Abstract
Studies in mice indicate that the gut microbiota promotes energy harvest and storage from components of the diet when these components are plentiful. Here we examine how the microbiota shapes host metabolic and physiologic adaptations to periods of nutrient deprivation. Germ-free (GF) mice and mice who had received a gut microbiota transplant from conventionally raised donors were compared in the fed and fasted states by using functional genomic, biochemical, and physiologic assays. A 24-h fast produces a marked change in gut microbial ecology. Short-chain fatty acids generated from microbial fermentation of available glycans are maintained at higher levels compared with GF controls. During fasting, a microbiota-dependent, Pparα-regulated increase in hepatic ketogenesis occurs, and myocardial metabolism is directed to ketone body utilization. Analyses of heart rate, hydraulic work, and output, mitochondrial morphology, number, and respiration, plus ketone body, fatty acid, and glucose oxidation in isolated perfused working hearts from GF and colonized animals (combined with in vivo assessments of myocardial physiology) revealed that the fasted GF heart is able to sustain its performance by increasing glucose utilization, but heart weight, measured echocardiographically or as wet mass and normalized to tibial length or lean body weight, is significantly reduced in both fasted and fed mice. This myocardial-mass phenotype is completely reversed in GF mice by consumption of a ketogenic diet. Together, these results illustrate benefits provided by the gut microbiota during periods of nutrient deprivation, and emphasize the importance of further exploring the relationship between gut microbes and cardiovascular health.
Keywords: energy homeostasis, gnotobiotic mice, gut–heart metabolic axis, host-microbial mutualism, isolated perfused working heart
Mammals harbor large and diverse communities of gut microbes, whose compositions are shaped by both diet and host phylogeny (1, 2). One important function of the gut microbiota is to break down dietary polysaccharides into end products, including short-chain fatty acids (SCFAs), that can be absorbed by the host (3). Without the glycoside hydrolases, polysaccharide lyases, and other components of microbial fermentation pathways, calories present in various classes of glycans would be lost to the host.
Studies of germ-free (GF) and colonized mice have revealed a dynamic relationship between the gut microbiota and host adiposity. Mice raised in the absence of microbes are leaner than their conventionally raised (CONV-R), microbe-laden counterparts, even though GF animals consume more food (4). Obesity caused by a null allele in the leptin gene (ob/ob), or by consumption of a high-fat, high-sugar “Western” diet, is associated with a shift in microbial ecology in the distal gut and an increase in the proportional representation of genes in the gut microbiome involved in processing dietary carbohydrates into SCFAs (5, 6). Transplantation of a distal gut microbiota from these obese mice to adult GF recipients causes a greater increase in adiposity than does a microbiota transplanted from lean donors. Moreover, switching from a Western diet to a reduced-calorie diet stabilizes or diminishes host adiposity, reverses the changes in gut microbial ecology and representation of genes involved in carbohydrate metabolic pathways seen in obese hosts, and is associated with diminished capacity of the gut microbial community to promote adiposity in GF recipients (5, 6). Studies of GF and CONV-R normal and knockout mice have also identified host genes that mediate the effects of the microbiota on host energy balance. These genes include Gpr41, a G protein-coupled receptor produced by intestinal enteroendocrine cells that is activated by SCFAs (7). Comparative metagenomic studies of the distal gut (fecal) microbiotas of adult monozygotic and dizygotic lean and obese twins have documented alterations in gut microbial ecology and the composition of the microbiome that are analogous to those seen in obese mice (8).
The fact that increased adiposity is associated with a change in the microbiota that allows it to be more efficient at harvesting calories seems somewhat paradoxical: What benefit would such a relationship have for the host? The capacity of the microbiota to promptly adjust its configuration in ways that allows it to increase energy harvest from a diet and promote energy storage in the host could be beneficial under conditions where there is only intermittent access to sources of nutrients/energy. A corollary hypothesis is that when nutrients are no longer available, the microbiota assumes another state or configuration that provides a different form or forms of benefit to the host. The nature of these latter postulated adjustments, and their potential benefits, are ill-defined. Intriguingly, after withdrawal of nutrients, GF mice die more rapidly than their CONV-R counterparts, despite losing weight at approximately the same rate (9).
Because the heart must maintain constant and high levels of ATP to support its mechanical and electrical functions, it has evolved the capacity to use different substrates for its energy-generating pathways, depending on their availability (10–12). Therefore, we hypothesized that this organ could be a sensitive reporter of adaptive changes during periods of nutrient deprivation that involve the gut microbiota. In the current report, we use gnotobiotic mice to identify a metabolic cross-talk between liver and heart that is influenced by the gut microbiota. This cross-talk, which is apparent during fasting, is centered around ketone body utilization and shapes myocardial metabolism and size.
Results and Discussion
Ketogenesis Is Reduced in Fasted GF Mice.
