Abstract
Freeze-fracture electron microscopy was used to study the structure of a human neuronal glutamate transporter (EAAT3). EAAT3 was expressed in Xenopus laevis oocytes, and its function was correlated with the total number of transporters in the plasma membrane of the same cells. Function was assayed as the maximum charge moved in response to a series of transmembrane voltage pulses. The number of transporters in the plasma membrane was determined from the density of a distinct 10-nm freeze-fracture particle, which appeared in the protoplasmic face only after EAAT3 expression. The linear correlation between EAAT3 maximum carrier-mediated charge and the total number of the 10-nm particles suggested that this particle represented functional EAAT3 in the plasma membrane. The cross-sectional area of EAAT3 in the plasma membrane (48 ± 5 nm2) predicted 35 ± 3 transmembrane α-helices in the transporter complex. This information along with secondary structure models (6–10 transmembrane α-helices) suggested an oligomeric state for EAAT3. EAAT3 particles were pentagonal in shape in which five domains could be identified. They exhibited fivefold symmetry because they appeared as equilateral pentagons and the angle at the vertices was 110°. Each domain appeared to contribute to an extracellular mass that projects ≈3 nm into the extracellular space. Projections from all five domains taper toward an axis passing through the center of the pentagon, giving the transporter complex the appearance of a penton-based pyramid. The pentameric structure of EAAT3 offers new insights into its function as both a glutamate transporter and a glutamate-gated chloride channel.
Glutamate transporters belong to a family of Na+- and K+-dependent transporters that are responsible for the transport of glutamate and aspartate into cells from bacteria to man (1–3). Several human isoforms have been cloned (excitatory amino acid transporters; EAAT1–5), and are found in the plasma membrane of neurons and/or glia throughout the central nervous system (4–7), and homologs are found in non-neuronal tissues (8). In the central nervous system, these transporters maintain low resting extracellular levels of glutamate/aspartate, thereby preventing the neurotoxic effects of high excitatory neurotransmitter levels, and during synaptic neurotransmission, they regulate the concentration of neurotransmitters at the synapse, leading to the modulation of the response of the postsynaptic cells (9–13).
The functional properties of the glutamate transporters have been studied extensively (14–20). These proteins function both as glutamate transporters and as glutamate-gated chloride channels. Structural studies of the glutamate transporters have been limited to unraveling the secondary structure, and several models have been proposed containing 6–10 transmembrane α-helices (21–24). The tertiary and quaternary structures of the glutamate transporters are unknown. Here, we have used freeze-fracture electron microscopy (25, 26) to examine the quaternary structure of the neuronal excitatory amino acid transporter-3 (EAAT3) (4). Our approach takes advantage of the ability of Xenopus laevis oocytes to express a large number of copies of functional integral membrane proteins in the plasma membrane, where their structure and function may be examined (27–33). We report that functional EAAT3 is a pentamer in the oocyte plasma membrane and contains an external domain that projects ≈3 nm into the extracellular space.
Experimental Procedures
Expression of EAAT3 in Xenopus Oocytes.
Stage V–VI Xenopus laevis oocytes were injected with 50 nl of water or cRNA for human EAAT3 (1 μg/μl) (4). Oocytes were maintained in Barth's medium at 18°C for 1–4 days until used in experiments. For all oocytes, the electrophysiological assay preceded the freeze-fracture studies.
Electrophysiological Assay of EAAT3.
Electrophysiological recordings were carried out using the two-electrode voltage clamp technique at 21 ± 1°C (34). EAAT3 function in the plasma membrane was assayed as the maximum carrier-mediated charge (Qmax) moved in response to a series of transmembrane voltage pulses (14, 31, 34). Whole-cell capacitance also was measured in these cells to estimate the total plasma membrane surface area (1 μF/cm2) (27–29).
Freeze-Fracture and Electron Microscopy.
After Qmax measurements, oocytes were immediately fixed in 3% glutaraldehyde in 0.2 M sodium cacodylate (pH 7.35), frozen by immersion in liquid propane, and stored in a liquid nitrogen freezer (27). Samples were fractured at −105 or −150°C in a JEOL RFD-9010CR freeze etch fracture system at ≤10−8 Torr. The fractured faces were replicated with platinum-carbon (Pt-C) at 80°, and the replica was stabilized with carbon at 90° (−150°C). Replicas were cleaned in sodium hypochlorite, washed in water, and placed directly on 400-mesh copper grids. Replicas were examined in a Zeiss 10C transmission electron microscope at 80 or 100 kV. Tilted images were obtained by using a goniometer stage.
