044-579X(02)00056-1. [DOI] [PubMed] [Google Scholar]
- Chan KY, Ozcelik H, Cheung AN, Ngan HY, Khoo US. Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 2002;62:4151–4156. [PubMed] [Google Scholar]
- Baylin SB. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin Cancer Biol. 2002;12:331–337. doi: 10.1016/S1044-579X(02)00053-6. [DOI] [PubMed] [Google Scholar]
- Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18:1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, Karlan BY. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60:5329–5333. [PubMed] [Google Scholar]
- Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- Suen TC, Goss PE. Identification of a novel transcriptional repressor element located in the first intron of the human BRCA1 gene. Oncogene. 2001;20:440–450. doi: 10.1038/sj.onc.1204078. [DOI] [PubMed] [Google Scholar]
- Thakur S, Nakamura T, Calin G, Russo A, Tamburrino JF, Shimizu M, Baldassarre G, Battista S, Fusco A, Wassell RP, Dubois G, Alder H, Croce CM. Regulation of BRCA1 Transcription by Specific Single-Stranded DNA Binding Factors. Mol Cell Biol. 2003;23:3774–3787. doi: 10.1128/MCB.23.11.3774-3787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Mueller CR. A CREB site in the BRCA1 proximal promoter acts as a constitutive transcriptional element. Oncogene. 2001;20:7110–7114. doi: 10.1038/sj.onc.1204890. [DOI] [PubMed] [Google Scholar]
- Mancini DN, Rodenhiser DI, Ainsworth PJ, O'Malley FP, Singh SM, Xing W, Archer TK. CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- DiNardo DN, Butcher DT, Robinson DP, Archer TK, Rodenhiser DI. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20:5331–5340. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- McCoy ML, Wu C, Mueller CR, Roskelley CD. Differential regulation of the BRCA1 promoter in normal ovarian surface epithelia and ovarian carcinoma cells [abstract] Proc American Ass'n Cancer Res. 2003;44:4850a. [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Whiskin K, Mueller CR. GA-binding protein alpha/beta is a critical regulator of the BRCA1 promoter. Oncogene. 2000;19:1933–1940. doi: 10.1038/sj.onc.1203516. [DOI] [PubMed] [Google Scholar]
- Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Smith AP, Desprez PY. Id proteins in epithelial cells. Exp Cell Res. 2003;285:131–45. doi: 10.1016/S0014-4827(03)00014-4. [DOI] [PubMed] [Google Scholar]
- Baker KM, Wei G, Schaffner AE, Ostrowski MC. Ets-2 and Components of Mammalian SWI/SNF Form a Repressor Complex That Negatively Regulates the BRCA1 Promoter. J Biol Chem. 2003;278:17876–17884. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]