Abstract
The accumulation of proteins damaged by reactive oxygen species (ROS), conventionally regarded as having pathological potentials, is associated with age-related diseases such as Alzheimer's, atherosclerosis, and cataractogenesis. Exposure of the aromatic amino acid phenylalanine to ROS-generating systems produces multiple isomers of tyrosine: m-tyrosine (m-Tyr), o-tyrosine (o-Tyr), and the standard p-tyrosine (Tyr). Previously it was demonstrated that exogenously supplied, oxidized amino acids could be incorporated into bacterial and eukaryotic proteins. It is, therefore, likely that in many cases, in vivo-damaged amino acids are available for de novo synthesis of proteins. Although the involvement of aminoacyl-tRNA synthetases in this process has been hypothesized, the specific pathway by which ROS-damaged amino acids are incorporated into proteins remains unclear. We provide herein evidence that mitochondrial and cytoplasmic phenylalanyl-tRNA synthetases (HsmtPheRS and HsctPheRS, respectively) catalyze direct attachment of m-Tyr to tRNAPhe, thereby opening the way for delivery of the misacylated tRNA to the ribosome and incorporation of ROS-damaged amino acid into eukaryotic proteins. Crystal complexes of mitochondrial and bacterial PheRSs with m-Tyr reveal the net of highly specific interactions within the synthetic and editing sites.
The aminoacyl-tRNA synthetases (aaRSs) play a central role in the translation of the genetic code, catalyzing the attachment of the correct amino acid to its cognate tRNA in a 2-step reaction (1). At the first step, the activation of the amino acid by ATP proceeds through the formation of an enzyme-bound aminoacyl adenylate. At the second step, the amino acid moiety is transferred to the 3′-terminal ribose of the cognate tRNA, leading to the synthesis of aminoacyl-tRNA (1). At the amino acid binding and recognition stage some aaRSs are unable to distinguish between amino acids having similar chemical structures. To ensure that the total level of accuracy of the protein synthesis is high enough, aaRSs developed an editing activity associated with the specific site where misacylated tRNA is hydrolyzed. Considerable progress has been made in understanding the structural basis and the mechanisms of the discrimination of noncognate amino acids (2). The proofreading activity of aaRSs has been shown to ensure the discrimination between natural and some artificial amino acids (3). However, it is well known that reactive oxygen species (ROS)-generating systems within the cell produce naturally occurring damaged amino acids having considerable pathological potential and associated with age-related diseases such as Alzheimer's, atherosclerosis, and cataractogenesis (4–6). Thus, it is important to follow the pathway of such amino acid incorporation into proteins. Notably, in human tissue proteins the concentrations of oxidized amino acids under normal and pathological circumstances differ to a small extent. For example, in the low-density lipoprotein, the physiological and pathological levels of m-Tyr (oxidized version of phenylalanine) are 62 and 105 pmol/mg, respectively (5). Although the involvement of components of the translation system in the incorporation of ROS-damaged amino acids into proteins has been hypothesized, the specific mechanism and the role of aaRSs in this process remain unclear (7, 8).
Phylogenetic and structural analyses revealed 3 major forms of phenylalanyl-tRNA synthetase (PheRS): the bacterial (αβ)2heterodimer, the archaeal/eukaryotic-cytosolic (αβ)2 heterodimer, and the mitochondrial monomer (9, 10). Recently, it was shown that cytoplasmic eukaryotic and prokaryotic PheRSs are able to misactivate tyrosine and hydrolyze the misacylated Tyr-tRNAPhe (11, 12). The monomeric mitochondrial PheRS also misactivates tyrosine, although its selectivity is 1 order of magnitude higher than that of the cytosolic PheRS (13). However, the mitochondrial PheRSs are unable to deacylate Tyr-tRNAPhe because of the absence of the structural module associated with editing activity (14). We show here that ROS-damaged phenylalanine (m-Tyr) may be incorporated into eukaryotic proteins via a specific tRNA-dependent pathway, using mitochondrial and possibly cytosolic PheRSs. For the lack of an editing domain from human mitochondrial (Hsmt)PheRS and enhanced selectivity of the synthetic active site of human cytosolic (Hsct)PheRS toward m-Tyr, we hypothesize the prevailing role of mitochondrial enzyme in this process.
Results
Discrimination Between Cognate and Noncognate Substrates by Various PheRSs.