Freeze-Fracture Data Analysis.
Twenty replicas from 15 EAAT3-expressing oocytes were examined for this study. Using these replicas, >500 negative images of plasma membrane fracture faces were obtained at ×50,000–×80,000. Similar to other recombinant membrane proteins expressed in oocytes, EAAT3 partitioned only to the protoplasmic (P) face of the plasma membrane (27–33) and, therefore, our study focused on the particles of the P fracture face. For EAAT3 particle density determinations, particles were sampled from six replicas from five oocytes. Images of P fracture faces were enlarged to a final magnification of ×150,000 and digitized. Particle densities were determined by counting P face particles from known areas of the membrane (image software, NIH). For each oocyte, a minimum of 4,000 particles were counted from a minimum of 5 μm2 of plasma membrane. For construction of the frequency histograms, particle diameter was measured directly from the negatives by using a comparator (Nikon, Model 6c) at a final magnification of ×1,000,000. The diameter measurements were plotted in a frequency histogram with a bin size of 0.5 nm and were fitted to a multiple Gaussian function (Fig. 2B) (32). All values are reported as mean ± SE.
Figure 2.
Quantitative analysis of EAAT3 expression in the plasma membrane. (A) Functional expression of EAAT3 in the plasma membrane was ascertained by measurement of the Qmax in response to step jumps in membrane voltage. In the same oocytes, the total number of EAAT3 particles in the plasma membrane was estimated (see text). The straight line is a linear regression through the data points with a slope of 7 × 10−19 C/EAAT3 particle. The effective valence per EAAT3 particle was 4 ± 1 elementary charges. In view of the pentameric structure (see text), the effective valence per monomer is ≈0.8 elementary charge. (B) Size frequency histogram of P face particles of EAAT3-expressing oocytes (n = 518). There were two particle populations: one had a mean diameter of 7.5 ± 0.4 nm corresponding to endogenous membrane proteins (hatched region), and the second had a mean diameter of 10.0 ± 0.3 nm and was present only in EAAT3-expressing oocytes. (C) The cross-sectional area of EAAT3 allows an estimation of the total number of membrane-spanning α-helices in the transporter complex. ○, The area of opsin, connexin50 (Cx50) and aquaporin-1 (AQP1) (adapted from ref. 32). The solid straight line is a linear regression through the open circles. The broken straight line is the same linear regression extrapolated beyond the known values. The cross-sectional area of EAAT3 predicted an integral membrane protein complex composed of 35 ± 3 transmembrane α-helices (●).
To study the structure of EAAT3 particles, the negatives were examined by using a comparator at ×1,000,000. Regions of interest were then photographically reproduced to yield ≈1500 prints of EAAT3 particles. From this pool, >10,000 EAAT3 particles were closely examined to ascertain the details of the proposed structural model. The dimensions of EAAT3 reported in Fig. 6 were obtained from ≈200 particles viewed at high angles (80–90°) and have been corrected for the thickness of the Pt-C coat. The area of individual EAAT3 particles was determined from digitized images by using NIH image. Fivefold symmetrization of the particle shown in Fig. 5 was performed with the imagic analysis program (Image Science, Berlin) (35). All other images shown are unprocessed images of individual EAAT3 particles in the plasma membrane.
Figure 6.
Dimensions of the neuronal glutamate transporter (EAAT3). The top and side views of the EAAT3 pentameric complex are shown. All reported values correspond to actual dimensions and were derived after correction for the thickness of the platinum-carbon coat. The dashed line in the side view indicates the position of the plane of the plasma membrane.
Figure 5.
