Abstract
Hepatic gluconeogenesis is a major contributing factor to hyperglycemia in the fasting and postprandial states in type 2 diabetes mellitus (T2DM). Because Sirtuin 1 (SirT1) induces hepatic gluconeogenesis during fasting through the induction of phosphoenolpyruvate carboxylase kinase (PEPCK), fructose-1,6-bisphosphatase (FBPase), and glucose-6-phosphatase (G6Pase) gene transcription, we hypothesized that reducing SirT1, by using an antisense oligonucleotide (ASO), would decrease fasting hyperglycemia in a rat model of T2DM. SirT1 ASO lowered both fasting glucose concentration and hepatic glucose production in the T2DM rat model. Whole body insulin sensitivity was also increased in the SirT1 ASO treated rats as reflected by a 25% increase in the glucose infusion rate required to maintain euglycemia during the hyperinsulinemic-euglycemic clamp and could entirely be attributed to increased suppression of hepatic glucose production by insulin. The reduction in basal and clamped rates of glucose production could in turn be attributed to decreased expression of PEPCK, FBPase, and G6Pase due to increased acetylation of signal transducer and activator of transcription 3 (STAT3), forkhead box O1 (FOXO1), and peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), known substrates of SirT1. In addition to the effects on glucose metabolism, SirT1 ASO decreased plasma total cholesterol, which was attributed to increased cholesterol uptake and export from the liver. These results indicate that inhibition of hepatic SirT1 may be an attractive approach for treatment of T2DM.
Keywords: gluconeogenesis, glucose 6 phosphatase, phosphoenolpyruvate carboxykinase, type 2 diabetes mellitus, hepatic insulin resistance
Type 2 diabetes mellitus (T2DM) is associated with an increased rate of hepatic glucose production that contributes to fasting hyperglycemia (1–3). Specifically, increased endogenous glucose production can be accounted for by increased rates of hepatic gluconeogenesis (4). Inhibition of hepatic gluconeogenesis has been shown to improve fasting plasma glucose levels and decrease endogenous glucose production in T2DM patients (5). Furthermore, inhibition of transcriptional gluconeogenic activators, such as forkhead box O1 (FOXO1), and peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), in turn improved hepatic insulin resistance in rodent models of diabetes (6, 7). Therefore inhibitors of gluconeogenesis are potentially excellent targets for treatment of poorly controlled T2DM.
Sirtuin1 (SirT1) is a NAD+-dependent deacetylase activated in response to fasting and caloric restriction (8). In the β-cells of the pancreas, SirT1 has been shown to increase insulin secretion through repression of uncoupling protein 2 (UCP2) (9). In adipose tissue, SirT1 has been shown to inhibit adipogenesis and decrease lipolysis through inhibition of peroxisome proliferator-activated receptor-γ (PPARγ) (10). In liver tissue, SirT1 activates gluconeogenesis transcription through deacetylation of PGC-1α, FOXO1, and signal transducer and activator of transcription 3 (STAT3) (8, 11, 12). SirT1 is induced by increased concentration of pyruvate, and the activity correlates with the redox state of the cell (8). Recent studies have identified novel endogenous regulators of SirT1. Active regulator of SirT1 (AROS) was determined to be an activator of SirT1 (13), and deleted in breast cancer-1 (DBC1) as an inhibitor of SirT1 (14, 15). However the role, if any, of these proteins in SirT1 regulation of hepatic gluconeogenesis has yet to be determined.
In the present study, we sought to investigate the effect of SirT1 knockdown in a T2DM rat model. To accomplish SirT1 knockdown, we used an ASO that causes specific reduction of SirT1 mRNA in liver and adipose tissue. Four weeks of fructose and high-fat feeding combined with an initial streptozotocin injection created a rat model that mimics T2DM (16). To evaluate peripheral and hepatic insulin sensitivity, we performed hyperinsulinemic-euglycemic clamps in combination within infusions of [6,6-2H] glucose and [2-14C] deoxyglucose to assess rates of whole-body glucose turnover and tissue-specific rates of glucose uptake. Because SirT1 plays a critical role in transcriptional regulation of key gluconeogenic genes, we hypothesized that knockdown of hepatic SirT1 would decrease endogenous and insulin-stimulated glucose production and reduce fasting hyperglycemia in this rat model of T2DM.