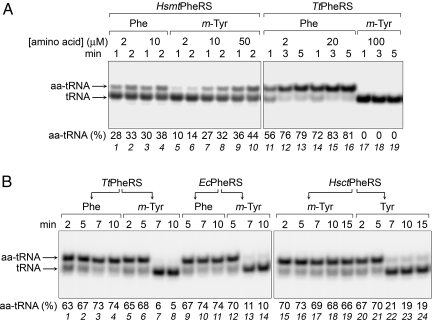
To understand the mechanism of the incorporation of ROS-damaged amino acids into the polypeptide chain, we studied the ability of PheRSs from various sources [bacterial Thermus thermophilus (Tt) and Escherichia coli (Ec), Hsct, and Hsmt] to activate m-Tyr and attach it to tRNAPhe. The m-Tyr is activated by these enzymes as assayed by ATP hydrolysis. Steady-state kinetic measurements of the aminoacylation (assayed by means of acidic gel electrophoresis) revealed that the mitochondrial enzyme is able to stably synthesize m-Tyr-tRNAPhe (Fig. 1A and Fig. S1). On the contrary, no mischarging of tRNAPhe with m-Tyr by the bacterial PheRS was detected. Analysis of kinetic parameters of the aminoacylation of tRNAPhe shows that the catalytic efficiency (kcat/Km) of the m-Tyr attachment by HsmtPheRS is only 5-fold lower than that of the correct amino acid, primarily because of a higher Km value (Table S1). At the same time, the aminoacylation of tRNAPhe with Tyr by this enzyme is 1,000-fold less efficient than the phenylalanylation. Mischarging of tRNAPhe with m-Tyr by HsctPheRS was detected by using yeast tRNAPhe transcript as an active substrate; a minor shift of the aminoacylated tRNA made it difficult to quantify the extent of charging. However, it was clear that m-Tyr-tRNAPhe is synthesized by the 2 eukaryotic enzymes (HsmtPheRS and HsctPheRS). To further compare the effectiveness of discrimination between the cognate and noncognate substrates by 3 PheRSs, we determined the apparent inhibition constants (Kiapp) for m-Tyr and Tyr in the aminoacylation reaction (Table S1). These data show that the bacterial and mitochondrial PheRSs bind m-Tyr in the aminoacylation site with almost the same affinity (13 and 13.6 μM, respectively), which is 1 order of magnitude higher than the affinity of human cytoplasmic PheRS (150 μM).
Fig. 1.
Aminoacylation of tRNAPhe with cognate and noncognate amino acids and specific deacylation of mischarged product. (A) Aminoacylation of E. coli tRNAPhe transcript (1.2 μM) with Phe or m-Tyr by HsmtPheRS (210 nM) or TtPheRS (24 nM) analyzed by electrophoresis in 8% denaturing gel at acidic conditions (0.1 M Na-acetate, pH 5). (B) Specific deacylation of m-Tyr-tRNAPhe. E. coli tRNAPhe transcript (1.2 μM) was aminoacylated with Phe (25 μM), m-Tyr (125 μM), or Tyr (1 mM) by HsmtPheRS (250 nM in experiments with Phe and m-Tyr, or 500 nM in experiments with Tyr) for 5 min; then the reaction was continued after addition (shown by arrows) of TtPheRS (16 nM), EcPheRS (48 nM), or HsctPheRS (32 nM).
Editing Activity of the Heterotetrameric PheRSs.
To test the ability of the heterotetrameric PheRSs to edit m-Tyr-tRNAPhe, the misaminoacylated product of the E. coli tRNAPhe was incubated with TtPheRS, EcPheRS, and HsctPheRS. Adding the bacterial enzymes TtPheRS and EcPheRS directly to the aminoacylation reaction mixture preincubated with HsmtPheRS resulted in the significant hydrolysis of m-Tyr-tRNAPhe (Fig. 1B). Under such conditions, Phe-tRNAPhe remains remarkably stable. These data indicate that both TtPheRS and EcPheRS are active in the transediting of the exogenous m-Tyr-tRNAPhe. However, HsctPheRS was unable to hydrolyze m-Tyr-tRNAPhe (Fig. 1B). To investigate whether this inability was caused by the lower selectivity of editing site, similar experiments were performed with Tyr-tRNAPhe, which showed that Tyr-tRNAPhe was efficiently deacylated (Fig. 1B), even though the E. coli tRNAPhe transcript is a worse substrate for HsctPheRS [owing to change of the recognition nucleotide at position 20 (9, 15)]. Taken together, these data demonstrate that the inability of bacterial PheRS to stably synthesize m-Tyr-tRNAPhe (which cannot be detected by steady-state kinetics) is caused by the editing of the mischarged product. In contrast, low, if any, deacylation of m-Tyr-tRNAPhe by HsctPheRS suggests that the more efficient discrimination of the synthetic site against m-Tyr contributes to the aminoacylation accuracy.