High-magnification images of EAAT3. Upper shows four EAAT3 particles viewed at different angles. Lower shows the corresponding interpretive model of EAAT3 at the appropriate viewing angle of the particle. The first particle from the left is a fivefold symmetrized EAAT3 particle. This image was obtained by applying five consecutive 72° rotational symmetry transformations about an axis passing through the central feature and perpendicular to the plane of the membrane. The raw image of this symmetrized particle is shown in Fig. 3 (≈90°). In the model, the plane of the lipid bilayer is shown for clarity (Lower), but it is emphasized that for particles in the P face, the outer lipid leaflet is removed during freeze-fracture exposing the hydrophobic core of the membrane. (Scale bar = 10 nm.)
Platinum-Carbon Grain Size.
To estimate the resolution of the freeze-fracture images, the size of the Pt-C grain was determined by using images of ice surfaces and hydrophobic fracture faces obtained at a defocus value of 250 nm (first minimum of the contrast transfer function at ≈1.2 nm−1). The diameter of the grain was 1.0 ± 0.1 nm (n = 100), and the grain center-to-center spacing was 1.2 ± 0.2 nm (n = 217). This limits the structural resolution of the replicas to ≈2.5 nm (36). The center-to-center spacing is in agreement with our previous estimate of the Pt-C film thickness of 1.2 ± 0.2 nm (32) and indicates that the replicas consist of a monolayer of Pt-C grains, where each grain is composed of 2–3 platinum atoms.
Results
The plasma membrane of native Xenopus laevis oocytes is unique among cells because it contains a low density of integral membrane proteins that appear as intramembrane particles in the P face (200–350/μm2) (Fig. 1A). Expression of recombinant plasma membrane proteins increases the density of the particles in the P face (27–33). For example, in the EAAT3-expressing oocyte of Fig. 1B, the total particle density increased to 546 ± 35/μm2 (n = 4186). After correction for background density, this corresponds to an increase in particle density of ≈230/μm2 and represents particles induced by EAAT3 expression. In the oocytes used in this study, the density of EAAT3 in the plasma membrane ranged from 200 to 800/μm2. Using the total membrane surface area obtained from whole-cell capacitance measured in the same cells, the density was used to estimate the total number of EAAT3 particles in the plasma membrane. In the oocyte shown, the total plasma membrane area was estimated from the capacitance (200 nF) to be ≈20 × 106 μm2 (using 1 μF/cm2). Therefore, there were ≈4.6 × 109 EAAT3 particles in the plasma membrane. In the oocytes studied, the total number of EAAT3 particles ranged from 4.5 × 109 to 2.5 ×1010 per cell.
Figure 1.
P face freeze-fracture micrographs of the plasma membrane of a control and an EAAT3-expressing oocyte. (A) The P face of control oocytes contains ≈200–350 particles/μm2, with a mean diameter of ≈7.5 nm (32). In the oocyte shown, the particle density was 275 ± 20/μm2 (n = 4953). (B) In oocytes expressing EAAT3, a new population of ≈10-nm diameter particles appeared in the P face. In the oocyte shown, the total particle density increased to 546 ± 35/μm2 (n = 4186). EAAT3 Qmax was 3.5 nC in this cell. Some endogenous and EAAT3 particles are enclosed in circles. Cyt, cytoplasm; ES, extracellular space. Scale bar A and B = 200 nm.
The number of particles induced by EAAT3 expression was directly proportional to EAAT3 Qmax measured in the same oocytes (Fig. 2A). Qmax represents the total amount of carrier-mediated charge movement, which results from conformational changes of the transporter in response to transmembrane voltage pulses (34, 37). It is directly proportional to the maximum rate of glutamate transport and is related to the total number of functional transporter molecules in the plasma membrane according to: Qmax = Nezδ, where N is the total number of EAAT3 particles in the plasma membrane, e is the elementary charge (1.6 × 10−19 C), and zδ is the effective valence of the transporter corresponding to a given number of elementary charges (z) moving across a given fraction (δ) of the membrane electric field (14, 27, 34). In EAAT3-expressing oocytes, Qmax ranged from 3 to 15 nC. The plot of Qmax vs. the number of EAAT3 particles in the same cells yielded a straight line and had a slope of 7 × 10−19 C (Fig. 2A), corresponding to an effective valence of 4 ± 1 elementary charges per EAAT3 particle. Similar values have been found for the Na+/glucose and Na+/iodide cotransporters (27, 31).