Results
SirT1 ASO Metabolic Parameters.
SirT1 ASO decreased body weight 17% (P < 0.05) on the normal chow diet and 12% (P < 0.05) in the T2DM rat model after 4 weeks of bi-weekly injections. The decreased body weight with SirT1 ASO can be explained by the 23% (P < 0.05) and 19% (P < 0.05) reductions in overnight food intake in the normal and T2DM rat model, respectively. To prevent confounding effects of differences in body weight, SirT1 ASO rats were pair-fed to minimize differences in food intake and body weight (Fig. 1A). SirT1 ASO decreased SirT1 mRNA expression by 77% in liver (Fig. 1B), and the drop in mRNA corresponded with a 68% decrease in SirT1 protein (Fig. 1C). Additionally, SirT1 ASO decreased SirT1 mRNA 91% (Fig. 1D) in the adipocyte. In addition, we found no difference in SirT1 mRNA in muscle (Fig. 1E) and a tendency of decreased SirT1 mRNA in brown adipose tissue (BAT) (Fig. 1F). ASOs have been previously shown not to distribute to muscle and not to cause reduction in gene expression in this tissue (6, 17, 18). Additionally, we tested the transcription of SirT1 in the hypothalamus as a possible explanation for the decreased food intake and found no differences in mRNA (control ASO 1.00 ± 0.14 vs. SirT1 ASO 1.08 ± 0.13) on the normal chow diet. There were no differences in plasma alanine transferase (ALT) concentrations (control ASO 59.0 ± 3.63 units/L vs. SirT1 51.5 ± 3.76 units/L) between control ASO and SirT1 ASO, indicating there was no toxicity associated with the ASO and indicating that the effect on food intake is specific to SirT1 peripheral knockdown. In addition, SirT1 ASO decreased epididymal white adipose tissue, plasma free fatty acids, leptin, and adiponectin concentrations (Table 1). The decreased epididymal white adipose tissue mass in SirT1 ASO could be attributed to a 77% and 72% reduction in peroxisome proliferator-activated receptor gamma (PPARγ) and PPARγ2 transcription respectively (Fig. 1 E and F). Consistent with these findings, CCAAT/enhancer binding protein alpha (C/EBPα) was reduced by 74% (P < 0.05) in adipose tissue. Because the western diet is rich in fructose and fat, we used 4 weeks of high-fructose combined with high-fat feeding, and a low dose of streptozotocin to decrease, but not eliminate, pancreatic β-cell function that mimics T2DM. Three weeks of high-fructose feeding has previously been shown to induce hepatic insulin resistance (19) and increased gluconeogenic gene expression (20). The T2DM model exhibited increased fasting glucose concentrations compared with normal chow-fed controls (Fig. 1I). In the T2DM rat model, SirT1 ASO had modest reductions in fasting plasma glucose (Fig. 1I), whereas insulin (Fig. 1J) and glucagon (Table 1) concentrations remain unchanged.
Fig. 1.
SirT1 ASO is effective and decreases basal glucose levels in T2DM. (A) Body weight of pair-fed rats. (B) SirT1 ASO decreased liver mRNA and (C) SirT1 hepatic protein (normalized to total STAT3). SirT1 ASO decreased WAT SirT1 (D), but not muscle SirT1 mRNA (E), and slightly decreased BAT SirT1 mRNA (F). In WAT, PPARγ (E) and PPARγ2 (F), mRNA were decreased with SirT1 ASO. Fasting basal glucose values in the T2DM rat model were decreased (H), but not basal insulin (I) or glucagon. (n = 6–19 per group.) *, P < 0.05; **, P < 0.005.
Table 1.