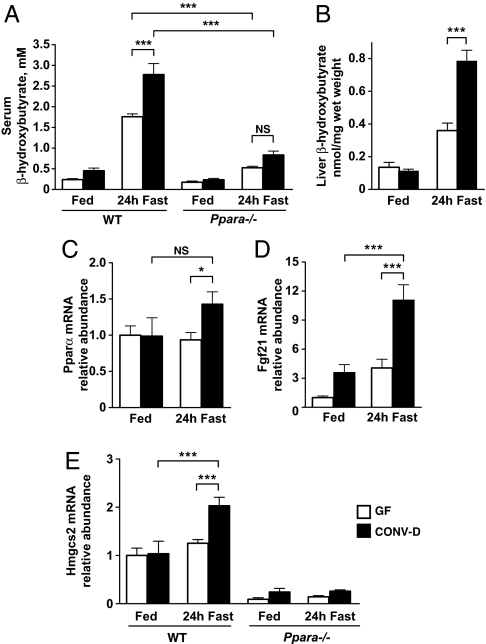
Nutrient deprivation in mammals induces a shift from carbohydrate to fat utilization (13). Free fatty acids, the products of lipolysis from peripheral adipose stores, are oxidized in the liver either to acetyl-CoA and ultimately CO2 via the tricarboxylic acid cycle, or partially to ketone bodies (acetoacetate, d-β-hydroxybutyrate, and acetone). Ketone bodies are used as carbon sources in the brain and other tissues (14, 15), including the heart where they form an avidly oxidized substrate pool (16). Therefore, we measured serum β-hydroxybutyrate levels in 6–10-week-old male C57BL/6J GF mice that were fed a standard low-fat, polysaccharide-rich chow (CARB) diet or fasted for 24 h, and in similarly aged fed or fasted mice that had received a microbiota transplant from the distal gut (ceca) of lean CARB-fed CONV-R animals 3 weeks before the experiment [transplant recipients are referred to as “conventionalized” (CONV-D)]. No significant difference in serum β-hydroxybutyrate levels was observed between fed GF and CONV-D mice. Levels were increased by fasting but were 37% lower in GF animals compared with their fasted, colonized counterparts (1.76 ± 0.07 mM vs. 2.78 ± 0.27 mM, n = 10 animals per group, P < 0.001) (Fig. 1A). There were no significant differences in serum insulin, glucose, free fatty acid, or triglyceride levels in fasted or fed GF versus CONV-D mice (Fig. S1 A–D).
Fig. 1.
Hepatic ketogenesis is enhanced during a 24-h fast by the gut microbiota. (A) Serum β-hydroxybutyrate levels in GF and CONV-D mice fed a standard CARB diet or after a 24-h fast. ***, P < 0.001; NS, not significant; n = 10 animals per condition. (B) Steady-state β-hydroxybutyrate levels in liver extracts. ***, P < 0.001; n = 6 animals per condition. (C–E) qRT-PCR assays of liver Pparα, Fgf21, and Hmgcs2 mRNA levels that are expressed relative to fed GF controls. *, P < 0.05; ***, P < 0.001 (n = 5–10 mice per condition; 2-way ANOVA with Bonferroni posthoc testing). Note that the difference in Fgf21 mRNA levels observed between GF- and CONV-D-fed animals does not reach statistical significance by ANOVA test.
The effect of a 24-h fast on adiposity was determined by measuring epididymal fat-pad-to-body-weight ratios. Fasting produced significant reductions in adiposity and significant increases in adipose triglyceride lipase gene (Pnpla2) expression in both GF and CONV-D animals, indicating that the capacity to mobilize peripheral fat stores is intact even in the absence of a microbiota (Fig. S1 E and F). In the fed state, CONV-D mice have increased hepatic triglyceride stores compared with GF animals. This difference between GF and CONV-D animals is dramatically enhanced with fasting (Fig. S1G), consistent with the notion that although mobilization of fat can occur in the absence of a microbiota, the presence of a gut community promotes hepatic triacylglyerol synthesis and storage as well as ketogenesis.
Peroxisome proliferator-activated receptor alpha (Pparα) is a fatty acyl-binding, nuclear receptor transcription factor required for utilization of fatty acids during fasting in a number of tissues including liver and heart. Pparα expression was significantly higher in the livers of fasting CONV-D animals compared with GF animals (Fig. 1C). The fasting-induced ketogenic response was blunted in CONV-D Pparα−/− mice (serum β-hydroxybutyrate levels were 0.84 ± 0.09 mM vs. 2.78 ± 0.27 mM in wild-type CONV-D controls, n = 10 animals per group, P < 0.001; Fig. 1A). Moreover, unlike wild-type mice, there was no significant difference in fasting serum β-hydroxybutyrate levels between GF and CONV-D Pparα−/− mice (Fig. 1A). Therefore, we concluded that the observed effect of the gut microbiota on fasting-induced ketosis involves Pparα.