Structural Basis of Molecular Recognition of m-Tyr.
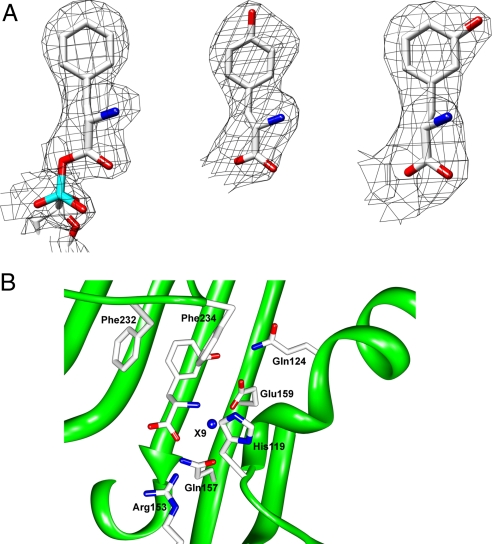
The modes of the molecular recognition of m-Tyr by TtPheRS and HsmtPheRS have been revealed by X-ray analysis of the corresponding complexes. Analysis of the unbiased difference (Fobs − Fcalc) electron density maps highlighted the electron densities that may be unambiguously attributed to the m-Tyr molecules located at the amino acid-binding pockets of both enzymes (Fig. 2; see also Fig. S2a). The electron density shaped by m-Tyr differs significantly from those shaped by Phe, Tyr, or p-Cl-Phe (Fig. 2A) (11). A distinctive topological feature of the PheRS active site is shown in the presence of a deep, phenylalanine-binding pocket. Specific recognition of m-Tyr in both enzymes is achieved through the interaction of the phenyl ring of the substrate with the phenyl rings of 2 phenylalanines located in the so-called “FPF loop” (Phe-232 and Phe-234 in HsmtPheRS; Pheα258 and Pheα260 in TtPheRS) (16). Three phenyl moieties thereby form a “network” of interactions, such that each aromatic pair makes an “edge-to-face” contact (see Fig. S3). The α-NH3+ group of m-Tyr is involved in hydrogen bonding with Ser-121 and His-119 of the mitochondrial enzyme (Serα180, Hisα178 in TtPheRS). The α-carboxylate of the ligand forms H bonds with the side chains of Gln-157 and with the class II invariant Arg-143 (Glnα218 and Argα204 in TtPheRS). Compared with the native phenylalanine substrate, m-Tyr is additionally stabilized by the hydrogen bonding of its OH group in metaposition with the Nε2 atom of Gln-124 (2.7 Å) and the Oε2 atom of Glu-159 (2.6 Å) (Glnα183 and Gluα220 in the bacterial enzyme). Thus, for both PheRSs containing identical sets of amino acids participating in the substrate binding (Fig. S2b), one would expect similar values for the m-Tyr affinities. As to the human cytoplasmic PheRS, comparative modeling studies indicate that amino acid residues involved in both phenylalanine and m-Tyr binding and recognition form a local environment that differs from those of HsmtPheRS and TtPheRS. These differences may lead to changes in the m-Tyr affinity observed in the kinetic experiments (see above). In the bacterial system, a second m-Tyr was identified at the editing module of PheRS (Fig. 3). The ability of TtPheRS to anchor the OH group of m-Tyr is achieved through its interactions with Oε1 of Gluβ334 and the main-chain amide of Glyβ315 (Fig. 3). A remarkable structural peculiarity of the editing site is revealed in the appearance of the invariant hydrophilic residue Gluβ334 in a fully hydrophobic environment formed by Pheβ213, Leuβ215, Pheβ263, Ileβ300, Alaβ314, Glyβ315, Alaβ336, and Pheβ338 (11). Consequently, the aromatic ring of m-Tyr is placed into the hydrophobic environment. Thus, Gluβ334 plays a critical role in the specific recognition of the m-Tyr moiety. As proposed in biochemical and structural studies of E. coli and T. thermophilus PheRSs the editing activity is directed against tyrosine only (11). However, our observation of the m-Tyr presence in the editing site demonstrates that plasticity is attributed not only to the synthetic active site, but also to the editing site, which is capable of binding ligands other than tyrosine.