Having established that the number of particles induced by EAAT3 expression was correlated with the function of the transporter (Qmax), we performed a size-analysis on the freeze-fracture particles of the P face of control and EAAT3-expressing oocytes. In control oocytes, the great majority (≈95%) of the P face endogenous integral membrane proteins have a mean diameter of ≈7.5 nm (32). The P face of EAAT3-expressing oocytes contained two particle populations (Fig. 2B): one with a mean diameter of 7.5 ± 0.4 nm and another with a mean diameter of 10.0 ± 0.3 nm (n = 518). The 7.5-nm particle population represents the endogenous plasma membrane integral proteins, as this population also is found in control cells (32). In the EAAT3-expressing oocyte of Fig. 2B, the relative frequency of this population (27% of total) predicted a ≈3.7-fold increase in particle density and was consistent with the measured increase in the density of P face particles in the same cell (≈1150/μm2; ≈3.7-fold higher than background) (32). The 10-nm particle appeared only after EAAT3 expression, and its total number in the plasma membrane was proportional to EAAT3 Qmax (Fig. 2A).
The cross-sectional area of the freeze-fracture particles of integral membrane proteins is proportional to the number of membrane-spanning α-helices. The relationship was established by an analysis of the freeze-fracture particles of proteins of known structure (opsin, aquaporin-1, and a connexin), and it was found that on average each transmembrane α-helix occupies 1.4 nm2 in the plane of the membrane (32). The cross-sectional area of EAAT3 was calculated either from the diameter obtained from the frequency histogram (Fig. 2B) (assuming circular cross-sectional geometry), or it was directly measured in ≈100 EAAT3 particles viewed at 80–90° (see Fig. 3). After correction for Pt-C film thickness, both area estimates yielded similar results (45 ± 5 vs. 48 ± 5 nm2; P > 0.05). Using 1.4 nm2/α-helix, the area corresponds to that of an integral membrane protein complex containing 35 ± 3 transmembrane α-helices (Fig. 2C).
Figure 3.
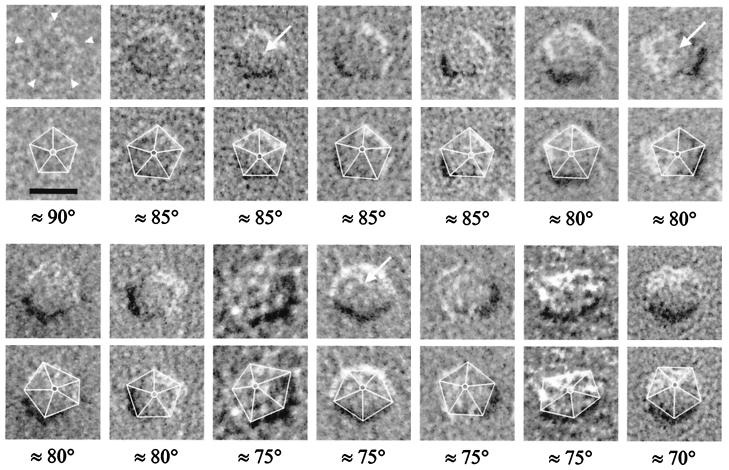
EAAT3 is pentagonal in the plane of the plasma membrane. Fourteen representative EAAT3 particles are shown in the P face of the plasma membrane. In many particles, five “domains” (arrowheads), and a central feature (arrows) were observed. Below each image, the pentagonal structure of the same particle is emphasized by superimposing the outline of the EAAT3 complex, as well as the center-to-vertex connections within the pentagonal arrangement. The viewing angle for each particle was estimated from the height and position of the central feature in the pentameric complex (see text). Angles are reported with respect to the plane of the plasma membrane (considered to be the horizontal). (Scale bar = 10 nm.)
We then examined the shape of individual EAAT3 particles in more detail. EAAT3 particles uniformly exhibited a pentagonal shape, in which five “domains” could be identified (arrowheads in Figs. 3 and 5). In each particle, five sides of the pentagon (6.9 ± 0.3 nm; n = 185) and five angles at the vertices (110 ± 5°; n = 185) could be measured. A central feature was present in many of the pentagonal EAAT3 particles (arrows in Fig. 3). EAAT3 particles exhibited fivefold symmetry because (i) the sides of the pentagon were equal in size (6.9 nm), (ii) the angle formed by the sides at the vertices was 110°; consistent with that expected for an equilateral pentagon (108°), and (iii) for particles viewed at ≈90° (see below), application of 72° rotational symmetry transformations to the particles along an axis passing through the center of the pentagon and perpendicular to the plane of the membrane produced identical views of the particles (see Figs. 3 and 5).