Metabolic profiles in control ASO and SirT1 ASO
| Metabolite and concentration | Normal chow |
T2DM rat model |
||
|---|---|---|---|---|
| Control ASO | SirT1 ASO | Control ASO | SirT1 ASO | |
| Plasma glucagon, pg/ml | n.d. | n.d. | 60.5 ± 4.6 | 56.5 ± 3.3 |
| Adiponectin, μg/ml | 2.8 ± 0.6 | 1.2 ± 0.2* | 2.9 ± 0.2 | 1.6 ± 0.2** |
| Leptin, ng/ml | 0.9 ± 0.2 | 0.3 ± 0.2* | 3.3 ± 0.5 | 1.4 ± 0.3* |
| Fatty acids, mEq/L | 0.79 ± 0.07 | 0.66 ± 0.06* | 0.85 ± 0.13 | 0.66 ± 0.08* |
| White adipose tissue, g | 1.8 ± 0.1 | 1.4 ± 0.1* | 3.1 ± 0.2 | 1.7 ± 0.2** |
| Plasma triglycerides, mg/dL | 22.5 ± 4.0 | 24.1 ± 3.9 | 31.4 ± 2.7 | 34.6 ± 2.7 |
| β-Hydroxybutyrate, mmol/L | 0.9 ± 0.1 | 0.9 ± 0.1 | 1.2 ± 0.1 | 0.9 ± 0.1* |
| Liver triglycerides, mg/g of tissue | n.d. | n.d. | 14.4 ± 1.2 | 11.8 ± 1.1 |
Data are presented as mean ± SEM (n = 6–19 per group); n.d., not determined.
*, P < 0.05;
**, P < 0.005; compared with control ASO
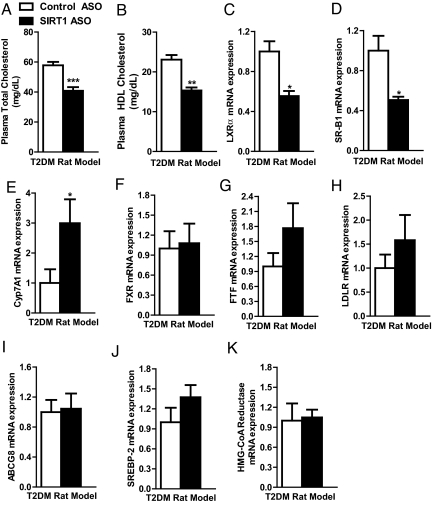
SirT1 ASO Decreases Plasma Cholesterol Concentration.
SirT1 ASO treatment did not affect plasma triglycerides (Table 1) or liver triglyceride content (Table 1). Plasma β-hydroxybutyrate concentrations were decreased 28% in the SirT1 ASO group (Table 1). SirT1 ASO reduced total plasma cholesterol 29% (Fig. 2A), which could be attributed to a 33% reduction in HDL cholesterol (Fig. 2B). SirT1 ASO down-regulated liver X receptor alpha (LXRα) transcription (Fig. 2C), an important factor in cholesterol transport and metabolism. Accordingly, the LXRα transcriptional target HDL uptake receptor (SR-B1) was also down-regulated (Fig. 2D). In contrast to previous reports, Cyp7A1 transcription was up-regulated with SirT1 ASO, indicating greater bile acid synthesis (Fig. 2E). In addition, Cyp7A1 repressors fetoprotein transcription factor (FTF) and farnesoid X receptor (FXR) were unchanged (Fig. 2 F and G). Cyp7A1 is a target of LXRα, so it is unclear at this time why Cyp7A1 is up-regulated in the SirT1 ASO group. However, this result could explain why SirT1 ASO reduces plasma cholesterol in this rodent model of T2DM. The low-density lipoprotein (LDL) receptor showed a tendency of increased transcription, and ATP binding cassette G8 (ABCG8), sterol response element-binding protein (SREBP-2), and HMG-CoA reductase were unchanged in the SirT1 ASO group (Fig. 2 H–K).
Fig. 2.
SirT1 ASO alters cholesterol metabolism. SirT1 ASO reduced both total plasma cholesterol (A) and HDL plasma cholesterol (B). Key cholesterol metabolism genes were assessed by RT-PCR (C–K). (n = 7–19 per group.) *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
SirT1 ASO Increases Hepatic Insulin Sensitivity.