Fig. 2.
Cognate and noncognate amino acids at the PheRS active site. (A) The unbiased electron density maps of phenylalanine moiety from HsmtPheRS complexed with Phe-AMP [Protein Data Bank (PDB) ID code 3CMQ] (Left), l- tyrosine from complex structure with TtPheRS (PDB ID code 2AMC) (Center), and m-Tyr from complex structure with HsmtPheRS (Right). (B) The active-site structure and residues of HsmtPheRS crucial for m-Tyr recognition. X9 is the well-ordered water molecule observed in almost all PheRS complexes.
Fig. 3.
The fragment of TtPheRS structure: editing site with bound m-Tyr. Amino acid residues making direct contacts with the ligand are shown. The electron density map is contoured at 2.5 σ.
Discussion
More is now known about the generation and the effects of ROS on biological systems. However, the pathways of ROS-damaged amino acids translation into cellular mechanisms are often unclear. We herein demonstrate that m-Tyr, a ROS-damaged amino acid, can be incorporated into proteins by using aaRSs and a tRNA-dependent pathway. The aaRSs show a remarkable specificity to their cognate amino acid substrates. As a result, the incorporation of non-natural amino acids often necessitates site-directed mutagenesis of the several residues within the active site area (17). However, despite the high level of the substrate stereo-specificity in the Phe system, structures of bacterial and mitochondrial PheRSs demonstrate a natural plasticity at the active site, thus enabling binding and specific aminoacylation of surrogates. It is commonly believed that there are no barriers for infiltration of surrogate amino acids into a growing polypeptide chain, when the aaRSs specificity is bypassed. However, in some studies it was hypothesized that the quality control provided by the editing activity of aaRSs may be supplemented by the discrimination of misaminoacylated tRNAs when they interact with the EF-Tu factor. In relation to PheRS, Ling et al. (18) showed that EF-Tu efficiently recognizes misaminoacylated tRNAPhe and that the ribosome does not discriminate between the cognate Phe-tRNAPhe and the noncognate Tyr-tRNAPhe. Thus, at least for the phenylalanine system, editing is the only barrier that blocks the penetration of ROS-damaged amino acids into the polypeptide chain. The editing activity of bacterial enzymes and the selectivity at the active site of the eukaryotic cytoplasmic enzyme with relation to m-Tyr suggest that incorporation of m-Tyr into the polypeptide chain is most likely associated with mitochondrial PheRS.
It is of interest that plants are uniquely sensitive to the m-Tyr incorporation. For a significant inhibition of the growth of plant cells, a 100-fold lower concentration of m-Tyr is needed when compared with mammals. This phenomenon could be explained by the protein synthesis activity in plants organelles, which is much higher than in mammals. Indeed, the mammal mitochondrion encodes only 13 proteins, whereas most plants encode >150 proteins in their organelles. Interestingly, the monomeric PheRS in plants, which is highly similar to HsmtPheRS, is dually targeted, i.e., occurring in both mitochondria and chloroplasts (19). These facts support the idea that organellar monomeric PheRS activity is highly required for plants.
It has been suggested that mitochondrial respiration is a major source of ROS and oxidized amino acids (20, 21). Under normal circumstances, the oxidized variant of phenylalanine seems to be produced at a rather low level. However, the identification of pathologies (including age-related) in which reactive species are generated by inflammatory processes indicate the possibility of a drastic accumulation of oxidized proteins (4). Indeed, accumulation of m-Tyr in rat mitochondrial proteins intensified aging (20). Although the molecular nature of most mitochondrial diseases is far from being completely understood, it is evident that some pathologies are related to the presence of free radicals (20). Patients with severe ailments, such as Leigh disease, cardiomyopathy with cataracts, and fatal infantile lactic acidosis showed a deficiency of complex I (NADH quinone oxidoreductase) and an elevated production of oxygen free radicals (22). Clearly, the activation of m-Tyr by PheRSs provides a link between tRNA and the ROS metabolism. Protection against mis-incorporation of oxidative-damaged amino acids could constitute a promising approach to the treatment of ROS-dependent pathologies.