The Effects of the Absence of a Gut Microbiota on Hepatic Ketogenesis.
The liver generates, but does not oxidize, ketone bodies because it does not express the key ketolytic enzyme, 3-oxoacid CoA transferase (17). To confirm that fasting-induced hepatic ketogenesis is reduced in the absence of a gut microbiota, we measured β-hydroxybutyrate levels and the expression of several regulators of the ketogenic response in the livers of fasted and fed GF and CONV-D mice. Although no significant difference in hepatic β-hydroxybutyrate levels were observed between fed GF and CONV-D wild-type animals, fasted GF mice had 50% lower concentrations of this ketone body (0.36 ± 0.05 vs. 0.79 ± 0.07 nmol/mg tissue, P < 0.001, n = 6 animals per group; Fig. 1B). Fgf21 is a Pparα-inducible target that stimulates ketone body production in the liver (18, 19). In the fed state, hepatic Fgf21 mRNA levels were not significantly different in wild-type CONV-D and GF mice. Levels rose 3.08 ± 0.8-fold after a 24-h fast in CONV-D mice (P < 0.001, n = 10) to a value 2.72 ± 0.7-fold greater than in fasted GF animals (P < 0.001, n = 10; Fig. 1D). Hepatic Fgf21 mRNA concentrations were at the lower limits of detection by qRT-PCR in the livers of both fed and fasting CONV-D and GF Pparα−/− animals, emphasizing the dependence of this gene's expression on Pparα. The mitochondrial form of 3-hydroxy-3-methylglutaryl-CoA synthase (Hmgcs2) is a critical ketogenic enzyme in the liver and a Pparα target (20). Hmgcs2 expression was unchanged by fasting in GF mice, but was induced 1.96 ± 0.09-fold in CONV-D animals (P < 0.05) (Fig. 1E). Together, these observations underscore the role of Pparα in regulating the augmentation in hepatic ketogenesis during fasting that is associated with the presence of a gut microbiota.
A colonized mouse has 2 sources for generating acetyl-CoA in the liver that are not well-represented in a GF animal. One source is the linkage of acetate, derived from microbial fermentation of polysaccharides in the gut, to CoA via acetyl-CoA synthetase (EC 6.2.1.1). The other source, as described above, is from oxidation of fatty acids derived from adipocyte triglyceride stores, which are larger in CONV-D animals (4). During states of high fatty acid oxidation, such as those encountered during nutrient deprivation, acetate generated by fermentation is absorbed from the gut and delivered to hepatocytes. In hepatocytes, acetate is either converted in the cytosol to acetyl-CoA for fatty acid synthesis (via the de novo lipogenesis pathway) or for mevalonate synthesis (21), or it is oxidized completely in mitochondria after conversion to acetyl-CoA (22). A fourth fate for hepatic acetyl-CoA is mitochondrial ketogenesis (see ref. 14 for further discussion).
Cecal acetate levels were 20-fold higher in fed CONV-D mice compared with GF wild-type mice (126.7 ± 7.5 vs. 6.3 ± 2.7 nmol/mg wet-weight cecal contents, P < 0.001, n = 5), a finding consistent with the fact that the gut microbiota ferments dietary polysaccharides to SCFAs, including acetate (23). Cecal acetate concentrations did not change significantly with fasting in GF animals. Levels in CONV-D mice dropped 3.5 ± 1.3-fold (P < 0.001) but remained significantly higher than those in fasted GF animals (36.0 ± 4.3 vs. 10.8 ± 2.0 nmol/mg cecal contents, P < 0.01, n = 5). These findings are compatible with the notion that acetate production, generated through fermentation of exogenous and endogenous polysaccharides by members of the gut microbiota, increases hepatocellular acetyl-CoA pools and thereby contributes to the increased hepatic ketogenic response observed in fasting CONV-D compared with GF animals.