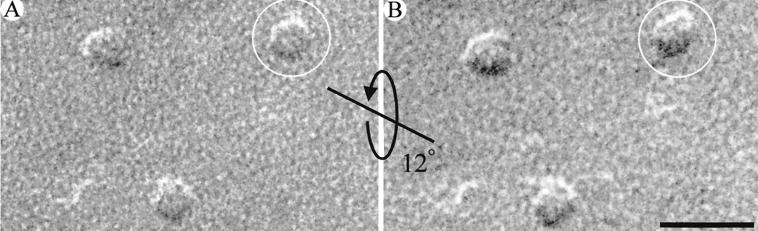
The pentagonal geometry was exhibited by the majority of EAAT3 particles. However, only a small fraction exhibited fivefold symmetry. Because the surface of the oocyte plasma membrane contains folds and microvilli (Fig. 1) (27), we reasoned that the replica is not flat and, thus, contains particles viewed at different angles depending on the local curvature of the plasma membrane. A goniometer stage was used to image individual EAAT3 particles at different tilt angles with respect to the incident electron beam. As the viewing angle of each particle was changed, there was a dramatic change in its appearance (Fig. 4). This experiment suggested that lack of fivefold symmetry in most of the particles was due to the orientation of the EAAT3 particles with respect to the electron beam. In addition, the loss of pentagonal appearance of the tilted particles suggested that the central feature within the pentagonal geometry was elevated with respect to the pentagon vertices (see below). Therefore, the absence of symmetry in most of the particles resulted from the height of the central feature and the wavy nature of the oocyte plasma membrane.
Figure 4.
Tilted EAAT3 particles. EAAT3 particles were tilted with respect to the incident electron beam. (A and B) The same particles at two different tilt angles separated by 12°. The pentagonal appearance of the particle enclosed by the circles was lost after tilting. The straight line represents the tilt axis, and the curved arrow represents the tilt direction. (Scale bar A and B = 20 nm.)
To estimate the height of the central feature of the EAAT3 particles (i.e., the distance it protrudes into the extracellular space), pairs of tilted images (12–24°) of individual particles were examined by using stereological methods (see Fig. 4) (38). By measuring the parallax difference between the central feature and one vertex of the pentagon in stereo pair images, the height of the central feature was estimated to be ≈3 nm (n = 20). Knowledge of the height of the external domain, and the realization that EAAT3 is a symmetrical pentagon in 90° projection, allowed us to estimate the viewing angle that could account for the observed projections of particles for which no tilt images were available (Fig. 3). Because of the projection of the central feature into the extracellular space, as the viewing angle approaches 90°, the fidelity of the pentagonal appearance of EAAT3 particles increases. Therefore, the apparent difference in the images of the particles results from the angle at which the particles are viewed and is not due to heterogeneity in size or shape of the transporter in the plasma membrane (Figs. 3–5).
Discussion
The combined electrophysiological and electron microscopic studies indicated that functional EAAT3 appeared as a distinct pentagonal 10-nm diameter particle in the plasma membrane of oocytes. The pentagonal 10-nm particle represents functional EAAT3 in the plasma membrane because (i) it was the only particle population that appeared in the P face of the plasma membrane after EAAT3 expression; (ii) its total number in the plasma membrane was directly correlated with EAAT3 Qmax measured in the same cells; (iii) control oocytes exhibited neither charge movement (Q) as that seen in EAAT3-expressing cells, nor a pentagonal 10-nm particle population in the plasma membrane; and (iv) expression of a wide variety of other recombinant membrane proteins does not give rise to pentagonal 10-nm particles, as each recombinant protein leads to the appearance of a distinct particle with a unique size and shape; examples include aquaporin-0 (tetramer), aquaporin-1 (tetramer), connexin50 (hexamer), opsin (monomer), epithelial Na+ channel (octamer), cystic fibrosis transmembrane conductance regulator (dimer), and Na+/glucose cotransporter (monomer) (29, 32, 33).