SirT1 knockout mice are smaller and hypermetabolic, making conclusions regarding differences in insulin sensitivity, independent of differences in body weight, difficult to assess (21, 22). To circumvent these difficulties, we used an ASO that allowed us to acutely knockdown SirT1 in an adult rodent model of T2DM. In the T2DM rat model basal hepatic glucose production was increased 15% compared with normal rats (Fig. 3A). This increase is comparable to the percentage increase seen in patients with T2DM (1, 23). SirT1 ASO decreased basal endogenous glucose production modestly in the T2DM rat model (Fig. 3A). The decrease in basal glucose production corresponded with the reductions in the basal plasma glucose concentration (Fig. 1I). The rate at which glucose was infused to maintain euglycemia during the clamp was 25% higher in the SirT1 ASO-treated group in the T2DM rat model (Fig. 3B). The increased glucose infusion rate could be attributed to a 46% decrease in hepatic glucose production during the clamp (Fig. 3C). In contrast, SirT1 ASO treatment had no effect on peripheral insulin-stimulated whole-body glucose metabolism in both the normal and T2DM rat models (Fig. 3D). Consistent with this finding, SirT1 ASO treatment did not affect the rate of glucose uptake into either the soleus muscle (Fig. 3E) or white adipose tissue (Fig. 3F).
Fig. 3.
Peripheral and hepatic insulin responsiveness was assessed by using the hyperinsulinemic-euglycemic clamp. Basal endogenous glucose production was decreased with SirT1 ASO in the T2DM rat model (A). Overall glucose infusion rate was increased in SirT1 ASO in the T2DM rat model (B) that can be attributed to the decrease in clamp hepatic glucose production (C). Insulin-stimulated whole-body glucose metabolism (D), glucose uptake in the soleus (E), and white adipose tissue (F) was similar between groups. (n = 9–21 per group.) *, P < 0.05.
SirT1 ASO Decreases Transcription of Key Gluconeogenic Enzymes.
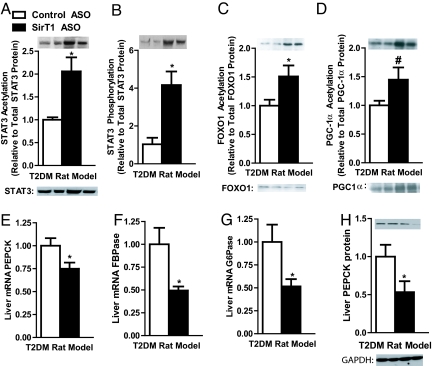
SirT1 ASO increased STAT3 acetylation more than two-fold over control ASO-treated rats (Fig. 4A) and STAT3 phosphorylation (Y705) more than three-fold compared with control ASO-treated rats (Fig. 4B). Phosphorylation is critical for STAT3 activation and translocation to the nucleus (24). Acetylation of STAT3 has been hypothesized to indirectly influence STAT3 phosphorylation (25, 26). In addition to STAT3, SirT1 ASO increased acetylation 51% and 45% in FOXO1 and PGC-1α, respectively (Fig. 4 C and D). The increased acetylation of these transcription factors was associated with 25%, 51%, and 49% decreases in phosphoenolpyruvate carboxylase kinase (PEPCK), fructose-1,6-bisphosphatase (FBPase), and glucose-6-phosphatase (G6Pase) transcription, respectively (Fig. 4 E–G). Western blot analysis also revealed a corresponding drop in PEPCK protein (Fig. 4E).
Fig. 4.
SirT1 ASO increased acetylation of key transcription factors for gluconeogenesis. STAT3 acetylation (A) and phosphorylation (B) were increased by SirT1 ASO. Foxo1 acetylation (C) and PGC-1α (D) acetylation levels were also increased with SirT1 ASO. The increased acetylation led to a decrease in transcription of PEPCK (E), FBPase (F), G6Pase (G), and PEPCK protein (H). (n = 4–8 per group.) #, P < 0.1; *, P < 0.05.