The gut microbiota of mammals, including mice and humans, is generally dominated by two phyla (divisions) of bacteria: the Bacteroidetes, whose members possess myriad glycoside hydrolases and polysaccharide lyases (www.cazy.org), and the Firmicutes (1). Studies in gnotobiotic mice colonized with saccharolytic Bacteroides have shown that these microbes are able to adaptively forage complex host glycans present in mucus and on the surface of gut epithelial cells when animals are deprived of dietary polysaccharides (24, 25). Therefore, we assessed the effects of a 24-h fast on gut microbial ecology, including the representation of the Bacteroidetes, by preparing DNA from the cecal contents of fasted and fed mice, and performing multiplex pyrosequencing of amplicons generated by PCR using primers that flank variable region 2 (V2) of bacterial 16S rRNA genes (26). Sequence reads were grouped into operational taxonomic units (OTUs) based on a threshold cutoff of ≥97% identity among the V2 16S rDNA reads (1). A phylogenetic tree was built by using 1 representative sequence from each OTU, and employed for UniFrac analysis [this metric uses the tree to measure the degree of similarity of any 2 communities based on the proportion of branch length (evolutionary history) that they share on the tree (27); see SI Text for additional details]. Although a 24-h fast did not result in a significant shift in bacterial diversity (P > 0.05; Fig. S2A) or a significant departure from fed communities in overall community composition (P > 0.05; Fig. S2B), fasting was associated with a significant increase in the proportional representation of the Bacteroidetes [from 20.6% (fed) to 42.3% (fasted); P = 0.01; 2-tailed Student t test with unequal variance], and a significant diminution in the Firmicutes [from 77.1% (fed) to 52.6% (fasted); P = 0.007] (n = 8 fasted; 13 controls; 2 independent experiments; Fig. S2C). This rapid and marked expansion in the representation of the Bacteroidetes, achieved after a 24-h fast, yields a microbiota whose configuration contrasts with that observed in mice who are switched from a CARB to more-calorically dense Western-style diet, or mice that become obese because they are leptin-deficient: In both these cases, development of obesity is accompanied by reduced proportional representation of the Bacteroidetes (6).
Functional Genomics Studies of the Effects of the Absence of a Gut Microbiota on Cardiac Metabolism.
Ingenuity Pathways Analysis (IPA) of GeneChip datasets generated from the hearts of GF and colonized animals revealed that the ketone body metabolic pathway was significantly enriched within the myocardial transcriptome of both CARB-fed CONV-D and GF wild-type mice compared with their Pparα−/− counterparts. Enrichment was also observed in CONV-D fed versus fasted wild-type mice but not in fed versus fasted GF animals (Tables S1 and S2).
To explore more fully the extent to which the microbiota influences substrate selection by the myocardium under fasting conditions, we used GeneChip and qRT-PCR assays to compare the levels of expression of genes involved in ketone body metabolism, fatty acid oxidation (FAO), and glucose uptake/oxidation. The results revealed that key FAO-associated genes increased in CONV-D and GF hearts after a 24-h fast (Fig. S3A), as did the ketogenic biomarker Hmgcs2 (Fig. S3B). Oxct1 encodes the ketolytic enzyme 3-oxoacid CoA transferase required for ketone body oxidation. Although there was no significant difference between Oxct1 expression in fed CONV-D and GF mice, fasting led to a significantly higher level of this mRNA in the myocardium of fasted CONV-D compared with fasted GF animals [P < 0.001, n = 5; Fig. S3C; note that differences between fasted and fed GF vs. CONV-D animals were not observed in their skeletal (gastrocnemius) muscle]. These results are consistent with a relative increase in myocardial ketone body oxidation in fasting CONV-D animals. Fasting was associated with a very modest, albeit statistically significant, 1.50 ± 0.1-fold higher level of expression of Glut1 (which encodes a critical noninsulin-dependent glucose transporter) in the hearts of GF animals but produced no significant change in CONV-D mice (Fig. S3 A and D). These findings, plus the microbiota-associated enhancement in ketone body delivery to the fasting myocardium, prompted us to test whether glucose utilization was also altered under these conditions.
Metabolic and Physiologic Studies of Isolated Perfused Working Hearts: A Compensatory Shift Toward Glucose Oxidation in the Fasted Germ-Free Myocardium.
To directly assay whether the presence of a gut microbiota altered substrate selection by actively functioning myocardial tissue, we turned to isolated working hearts prepared from GF and CONV-D mice that had just completed a 24-h fast (n = 4 or 5 animals per group). Hearts were excised immediately after animals were killed, and perfused ex vivo in the working mode by using a buffer that included labeled and unlabeled energy substrates whose final concentrations mimicked the fasting serum levels of glucose, fatty acids, and ketone bodies (i.e., 5 mM glucose, 1.2 mM palmitate, and 1.7 mM d-β-hydroxybutyrate).
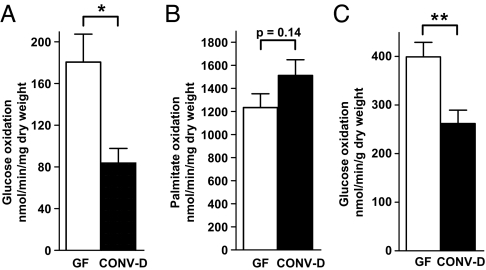
Myocardial ketone body oxidation in the isolated working heart model is known to be proportional to the concentration of ketone bodies delivered in the perfusate (16, 28–30). By keeping the input ketone body concentration constant for both perfused fasted GF and CONV-D hearts, we were able to document that ketone body oxidation rates remained constant (773.3 ± 157 and 779.5 ± 80 nmol β-hydroxybutyrate/min/mg dry weight for fasted GF and CONV-D myocardium, respectively), thereby allowing us to evaluate the relative roles of glucose versus fatty acid oxidation on myocardial energetics.