Materials and Methods
Chemicals, Proteins, and tRNAs.
dl-m-Tyr, l- p-tyrosine, and l-phenylalanine were purchased from Sigma–Aldrich. PheRSs from natural or overexpressed sources were purified as described (23–25). The tRNAPhes were synthesized by using runoff transcription of synthetic genes with T7 RNA polymerase followed by electrophoretic isolation of the correct-length transcripts as described (26). To prepare labeled tRNAs, transcriptions were run in the presence of α-[32P]ATP (Amersham Biosciences). Yeast tRNAPhe was purchased from Roche Molecular Biochemicals.
tRNA Aminoacylation.
Aminoacylation reaction was performed in optimal conditions described for different PheRSs (24–26). In experiments with T. thermophilus and human mitochondrial enzymes performed at 37 °C the reaction mixtures contained 50 mM Tris·HCl (pH 8.5), 9 mM MgCl2, 5 mM ATP, and 1.2–2 μM E. coli tRNAPhe transcript [an excellent substrate of TtPheRS (26)] or yeast tRNAPhe (native or prepared by transcription). All 3 tRNAs were aminoacylated by HsmtPheRS with similar catalytic efficiencies. The activity of human cytoplasmic PheRS was tested (generally at 30 °C) in mixtures containing 50 mM Tris·HCl (pH 8.0), 30 mM MgCl2, 20 mM KCl, 10 mM 2-mercaptoethanol, 1.5 μM yeast tRNAPhe, and 5 mM ATP. The amino acid concentrations used as variable substrates were 0.2–25 μM for Phe, 1–125 μM for m-Tyr, and 0.05–2 mM for Tyr. In inhibition analyses the concentrations of m-Tyr and Tyr were varied in the range of 10 to 200 μM and 0.1 to 4 mM, respectively. Attachment of 14C/3H-labeled Phe to tRNAPhe was measured by trichloroacetic acid precipitation. Aminoacylation of 32P-labeled tRNAPhe with m-Tyr or other unlabeled amino acids was tested by an acidic gel electrophoretic method (27). Gels were run at 4 °C for 12 h at 6 V/cm (18) for an 11-cm migration (16) of xylene cyanol, dried, and quantified by Phosphor imaging. Stability of the aminoacyl bond during electrophoresis was examined by comparison of the aminoacylation extents measured in identical conditions by the 2 assays. The kinetic parameters kcat, Km, and Kmapp = Km(1 + I/Ki) for competitive inhibitor (where I is concentration of the inhibitor) were calculated by using the Microcal Origin 4.10 program. The reported kcat, Km, and Ki values represent the average of at least 2 determinations with experimental errors within 15% of the indicated values.
Crystallization and Structure Determination.
Crystallization of TtPheRS and HsmtPheRS with various ligands has been described in detail (11, 25). Crystal soaking with m-Tyr for TtPheRS and HsmtPheRS was carried out under conditions similar to those described. Complete X-ray datasets were collected from single crystals on an European Synchrotron Radiation Facility BM-14 station. The structures of TtPheRS and HsmtPheRS complexed with m-Tyr were isomorphous to the native ones. After rigid-body refinement (REFMAC5) and cycles of simulated annealing and conjugate gradient minimization (CNS), an unbiased difference Fourier map with coefficients (Fobs − Fcalc) was calculated (28, 29). Water molecules were added by using Arp/Warp (30). The manual refinement of the model building was done with O and Coot (31, 32). The models were refined to Rwork/Rfree of 19.4%/23.9% and 24.2%/27.3% for HsmtPheRS and TtPheRS, respectively.
Supplementary Material
Acknowledgments.
We thank P. Schimmel for critical reading of the manuscript and valuable comments. This work was supported by Binational Science Foundation Grant 2005209 (to M.G.S.), the Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (M.G.S.), and Russian Foundation of Basic Research Grant 06-04-48798 (to N.M.). L.K. is supported by a doctoral research fellowship from the Ori Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3HFZ and 3HFV).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905212106/DCSupplemental.