Discussion
We found that knockdown of SirT1 expression in liver by a SirT1 ASO decreased fasting plasma glucose concentrations and normalized rates of basal hepatic glucose production in a T2DM rat model. Furthermore, these changes were associated with increased hepatic insulin responsiveness. These results are consistent with a recent mouse study that showed decreased hepatic SirT1 expression, using a SirT1 small hairpin RNA (shRNA), led to improved whole body insulin sensitivity as assessed by an insulin tolerance test (11). To better assess the mechanism for this improved response to insulin, we used the hyperinsulinemic-euglycemic clamp in conjunction with stable and radiolabeled glucose isotopes to determine which tissues are responsible for the increased insulin responsiveness. By using this approach, we found that knockdown of hepatic SirT1 effectively reduces fasting hyperglycemia by decreasing basal glucose production. In addition, SirT1 ASO treatment also increases hepatic insulin sensitivity as shown by the decreased insulin-suppressed hepatic glucose production. In our model, SirT1 knockdown increased acetylation and phosphorylation of the critical transcriptional inhibitor of gluconeogenesis STAT3. Moreover, SirT1 ASO increased acetylation of PGC-1α and FOXO1. The regulation of these 3 transcriptional factors contributes to the decreased transcription of the key gluconeogenic enzymes. The decreased gluconeogenic gene expression from SirT1 knockdown differs from 2 recent papers (27, 28), possibly because of the length of fasting. The decreased transcription of key gluconeogenic genes with SirT1 ASO resulted in decreased basal glucose production and increased hepatic insulin sensitivity as assessed by the hyperinsulinemic-euglycemic clamp.
STAT3 is a potent transcriptional inhibitor of gluconeogenesis and has been shown to lower gene transcription of Pck1 and G6pc in vivo (29). Furthermore, STAT3 phosphorylation has been shown to be regulated indirectly by SirT1 deacetylation (25). SirT1 deacetylates STAT3 during fasting conditions, which is coupled to decreased STAT3 phosphorylation (25). The decreased phosphorylation of STAT3 decreases translocation to the nucleus and thus decreases STAT3 inhibition of gluconeogenesis. Knockdown of SirT1 disrupts the process of STAT3 deactivation. In addition to its effect on gluconeogenesis, recent studies have pointed to the role of STAT3 in sensitizing insulin signaling by down-regulating the Akt2 inhibitor glycogen synthase kinase-3 beta (GSK-3β) (30). This observation could also contribute to the increased hepatic insulin sensitivity seen during the hyperinsulinemic-euglycemic clamp.
SirT1 ASO decreased transcription of PPARγ, the master regulator of adipogenesis. The decreased PPARγ expression is consistent with the decreased fat mass, plasma leptin, and adiponectin concentrations found after SirT1 ASO treatment; however, this finding is in contrast with a previous study that showed activation of SirT1 repressed PPARγ (10). In rats that were not pair-fed, the differences in food intake can explain the differences in adipocyte mass. However, in pair-fed body-weight-matched rats, their adipose tissue mass was lower, indicating that SirT1 regulates adipocyte formation in a specific manner.
Recently, Milne et al. have shown that SirT1 activators improved whole body insulin sensitivity and liver insulin responsiveness in Zucker fa/fa rats (31). In contrast to these results, we found that SirT1 inhibition decreased fasting hyperglycemia and improved insulin sensitivity because of increased hepatic insulin responsiveness and decreased hepatic glucose production. In addition to the activators, mice over-expressing SirT1 showed greater hepatic insulin sensitivity (32). The authors speculated that increased plasma adiponectin increased hepatic insulin sensitivity (32). In our study, we found a reduction of plasma adiponectin concentrations, but the reduced adiponectin did not affect hepatic or peripheral insulin sensitivity. Another recent study found that SirT1 activators increased hepatic fatty acid oxidation and decreased hepatic triglyceride content, which might explain the increased hepatic insulin sensitivity that has been observed with SirT1 activators (33). However, in our model of SirT1 knockdown, we saw no differences in hepatic lipid content. Taken together these studies suggest that different mechanisms predominate to reverse hepatic insulin resistance depending on whether SirT1 is activated or inhibited.