The rate of glucose oxidation was significantly increased in the isolated working hearts of fasted GF mice (180.6 ± 26.8 vs. 83.8 ± 14.0 nmol/min/mg dry weight (CONV-D), P < 0.02, n = 5 animals per group; Fig. 2A), whereas fatty acid oxidation was not significantly altered (Fig. 2B). Glucose utilization was also significantly greater in GF hearts in the absence of β-hydroxybutyrate in the perfusion buffer (Fig. 2C). Thus, an increased glucose utilization phenotype is “imprinted” in the fasted isolated working GF heart and is indicative of how, in the absence of a microbiota-dependent increase in hepatic ketogenesis, the heart undergoes a pronounced shift in its metabolism. Stated another way, because ketone bodies suppress myocardial glucose utilization (31), with reduced delivery of ketone bodies, the GF myocardium is able to turn to glucose as a prime energy source.
Fig. 2.
Influence of the gut microbiota on myocardial substrate selection. Hearts from fasted WT GF or CONV-D animals were perfused for 60 min in the working mode by using Krebs–Henseleit buffer containing 1.7 mM β-hydroxybutyrate, 5 mM glucose, and 1.2 mM palmitate. (A) Oxidation of d-[14C(U)]-glucose; n = 5 animals per group. (B) Oxidation of [9,10]-3H-palmitate; n = 8 animals per group. (C) Oxidation of d-[14C(U)]-glucose in the absence of added β-hydroxybutyrate, n = 5 animals per group. *, P < 0.05; **, P < 0.01 (Student's t test).
This increase in glucose utilization was not accompanied by alterations in steady state levels of phosphorylated Akt or phosphorylated AMPK-α in the fasted GF compared with CONV-D myocardium (Fig. S4 A and B). Glycogen levels were significantly reduced in the fasting GF versus fasting CONV-D myocardium (20.9 ± 3.5 vs. 41.0 ± 6.0 nmol/mg protein, n = 5/group, P = 0.02), consistent with increased glucose utilization in the GF state. These results are also consistent with studies performed in conventionally raised animals where enhanced β-hydroxybutyrate delivery is known to suppress myocardial glucose oxidation, and thus increase glycogen storage (32).
Heart rate, cardiac hydraulic work, and cardiac output were not statistically different between isolated working hearts prepared from fasted GF versus CONV-D mice (Fig. S4 C–E) despite decreased glucose utilization in CONV-D hearts. There were no detectable differences in mitochondrial morphology or number (Fig. S5 A and B) or in the results of in vitro assays of state 2, state 3, or state 4 mitochondrial respiration (Fig. S5C). In vivo echocardiographic assessments indicated that heart rate and fractional shortening decreased to a comparable degree in both groups of animals after a 24-h fast (Fig. S6 A–C). Thus, in the absence of a microbiota-associated, fasting-induced shift in myocardial metabolism toward ketone body utilization, the (fasting) GF heart is able to sustain its performance, at least in part, by increasing glucose utilization.
An Effect of the Gut Microbiota on Myocardial Mass.
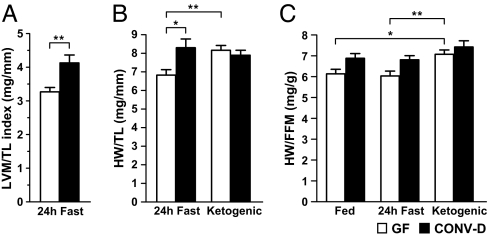
Intriguingly, heart weight, whether measured echocardiographically or as wet mass, and whether normalized to tibial length or lean body mass, was significantly reduced in fasted GF versus CONV-D mice (Fig. 3 A–C). This difference was also evident in CARB-fed animals (HW/TL = 7.28 ± 0.21 in CONV-D vs. 6.65 ± 0.18 in GF animals; P < 0.03 according to Student's t test). IPA analysis of GeneChip datasets indicated that the increased myocardial mass observed in fed and fasted CONV-D compared with GF mice was not associated with an enrichment in the representation of signaling pathways that mediate pathological hypertrophy (for further details see SI Text and Fig. S7).
Fig. 3.
Influence of the presence or absence of the gut microbiota on myocardial mass. (A) Echocardiographic measurement of left ventricular mass (LVM) in the fasting state normalized to tibial length (TL) in mm (n = 5 animals per group). (B and C). The reduction in myocardial mass [heart weight (HW)] normalized to tibial length (B), or to lean body mass [fat-free mass (FFM), as defined by MRI] (C), that is observed in GF mice fasted for 24 h after being maintained on a CARB diet, is abrogated in GF animals maintained on a ketogenic diet. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (n = 8 animals in each fasting and ketogenic diet group, and 6 animals in each CARB-fed group; 2-way ANOVA with Bonferroni posthoc testing).
Endurance Training Produces Physiological Hypertrophy of the Heart.