The cross-sectional area of integral membrane proteins can be estimated from the area of their freeze-fracture particles, and it can be used to estimate the number of membrane-spanning α-helices (32). The EAAT3 cross-sectional area was 48 ± 5 nm2, corresponding to 35 ± 3 transmembrane α-helices (using 1.4 nm2/helix). Topological models of the glutamate transporters predict 6–10 transmembrane α-helices (21–24). Despite uncertainties about the secondary structure, the large protein complex seen in the freeze-fracture images is suggestive of an oligomeric state for EAAT3, where 3–6 subunits are present in the transporter complex.
High-magnification freeze-fracture images further supported the oligomeric structure and, in addition, showed EAAT3 to be a symmetrical pentamer in the plasma membrane. The EAAT3 particle is characterized by five domains (subunits?) (Fig. 6). Each domain projects ≈3 nm into the extracellular space and tapers toward an axis passing through the center of the pentameric complex. The tapered projections from all five domains give rise to the observed central feature. The entire complex rising out of the plane of the plasma membrane and reaching into the extracellular space resembles a penton-based pyramid (Fig. 5 and 6). The transporter exhibits fivefold symmetry in 90° projections of the particles; however, as the viewing angle deviates significantly from normal, the pentagonal appearance is obscured because of the prominent external mass.
Although particle deformation caused by freezing, fracturing, and replication may lead to errors in the shape and estimated height of the external domain, that the estimated dimension (≈3 nm) is within the resolution of the replicas (≈2.5 nm) suggests that the images may be used to make predictions about the tertiary structure of EAAT3. The EAAT3 external domain inferred from the images is estimated to occupy ≈50 nm3, corresponding to a mass of ≈40 kDa (using 0.73 cm3/g). A large extracellular loop predicted to connect transmembrane domains 3 and 4 may contribute to this extracellular mass (1–6). It contains ≈94 amino acids, leading to a mass of ≈12 kDa (4), and the combined mass from five subunits would be ≈60 kDa. N-linked glycosylation at two potential sites may lead to an additional mass of 5–10 kDa (39). Thus, this loop may account for the external domain seen in the images. Whether this external mass is involved in ligand recognition and binding remains to be determined.
In view of the pentameric structure of the glutamate transporter, two questions need to be considered. First, does EAAT3 function as a pentamer? Second, is the oligomeric assembly required for secondary active transport? The first question can be addressed because images of the pentameric complex were collected from the very oocytes in which EAAT3 functional measurements were obtained. Results from radiation inactivation studies also suggest that the neuronal (EAAT1–3) and the intestinal glutamate transporters function as oligomeric assemblies (40, 41). Furthermore, recent functional studies of EAAT3 in oocytes coexpressing various ratios of wild-type and cysteine mutants (sensitive to methanethiosulfonate reagents) indicate and this transporter is a functional oligomer (J. M. Hubbard and M.P.K., unpublished data). Altogether, the data are suggestive of EAAT3 as a functional oligomer in the plasma membrane, where it is most likely composed of five identical subunits.
The second question involves the functional unit of EAAT3. Is the functional unit the pentameric complex, or is each EAAT3 monomer sufficient to perform secondary active transport? This question is more difficult to address based on the data obtained here; however, evidence from other ion-driven transporters indicates that oligomeric assembly per se is not necessary for secondary active transport. The mammalian Na+/glucose cotransporter and Escherichia coli lactose permease both function as monomeric units (32, 42). What then is the significance of the pentameric assembly in the neuronal glutamate transporter? It has been shown that in addition to their function as secondary active transporters of glutamate, these proteins behave as glutamate-activated chloride channels (5). Chloride flux through the channel is not thermodynamically coupled to glutamate transport (1, 2). It is possible that the EAAT3 monomer can perform secondary active transport, but the chloride channel mode seen in these transporters is a consequence of the oligomeric assembly. In many multimeric ion channels, the subunits surround and contribute to the lining of the ion permeation pathway. This also may be the case for the EAAT3 chloride channel.