In summary, this study demonstrates that knockdown of hepatic SirT1 expression prevents fasting hyperglycemia by decreasing hepatic glucose production and increasing hepatic insulin responsiveness in a rat model of T2DM. Because increased hepatic glucose production, due to increased hepatic gluconeogenesis, associated with hepatic insulin resistance are major factors in the pathogenesis of fasting and postprandial hyperglycemia in T2DM, novel SirT1 inhibitors targeted to liver could prove beneficial in the treatment of T2DM.
Materials and Methods
Animals.
The Institutional Animal Care and Use Committee (IACUC) of Yale University approved all procedures. To induce mild controlled diabetes, male, 150-g Sprague-Dawley rats (Charles River Laboratories) received i.p. injections of a 175 mg/kg dose of nicotinamide around 5 p.m. After 15 min, rats were i.p. injected with streptozotocin dissolved in saline at a dose of 65 mg/kg. After 3 days of recovery, afternoon blood-glucose values were determined, and the rats were randomized based on blood-glucose values before the first ASO injection. Rats were individually housed and were on a 12:12-h light/dark cycle. SirT1 ASO and control ASO solutions were prepared in normal saline and injected i.p. twice a week at a concentration of 37.5 mg/kg. During the treatment period, rats were pair-fed a combination of a high-fructose diet (60% of the calories from fructose) (TD89247; Harlan Teklad) and a high-fat diet (TD93075; Harlan Teklad).
Selection of Rat SirT1 ASO.
To identify rat SIRT antisense inhibitors, rapid-throughput screens were performed in primary rat hepatocytes. In brief, 80 ASOs were designed to the SirT1 mRNA sequence, all of which targeted a binding site within the coding region of the SirT1 mRNA. The reduction of target gene expression was analyzed with real-time quantitative RT-PCR after transfection of the cells with ASOs for 24 h. Based on target reduction, 8 ASOs were selected and further characterized in a dose-response screen. The 2 most potent ASOs from the screen were chosen, and their in vivo activity was confirmed in lean Sprague-Dawley rats. The most potent ASO was chosen as the SirT1 ASO for subsequent studies. All of the ASOs screened have a uniform phosphorothioate backbone and a 20-base chimeric design with a 2′-O-(methoxy)-ethyl (2′-MOE) modification on the first 5 and the last 5 bases. This modification enhances their binding affinity to complementary sequences and their resistance to the action of nucleases. A negative control ASO (ISIS 141923), which has the same chemical composition as the SirT1 ASO but no complementarity to any known gene sequence, was also included in the studies.
Hyperinsulinemic-Euglycemic Clamp Studies.
Before the clamp studies, catheters were inserted into the right internal jugular vein, extending to the right atrium, and left carotid artery, extending into the aortic arch. Rats were given 1 week to recover from the surgery. At 6 p.m., rats were fasted overnight and in the morning (6 a.m.), rats were infused with 99% [6,6-2H] glucose (1.1 mg/kg prime, 0.1 mg/kg) to assess the basal glucose turnover. All plasma parameters were measured after the basal glucose turnover period before the hyperinsulinemic-euglycemic portion. After the basal period, the hyperinsulinemic-euglycemic clamp was conducted for 140 min with a primed/continuous infusion of insulin (400 mU/kg primed over 5 min and 4 mU/kg per min constant infusion) and a variable infusion of 20% dextrose spiked with ≈2.5% [6,6-2H]glucose to maintain euglycemia. Once rats maintained euglycemia for 30 min, plasma samples were taken for clamp calculations. The hepatic glucose production was calculated by using the rate of infusion of [6,6-2H]glucose over the atom percent excess in the plasma minus the rate of glucose being infused. The insulin-stimulated whole-body glucose uptake was calculated by adding the total glucose infusion rate plus the hepatic glucose production. After the completion of the clamp, sodium pentobarbital was injected via the venous catheter administered at 150 mg/kg. After rats were completely anesthetized, tissues were extracted and frozen with the use of liquid cooled N2 tongs. The samples were stored at −80 °C until further analysis.