To determine whether the GF heart is able to manifest the features of the normal physiological hypertrophic response to training, GF and CONV-D animals were exercised in swimming tanks placed in each of their respective gnotobiotic isolators (for further details concerning these “gnotatoria” and the exercise regimen, see SI Text). After the 30-day training period, echocardiograms were performed: Heart rate was decreased in GF animals to a level that was not statistically different from CONV-D mice (Fig. S7C). However, the hearts of trained GF mice remained smaller than the hearts of the trained CONV-D mice (Fig. S7 D and E). IPA-based comparisons of the myocardial transcriptomes of untrained versus trained CARB-fed GF mice indicated that exercise resulted in gene expression changes in the IGF-1 and ERK/MAPK signaling cascades, signal transduction pathways that in CONV-D mice are associated with “physiological hypertrophy”, plus enrichment for mRNAs encoding mediators of oxidative phosphorylation (Fig. S7F; Tables S3 and S4). However, unlike the transcriptional changes that occur in the hearts of trained CONV-D mice, trained GF mice lack significant enrichment in a subset of pathways that includes ketone body metabolism (Table S3). Together, these findings suggest that diminished ketone metabolism during exercise may contribute to the inability of endurance training to rescue the reduced-myocardial-mass phenotype in GF animals.
A Ketone Body-Dependent Effect on Myocardial Mass in the GF Mouse.
To further test the notion that the myocardial metabolic and morphologic phenotypes observed in fasting GF mice reflect the consequences of decreased hepatic ketogenesis, untrained GF and CONV-D mice were placed on a very low carbohydrate, ketogenic diet (0.4% of calories as CARB, 95% as fat, 4.5% as protein). A ketogenic diet did not produce a notable change in the ratio of Bacteroidetes to Firmicutes in the cecal microbiota compared with CARB-fed, untrained (or trained) controls (Fig. S2C). However, unlike fasting, a ketogenic diet was associated with a significant decrease in bacterial diversity (P < 0.001; Fig. S2A). Principal coordinates analysis (PCoA), based on UniFrac distances, also revealed clustering of the gut communities of ketogenic diet-fed animals and a significant difference in the overall composition of their cecal microbiota versus mice fed the CARB diet (P < 0.001 along PC1, Fig. S2B).
Nonfasting GF animals that consumed the ketogenic diet for 30 days were able to mount a hepatic ketogenic response equivalent to that seen in their ketogenic diet-fed CONV-D counterparts (Fig. S8 A–D). Correspondingly, this diet produced a shift in the transcriptome toward ketone body metabolism in the hearts of GF animals that was equivalent to CONV-D mice (see Fig. S8E). Remarkably, the reduction in heart weight observed in GF animals was completely abrogated (Fig. 3 B and C). Together, these results support the notion that myocardial ketone body metabolism contributes to the myocardial-mass phenotype observed in GF mice.
We subsequently placed Pparα−/− mice on the ketogenic diet. However, in keeping with their deficiency in hepatic fatty acid oxidative capacity (33, 34), this diet led to ≈50% weight loss, a moribund appearance within 2 weeks, and overt hepatic steatosis at the time of euthanasia. Therefore, we were not able to use these animals to perform a direct genetic test of the role played by Pparα in mediating the effect of the ketogenic diet on the GF myocardial-mass phenotype.
Prospectus.
Although considerable progress has been made in characterizing the mechanisms regulating fatty acid and glucose metabolism in normal, diabetic, and failing heart by Ppars and insulin, substantially less is known about the relationship between ketone body metabolism and cardiovascular health. The results presented here indicate that during fasting, the presence of a gut microbiota is able to enhance the supply of a nutrient source (ketone bodies) that the heart avidly oxidizes. In the absence of a microbiota, the relative paucity of ketone bodies is associated with a compensatory increase in glucose utilization. As such, the present study expands our view of the benefits provided by a gut microbiota, both during periods of exposure to nutrient excess, where harvest and storage of energy is promoted, and during periods of nutrient deprivation, where substrate selection in the heart and myocardial mass are modified through effects on systemic ketone body metabolism.
We anticipate that using metagenomic methods to define the composition and operations of the gut microbial community will result in better understanding of interrelationships between diet, nutritional status, and the determinants of cardiovascular health. Moreover, comparative metagenomic studies of the gut microbiota of a variety of host species may be particularly informative, e.g., Burmese pythons increase their heart mass by 40% in 48 h after breaking a long fast (35).
Methods
Animals.
All protocols involving animals used in this study were approved by the Washington University Animal Studies Committee. GF wild-type C57BL/6J mice were maintained in flexible plastic gnotobiotic isolators under a strict 12-h light cycle (lights on at 0600), and fed an autoclaved CARB diet (B & K Universal) ad libitum. GF C57BL/6J Pparα−/− mice were generated as previously described (4).