In summary, using freeze-fracture electron microscopy, we have shown that EAAT3 exists as a functional pentamer in the plasma membrane. The transporter complex resembles a penton-based pyramid that projects ≈3 nm into the extracellular space. The oligomeric structure of EAAT3 may be responsible for its dual function as a secondary active transporter and an ion channel.
Acknowledgments
We thank D. Leung, M. Lai, and L. Zampighi for technical assistance. Supported by National Institute of Health Grants DK-19567 and DK-44602 (E.M.W.), EY-04110 (G.A.Z.), and NS-33270 (M.P.K.). S.E. is supported by a postdoctoral fellowship from a National Institutes of Health Cellular Neurobiology Training grant (T32 NS-07101–22).
Abbreviations
- EAAT
excitatory amino acid transporter
- P
protoplasmic
- Qmax
maximum carrier-mediated charge
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Masson J, Sagné C, Hamon M, El Mestikawy S. Pharmacol Rev. 1999;51:439–464. [PubMed] [Google Scholar]
- 2.Seal R P, Amara S G. Annu Rev Pharmacol Toxicol. 1999;39:431–456. doi: 10.1146/annurev.pharmtox.39.1.431. [DOI] [PubMed] [Google Scholar]
- 3.Slotboom D J, Konings W N, Lolkema J S. Microbiol Mol Biol Rev. 1999;63:293–307. doi: 10.1128/mmbr.63.2.293-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arriza J L, Fairman W A, Wadiche J I, Murdoch G H, Kavanaugh M P, Amara S G. J Neurosci. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairman W A, Vandenberg R J, Arriza J L, Kavanaugh M P, Amara S G. Nature (London) 1995;375:599–603. doi: 10.1038/375599a0. [DOI] [PubMed] [Google Scholar]
- 6.Arriza J L, Eliasof S, Kavanaugh M P, Amara S G. Proc Natl Acad Sci USA. 1997;94:4155–4160. doi: 10.1073/pnas.94.8.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danbolt N C, Chaudhry F A, Dehnes Y, Lehre K P, Levy L M, Ullensvang K, Storm-Mathisen J. Prog Brain Res. 1998;116:23–43. doi: 10.1016/s0079-6123(08)60428-8. [DOI] [PubMed] [Google Scholar]
- 8.Hediger M A. Am J Physiol. 1999;277:F487–F492. doi: 10.1152/ajprenal.1999.277.4.F487. [DOI] [PubMed] [Google Scholar]
- 9.Diamond J S, Jahr C E. J Neurosci. 1997;17:4672–4687. doi: 10.1523/JNEUROSCI.17-12-04672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otis T S, Kavanaugh M P, Jahr C E. Science. 1997;277:1515–1518. doi: 10.1126/science.277.5331.1515. [DOI] [PubMed] [Google Scholar]
- 11.Bergles D E, Jahr C E. Neuron. 1997;19:1297–1308. doi: 10.1016/s0896-6273(00)80420-1. [DOI] [PubMed] [Google Scholar]
- 12.Gaal L, Roska B, Picaud S A, Wu S M, Marc R, Werblin F S. J Neurophysiol. 1998;79:190–196. doi: 10.1152/jn.1998.79.1.190. [DOI] [PubMed] [Google Scholar]
- 13.Billups B, Rossi D, Oshima T, Warr O, Takahashi M, Sarantis M, Szatkowski M, Attwell D. Prog Brain Res. 1998;116:45–57. doi: 10.1016/s0079-6123(08)60429-x. [DOI] [PubMed] [Google Scholar]
- 14.Wadiche J I, Arriza J L, Amara S G, Kavanaugh M P. Neuron. 1995;14:1019–1027. doi: 10.1016/0896-6273(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 15.Wadiche J I, Amara S G, Kavanaugh M P. Neuron. 1995;15:721–728. doi: 10.1016/0896-6273(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 16.Billups B, Attwell D. Nature (London) 1996;379:171–174. doi: 10.1038/379171a0. [DOI] [PubMed] [Google Scholar]