Biochemical Analysis and Calculations.
Plasma glucose values were determined by using a glucose oxidase method (Beckman Glucose Analyzer II; Beckman Coulter). Plasma insulin, glucagon, leptin, and adiponectin concentrations were determined by a RIA Assay system (Linco). The enrichment of plasma glucose was determined by using 30 μl of the designated samples that were deproteinized in 150 μl of 100% methanol. The samples were then dried overnight and derivatized with 1:1 acetic anhydride/pyridine to produce the pentacetate derivative of glucose. The atom percentage enrichment of 6,6–2H glucose was then measured by gas chromatographic/mass spectrometric analysis by using a Hewlett-Packard 5890 gas chromatograph interfaced to a Hewlett-Packard 5971A mass-selective detector, operating in the electron ionization mode (34). The atom percent excess of glucosem + 2 was determined from the m/z ratio 202:200. The adipose and muscle 14C-2-deoxyglucose uptake was determined as previously described (17).
Liver Triglycerides.
Triglycerides were extracted by using the method of Bligh and Dyer (35) and measured with the use of a commercially available triglyceride kit (DCL Triglyceride Reagent; Diagnostic Chemicals Ltd.).
Total RNA Preparation, Real-Time Quantitative RT-PCR Analysis.
RNA was extracted by using a QIAGEN RNAeasy kit (QIAGEN). The mRNA was transcribed to cDNA by using MuLV reverse transcriptase (New England Bio Labs). The abundance of transcripts was assessed by real-time PCR on a 7500 Fast Real-Time PCR System (Applied Biosystems). Each run was evaluated in duplicate for both the gene of interest and 18s (or actin) as a control. The expression data for the gene of interest and 18s (or actin) were normalized for the efficiency of amplification determined by the standard curve included for each data acquisition.
Immunoblots.
Liver was ground with a mortar and pestle and mixed with 1 ml of lysis buffer [50 mM Tris-HCl buffer (pH 7.5 at 4 °C), 50 mM NaF, 5 mM NaPPi, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM benzamidine, 1 mM phenylmethanesulfonyl fluoride (PMSF), glycerol (10% vol/vol), Triton X-100 (1% vol/vol), 1 μM TSA, and 50 mM nicotinamide] and homogenized for 30 s. Homogenates were spun at 20,800 × g for 10 min at 4 °C, and protein concentrations were determined. PGC-1α (Cell Signaling) was immunoprecipitated overnight from cell lysates containing 200 μg of total protein. For PGC-1α the TrueBlot Western Blot Kit (eBioscience) was used to remove any heavy- and light-chain Ig proteins. Immunoprecipitation lysates were washed 3× before SDS-Page. SDS gel electrophoresis was performed by using precast Bis-Tris 4%–12% gradient polyacrylamide gels in the Mops buffer system (Invitrogen). After transfer to nitrocellulose membranes, membranes were incubated in blocking buffer (5% milk) for 1 h and immunoblotted with antiacetylated lysine (Cell Signaling), antiacetyl FOXO1 (Santa Cruz Biotechnology), FOXO1 (Cell Signaling), Phospho-STAT3 (Y705) (Cell Signaling), antiacetyl STAT3 Lys-685 (Cell Signaling), STAT3 (Santa Cruz Biotechnology), PGC-1α (Cell Signaling), GAPDH (Cell Signaling), PEPCK, or SirT1 (Upstate Biotechnology). After the incubation with the primary antibody, the membranes were washed 3 times for 15 min with TBS [10 mM Tris-HCl (pH 7.4), 0.5 M NaCl] plus Tween 20 (0.2% vol/vol) (TBST). The membranes were immersed in blocking buffer and a corresponding IgG-conjugated secondary antibody and were incubated for 2 h. The membranes were then washed 3 times for 5 min using TBST. Proteins were then detected with enhanced chemiluminescence (Thermo Scientific), and autoradiographs were quantified by using densitometry (Scion Image).