Male GF mice were colonized at 6–10 weeks with a single gavage of cecal contents that had been harvested from adult CONV-R donors and resuspended in PBS (5 mL per donor; 500 μL of this suspension administered per recipient). Recipients were subsequently kept in gnotobiotic isolators and maintained for 2–3 weeks on a CARB diet. Animals were then either fasted for 24 h or continued on the CARB diet. A subset of GF and CONV-D mice was fed an irradiated sterile ketogenic diet (95.1% fat, 4.5% protein, 0.1% carbohydrate, Bio-Serv AIN-76A) for 30 days. All animals in all treatment groups were killed in mid-morning.
16S rRNA-Based Enumeration Studies of the Gut Microbiota.
Isolation of DNA from cecal contents, PCR of the V2 region of bacterial 16S rDNA genes, multiplex sequencing of the resulting bar-coded amplicons, and data analysis were performed by using methods described previously (1, 26, 27); see SI Text for more details.
Metabolite Analyses.
β-hydroxybutyrate was measured in serum or liver extracts by GC-MS, using DL-3-Hydroxybutyric acid-13C4 sodium salt (Sigma) as the internal standard (5 nmol added per μL serum; 100 nmol per 750 μL of liver extract). Liver extracts were prepared by homogenizing a 100-mg sample of liver (frozen in liquid nitrogen at tissue harvest and stored at −80 °C until use) in 750 μL of chloroform/methanol (2:1 ratio). Protein concentrations in liver extracts were measured by using a bicinchoninic acid assay (Pierce) and were not significantly different among fasted and fed GF and CONV-D animals.
To quantify acetate in cecal contents, 100 nmol sodium acetate-13C2,d3 (Sigma) was added to snap-frozen cecal contents (100 mg).The material was emulsified by adding 50 μL of 37% HCl, extracted with diethyl ether, and derivitized with N-(butyl-dimethyl-silyl)-2,2,2-trifluoro-N-methyl-acetamide (Sigma). Derivatized samples were analyzed on a Hewlett Packard 6890 gas chromatograph interfaced to an Agilent 5973 mass spectrometer.
GeneChip Analyses.
Mammalian cRNA targets were prepared and hybridized to Affymetrix MOE 430 2.0 GeneChips. Five hearts were analyzed individually for each condition. Raw data were scaled with the Affymetrix Microarray Suite (5.0) software to an intensity of 500 and subsequently analyzed with dChip (http://biosun1.harvard.edu/complab/dchip/). Selection criteria for differentially expressed genes were as follows: (i) fold-difference in expression (lower-bound 90% confidence interval) > 2; (ii) P value (t test) < 0.05; (iii) 100% present call in the condition with higher expression; (iv) minimal absolute intensity >100; (v) false discovery rate <1%. Datasets generated in dChip using these criteria were organized into canonical signaling and metabolic pathways by using IPA software (www.ingenuity.com). For more information on the genes used in dChip analysis, see SI Text and Table S5.
qRT-PCR.
Total cellular RNA was isolated from snap-frozen heart and liver samples (4). qRT-PCR assays were performed by using SYBR-green detection with an Mx3000P Stratagene Thermocycler. Data were normalized to Rpl32 mRNA and analyzed by using the ΔΔCΤ method. PCR primers are listed in Table S6.
Isolated Working Heart.
Mice received 100 units heparin by i.p. injection and 10 min later were anesthetized with an i.p. injection of 10 mg of sodium pentobarbital. Hearts were excised and placed in ice-cold Krebs–Henseleit bicarbonate solution (118 mM NaCl, 25 mM NaHCO3, 4.7 mM KCl, 0.4 mM KH2PO4, 2.5 mM CaCl2, pH 7.4) supplemented with 5 mM glucose, 1.2 mM palmitic acid (bound to 3% fatty acid-free BSA), and 1.7 mM d-β-hydroxybutyrate. Hearts were cannulated via the aorta and temporarily perfused, in a retrograde fashion, by using the Langendorff mode, with continuous bubbling of a 95% O2/5% CO2 gas mixture into the buffer reservoir to maintain tissue oxygenation during subsequent left atrial cannulation. Following cannulation, the perfusion circuit was changed to the antegrade working mode at 37 °C. Samples of the perfusate were collected every 10 min for measurement of (i) 14CO2 (trapped in 1 M hyamine hydroxide solution), generated from oxidation of glucose or β-hydroxybutyrate, and (ii) 3H2O, released into the buffer as a result of palmitate oxidation (36).
Measurements of cardiac output, aortic flow, peak systolic pressure, and heart rate were acquired every 10 min for 10 sec by using inline flow probes (Transonic Systems, Inc), the MP100 system from AcqKnowledge (BIOPAC Systems, Inc), and a pressure transducer (TSD 104A, BIOPAC System, Inc). Cardiac work was calculated as the product of peak systolic pressure and cardiac output. At the end of each perfusion, the dry weight of the ventricles was defined after desiccation in a vacuum oven.