- 17.Zerangue N, Kavanaugh M P. Nature (London) 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 18.Otis T S, Jahr C E. J Neurosci. 1998;15:7099–7110. doi: 10.1523/JNEUROSCI.18-18-07099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadiche J I, Kavanaugh M P. J Neurosci. 1998;18:7650–7661. doi: 10.1523/JNEUROSCI.18-19-07650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy L M, Warr O, Attwell D. J Neurosci. 1998;18:9620–9628. doi: 10.1523/JNEUROSCI.18-23-09620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahle S, Stoffel W. J Cell Biol. 1996;135:1867–1877. doi: 10.1083/jcb.135.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slotboom D J, Lolkema J S, Konings W N. J Biol Chem. 1996;271:31317–31321. doi: 10.1074/jbc.271.49.31317. [DOI] [PubMed] [Google Scholar]
- 23.Grunewald M, Bendahan A, Kanner B I. Neuron. 1998;21:623–632. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 24.Seal R P, Amara S G. Neuron. 1998;21:1487–1498. doi: 10.1016/s0896-6273(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 25.Branton D. Proc Natl Acad Sci USA. 1966;55:1048–1056. doi: 10.1073/pnas.55.5.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto da Silva P, Branton D. J Cell Biol. 1970;45:598–605. doi: 10.1083/jcb.45.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zampighi G A, Kreman M, Boorer K J, Loo D D F, Bezanilla F, Chandy G, Hall J E, Wright E M. J Membr Biol. 1995;148:65–78. doi: 10.1007/BF00234157. [DOI] [PubMed] [Google Scholar]
- 28.Chandy G, Zampighi G A, Kreman M, Hall J E. J Membr Biol. 1997;159:29–39. doi: 10.1007/s002329900266. [DOI] [PubMed] [Google Scholar]
- 29.Zampighi G A, Loo D D F, Kreman M, Eskandari S, Wright E M. J Gen Physiol. 1999;113:507–523. doi: 10.1085/jgp.113.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright E M, Hirsch J R, Loo D D F, Zampighi G A. J Exp Biol. 1997;200:287–293. doi: 10.1242/jeb.200.2.287. [DOI] [PubMed] [Google Scholar]
- 31.Eskandari S, Loo D D F, Dai G, Levy O, Wright E M, Carrasco N. J Biol Chem. 1997;272:27230–27238. doi: 10.1074/jbc.272.43.27230. [DOI] [PubMed] [Google Scholar]
- 32.Eskandari S, Wright E M, Kreman M, Starace D M, Zampighi G A. Proc Natl Acad Sci USA. 1998;95:11235–11240. doi: 10.1073/pnas.95.19.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eskandari S, Snyder P M, Kreman M, Zampighi G A, Welsh M J, Wright E M. J Biol Chem. 1999;274:27281–27286. doi: 10.1074/jbc.274.38.27281. [DOI] [PubMed] [Google Scholar]
- 34.Loo D D F, Hazama A, Supplisson S, Turk E, Wright E M. Proc Natl Acad Sci USA. 1993;90:5767–5771. doi: 10.1073/pnas.90.12.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel M, Harauz G, Orlova E V. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 36.Willison J H M, Rowe A J. Replica, Shadowing and Freeze-Etching Techniques. Amsterdam: Elsevier; 1980. [Google Scholar]
- 37.Loo D D F, Hirayama B A, Gallardo E M, Lam J T, Turk E, Wright E M. Proc Natl Acad Sci USA. 1998;95:7789–7794. doi: 10.1073/pnas.95.13.7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson B. J Microsc (Oxford) 1973;98:396–401. [Google Scholar]
- 39.Conradt M, Storck T, Stoffel W. Eur J Biochem. 1995;229:682–687. doi: 10.1111/j.1432-1033.1995.tb20514.x. [DOI] [PubMed] [Google Scholar]
- 40.Haugeto O, Ullensvang K, Levy L M, Chaudhry F A, Honoré T, Nielsen M, Lehre K P, Danbolt N C. J Biol Chem. 1996;271:27715–27722. doi: 10.1074/jbc.271.44.27715. [DOI] [PubMed] [Google Scholar]
- 41.Béliveau R, Demeule M, Jetté M, Potier M. Biochem J. 1990;268:195–200. doi: 10.1042/bj2680195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahin-Tóth M, Lawrence M C, Kaback H R. Proc Natl Acad Sci USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]