References
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Jakubowski H. Accuracy of aminoacyl-tRNA synthetases: Proofreading of amino acids. In: Ibba M, Francklyn C, Cusack S, editors. Aminoacyl-tRNA Synthetases. Austin, TX: Landes Bioscience; 2005. pp. 384–396. [Google Scholar]
- 3.Hendrickson TL, de Crecy-Lagard V, Schimmel P. Incorporation of nonnatural amino acids into proteins. Annu Rev Biochem. 2004;73:147–176. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 4.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 5.Dean RT, Fu S, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu S, Davies MJ, Stocker R, Dean RT. Evidence for roles of radicals in protein oxidation in advanced human atherosclerotic plaque. Biochem J. 1998;333:519–525. doi: 10.1042/bj3330519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurer-Orhan H, et al. Misincorporation of free m-tyrosine into cellular proteins: A potential cytotoxic mechanism for oxidized amino acids. Biochem J. 2006;395:277–284. doi: 10.1042/BJ20051964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlop RA, Dean RT, Rodgers KJ. The impact of specific oxidized amino acids on protein turnover in J774 cells. Biochem J. 2008;410:131–140. doi: 10.1042/BJ20070161. [DOI] [PubMed] [Google Scholar]
- 9.Safro MG, Moor N, Lavrik O. Phenylalanyl-tRNA synthetase. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Austin, TX: Landes Bioscience; 2005. pp. 250–265. [Google Scholar]
- 10.Mosyak L, Reshetnikova L, Goldgur Y, Delarue M, Safro MG. Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus. Nat Struct Biol. 1995;2:537–547. doi: 10.1038/nsb0795-537. [DOI] [PubMed] [Google Scholar]
- 11.Kotik-Kogan O, Moor N, Tworowski D, Safro M. Structural basis for discrimination of l-phenylalanine from l-tyrosine by phenylalanyl-tRNA synthetase. Structure (London) 2005;13:1799–1807. doi: 10.1016/j.str.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the β-subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 14.Klipcan L, et al. The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase. Structure (London) 2008;16:1095–1104. doi: 10.1016/j.str.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Nazarenko IA, Peterson ET, Zakharova OD, Lavrik OI, Uhlenbeck OC. Recognition nucleotides for human phenylalanyl-tRNA synthetase. Nucleic Acids Res. 1992;20:475–478. doi: 10.1093/nar/20.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishman R, Ankilova V, Moor N, Safro M. Structure at 2.6-Å resolution of phenylalanyl-tRNA synthetase complexed with phenylalanyl-adenylate in the presence of manganese. Acta Crystallogr D. 2001;57:1534–1544. doi: 10.1107/s090744490101321x. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 18.Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA. 2007;13:1881–1886. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duchene AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies SM, Poljak A, Duncan MW, Smythe GA, Murphy MP. Measurements of protein carbonyls, ortho- and meta-tyrosine and oxidative phosphorylation complex activity in mitochondria from young and old rats. Free Radical Biol Med. 2001;31:181–190. doi: 10.1016/s0891-5849(01)00576-7. [DOI] [PubMed] [Google Scholar]
- 21.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 22.Raha S, Robinson BH. Mitochondria, oxygen free radicals, disease, and aging. Trends Biochem Sci. 2000;25:502–508. doi: 10.1016/s0968-0004(00)01674-1. [DOI] [PubMed] [Google Scholar]
- 23.Chernaya MM, Korolev SV, Reshetnikova LS, Safro MG. Preliminary crystallographic study of the phenylalanyl-tRNA synthetase from Thermus thermophilus HB8. J Mol Biol. 1987;198:555–556. doi: 10.1016/0022-2836(87)90301-9. [DOI] [PubMed] [Google Scholar]
- 24.Moor N, Linshiz G, Safro M. Cloning and expression of human phenylalanyl-tRNA synthetase in Escherichia coli: Comparative study of purified recombinant enzymes. Protein Expression Purif. 2002;24:260–267. doi: 10.1006/prep.2001.1560. [DOI] [PubMed] [Google Scholar]
- 25.Levin I, et al. Purification, crystallization, and preliminary X-ray characterization of a human mitochondrial phenylalanyl-tRNA synthetase. Acta Crystallogr F. 2007;63:761–764. doi: 10.1107/S1744309107038651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasil'eva IA, Ankilova VN, Lavrik OI, Moor NA. tRNA discrimination by T. thermophilus phenylalanyl-tRNA synthetase at the binding step. J Mol Recognit. 2002;15:188–196. doi: 10.1002/jmr.575. [DOI] [PubMed] [Google Scholar]
- 27.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 28.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Lamzin VS, Perrakis A current state of automated crystallographic data analysis. Nat Struct Biol. 2000;7(Suppl):978–981. doi: 10.1038/80763. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.