Statistical Analysis.
The approach used for analysis of GeneChip datasets is described within GeneChip Analyses above. Other analytes were compared by Student's t test or 2-way ANOVA with Bonferroni posthoc testing, as appropriate, by using GraphPad Prism software. Mean values ± SEM are shown for all bar graph plots.
Supplementary Material
Acknowledgments.
We thank Maria Karlsson, David O'Donnell, Sabrina Wagoner, Jill Manchester, Howard Wynder, Sandeep Mahajan, Naveen Reddy, and Attila Kovacs for superb technical assistance; Jean Schaffer, Dan Ory, and Clay Semenkovich for critical review of the manuscript; and Laura Kyro for assistance with graphics. This work was supported in part by National Institutes of Health Grants DK073282, DK70977, DK020579, DK56341, and DK052574.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The GeneChip data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14929). The 16S rRNA sequence data reported in this paper have been deposited in the GenBank database (accession no. SRA008712.2).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902366106/DCSupplemental.
References
- 1.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flint HJ, et al. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 4.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuel BS, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tennant B, Malm OJ, Horowitz RE, Levenson SM. Response of germfree, conventional, conventionalized and E. coli monocontaminated mice to starvation. J Nutr. 1968;94:151–160. doi: 10.1093/jn/94.2.151. [DOI] [PubMed] [Google Scholar]
- 10.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: A question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopaschuk GD. Optimizing cardiac fatty acid and glucose metabolism as an approach to treating heart failure. Semin Cardiothorac Vasc Anesth. 2006;10:228–230. doi: 10.1177/1089253206291150. [DOI] [PubMed] [Google Scholar]
- 12.Kodde IF, van der Stok J, Smolenski RT, de Jong JW. Metabolic and genetic regulation of cardiac energy substrate preference. Comp Biochem Physiol A Physiol. 2007;146:26–39. doi: 10.1016/j.cbpa.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Cahill GF, Jr, et al. Hormone–fuel interrelationships during fasting. J Clin Invest. 1966;45:1751–1769. doi: 10.1172/JCI105481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 15.Cahill GF., Jr Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 16.Jeffrey FM, Diczku V, Sherry AD, Malloy CR. Substrate selection in the isolated working rat heart: Effects of reperfusion, afterload, and concentration. Basic Res Cardiol. 1995;90:388–396. doi: 10.1007/BF00788500. [DOI] [PubMed] [Google Scholar]
- 17.Fukao T, et al. Enzymes of ketone body utilization in human tissues: Protein and messenger RNA levels of succinyl-coenzyme A (CoA):3-ketoacid CoA transferase and mitochondrial and cytosolic acetoacetyl-CoA thiolases. Pediatr Res. 1997;42:498–502. doi: 10.1203/00006450-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 18.Badman MK, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: A control enzyme in ketogenesis. Biochem J. 1999;338:569–582. [PMC free article] [PubMed] [Google Scholar]
- 21.Fujino T, et al. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 22.Pouteau E, et al. Kinetic aspects of acetate metabolism in healthy humans using [1–13C] acetate. Am J Physiol. 1996;271:E58–E64. doi: 10.1152/ajpendo.1996.271.1.E58. [DOI] [PubMed] [Google Scholar]
- 23.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 24.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 25.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamady M, et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaffe SR, Gold AJ. Effect of prolonged starvation on substrate uptake in the isolated perfused rat heart. J Nutr. 1979;109:2140–2145. doi: 10.1093/jn/109.12.2140. [DOI] [PubMed] [Google Scholar]
- 29.Sultan AM. Effects of diabetes and insulin on ketone bodies metabolism in heart. Mol Cell Biochem. 1992;110:17–23. doi: 10.1007/BF02385001. [DOI] [PubMed] [Google Scholar]
- 30.Fukao T, Lopaschuk GD, Mitchell GA. Pathways and control of ketone body metabolism: On the fringe of lipid biochemistry. Prostaglandins Leukot Essent Fatty Acids. 2004;70:243–251. doi: 10.1016/j.plefa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier A, Coderre L. Ketone bodies alter dinitrophenol-induced glucose uptake through AMPK inhibition and oxidative stress generation in adult cardiomyocytes. Am J Physiol. 2007;292:E1325–E1332. doi: 10.1152/ajpendo.00186.2006. [DOI] [PubMed] [Google Scholar]
- 32.Goodwin GW, Taegtmeyer H. Metabolic recovery of isolated working rat heart after brief global ischemia. Am J Physiol. 1994;267:H462–H470. doi: 10.1152/ajpheart.1994.267.2.H462. [DOI] [PubMed] [Google Scholar]
- 33.Kersten S, et al. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen JB, et al. Physiology: Postprandial cardiac hypertrophy in pythons. Nature. 2005;434:37–38. doi: 10.1038/434037a. [DOI] [PubMed] [Google Scholar]
- 36.Finck BN, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.