Statistical Analysis.
All values are expressed as the mean ± SEM. The significance between the mean values was evaluated by two-tailed unpaired Student's t test.
Acknowledgments.
We thank Dr. Gary Cline, Jianying Dong, Yanna Kosover, and Todd May for their expert technical assistance with the studies; Aida Groszmann for performing the hormone assays; and Thanh Vu for insightful commentary. The PEPCK antibody was a gift from Dr. Daryl Granner (Vanderbilt Medical Center, Nashville, TN). This work was supported by U.S. Public Health Service Grants R01 DK-40936 and P30 DK-45735 (to G.I.S.).
Footnotes
Conflict of interest statement: X.X.Y., S.F.M., S.B., and B.P.M. own stock and/or hold stock options in Isis Pharmaceuticals.
References
- 1.Maggs DG, et al. Metabolic effects of troglitazone monotherapy in type 2 diabetes mellitus. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 1998;128:176–185. doi: 10.7326/0003-4819-128-3-199802010-00002. [DOI] [PubMed] [Google Scholar]
- 2.Gastaldelli A, et al. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology. 2007;133:496–506. doi: 10.1053/j.gastro.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA. Pharmacologic therapy for type 2 diabetes mellitus. Ann Intern Med. 1999;131:281–303. doi: 10.7326/0003-4819-131-4-199908170-00008. [DOI] [PubMed] [Google Scholar]
- 4.Magnusson I, Rothman DL, Katz LD, Shulman RG, Shulman GI. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest. 1992;90:1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hundal RS, et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49:2063–2069. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel VT, et al. Targeting foxo1 in mice using antisense oligonucleotide improves hepatic and peripheral insulin action. Diabetes. 2006;55:2042–2050. doi: 10.2337/db05-0705. [DOI] [PubMed] [Google Scholar]
- 7.Koo SH, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 8.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 9.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 13.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masiello P, et al. Experimental NIDDM: Development of a new model in adult rats administered streptozotocin and nicotinamide. Diabetes. 1998;47:224–229. doi: 10.2337/diab.47.2.224. [DOI] [PubMed] [Google Scholar]
- 17.Samuel VT, et al. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J Clin Invest. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi CS, et al. Suppression of diacylglycerol acyltransferase-2 (DGAT2), but not DGAT1, with antisense oligonucleotides reverses diet-induced hepatic steatosis and insulin resistance. J Biol Chem. 2007;282:22678–22688. doi: 10.1074/jbc.M704213200. [DOI] [PubMed] [Google Scholar]
- 19.Lee MK, et al. Metabolic effects of troglitazone on fructose-induced insulin resistance in the rat. Diabetes. 1994;43:1435–1439. doi: 10.2337/diab.43.12.1435. [DOI] [PubMed] [Google Scholar]
- 20.Pagliassotti MJ, Wei Y, Bizeau ME. Glucose-6-phosphatase activity is not suppressed but the mRNA level is increased by a sucrose-enriched meal in rats. J Nutr. 2003;133:32–37. doi: 10.1093/jn/133.1.32. [DOI] [PubMed] [Google Scholar]
- 21.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen KF, et al. Reversal of nonalcoholic hepatic steatosis, hepatic insulin resistance, and hyperglycemia by moderate weight reduction in patients with type 2 diabetes. Diabetes. 2005;54:603–608. doi: 10.2337/diabetes.54.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Z, Wen Z, Darnell JE., Jr. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 25.Nie Y, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue H, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 30.Moh A, et al. STAT3 sensitizes insulin signaling by negatively regulating glycogen synthase kinase-3 beta. Diabetes. 2008;57:1227–1235. doi: 10.2337/db06-1582. [DOI] [PubMed] [Google Scholar]
- 31.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Hundal RS, et al. Mechanism by which high-dose aspirin improves glucose metabolism in type 2 diabetes. J Clin Invest. 2002;109:1321–1326. doi: 10.1172/JCI14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]