Abstract
Microsomal epoxide hydrolase (EPHX1) biotransforms epoxide derivatives of pharmaceuticals, including metabolites of certain antiepileptic medications, such as phenytoin and carbamazepine, and many environmental epoxides, such as those derived from butadiene, benzene, and carcinogenic polyaromatic hydrocarbons. We previously identified a far upstream promoter region, designated E1-b, in the EPHX1 gene that directs expression of an alternatively spliced EPHX1 mRNA transcript in human tissues. In this investigation, we characterized the structural features and expression character of the E1-b promoter region. Results of quantitative real-time polymerase chain reaction analyses demonstrated that the E1-b variant transcript is preferentially and broadly expressed in most tissues, such that it accounts for the majority of total EPHX1 transcript in vivo. Comparative genomic sequence comparisons indicated that the human EPHX1 E1-b gene regulatory region is primate-specific. Direct sequencing and genotyping approaches in 450 individuals demonstrated that the E1-b promoter region harbors a series of transposable element cassettes, including a polymorphic double Alu insertion. Results of reporter assays conducted in several human cell lines demonstrated that the inclusion of the Alu(+/+) insertion significantly decreases basal transcriptional activities. Furthermore, using haplotype block analyses, we determined that the E1-b polymorphic promoter region was not in linkage disequilibrium with two previously identified nonsynonomous single nucleotide polymorphisms (SNPs) in the coding region or with functional SNPs previously identified in the proximal promoter region of the gene. These results demonstrate that the upstream E1-b promoter is the major regulator of EPHX1 expression in human tissues and that polymorphism in this region may contribute an interindividual risk determinant to xenobiotic-induced toxicities.
Although xenobiotic metabolism often results in detoxication, in certain instances, both in cases of pharmaceutical and environmental chemical metabolism, bioactivated and highly toxic intermediates are generated. In particular, cellular levels of epoxide moieties resulting from chemical metabolism appear to be critical initiators of toxic damage, including genetic mutation. Frequently reactive and unstable, epoxide metabolites, formed via the action of the cytochrome P450 monooxygenases, have been identified as ultimate carcinogenic and cytotoxic reaction products (Sayer et al., 1985; Thakker et al., 1986; Fretland and Omiecinski, 2000). Ultimately, the overall balance between bioactivation and detoxication pathways will determine the kinetics and fate of reactive intermediates within target cells. It seems likely that interindividual differences in susceptibility to toxic sequelae, including cancer incidence, may be associated with an altered genetic predisposition to detoxify epoxides. Tissues have developed the capacity to metabolize xenobiotic epoxides through several pathways. Prominent among these is the microsomal epoxide hydrolase (EPHX) 1 pathway.
A wide variation of interindividual EPHX1 activities have been reported across human tissues (Mertes et al., 1985; Omiecinski et al., 1993; Hassett et al., 1997). We previously characterized two structural polymorphisms, an exon 3 polymorphism corresponding to amino acid position 113, with a resulting Tyr to His substitution (rs1051740), and an exon 4 polymorphism at position 139, coding for a His-to-Arg substitution (rs2234922) (Hassett et al., 1994a). Tyr is the predominant amino acid at the 113 position in white populations, with His most common at 139. A large number of investigations have been published now examining the potential association of these coding region polymorphisms with altered xenobiotic disposition and disease incidence. Reports include potential associations with EPXH1 genetics and warfarin dose requirements (Loebstein et al., 2005), estrogen production (Hattori et al., 2000), and anticonvulsant disposition (Robbins et al., 1990). Epidemiological studies focusing on cancer susceptibility have reported associations with the H113H genotype of EPHX1 and a decreased risk of lung cancer (Kiyohara et al., 2006). Other reports have indicated an increased risk of chronic obstructive pulmonary disease (Park et al., 2005; Brøgger et al., 2006) and emphysema (Smith and Harrison, 1997) with the H113H genotype and a protective association of the Y113H genotype with chronic obstructive pulmonary disease (Brøgger et al., 2006). Certain of these results may be contributed by differential activities of the various EPHX1 allelic proteins on specific substrates involved in diverse disease etiologies.
It is well established that the RNA transcriptome in mammals is expanded markedly through the use of alternative promoters and differential RNA splicing mechanisms and that the use of alternative promoters is an important source of generating protein and regulatory multiplicity (Kimura et al., 2006; Davuluri et al., 2008). We reported recently that expression of the human EPHX1 gene is driven by an alternative promoter region, termed the E1-b promoter, that is localized ∼18.5 kb 5′ upstream from the structural region of the EPHX1 gene and is responsible for driving expression of EPHX1 mRNA transcripts in many tissues from both adult and fetal sources (Liang et al., 2005). Before this discovery, only the proximal E1 promoter was recognized, which lies directly upstream of the EPHX1 coding region (Skoda et al., 1988; Hassett et al., 1994b). However, the E1 promoter seemed to drive EPHX1 expression selectively in the liver (Liang et al., 2005).
Given the importance of the upstream EPHX1 promoter as a regulator of its functional expression in most human tissues, in this study, we conducted comparative genomic analyses on this region and identified the variable presence of polymorphic repetitive elements of the Alu class among humans within a ∼3-kb region of the E1-b promoter. In vitro promoter activity assays conducted in several human cell lines demonstrated that these polymorphisms were associated with reduced transcriptional activities and, therefore, may represent important contributors to interindividual differences in EPHX1 activity within tissues targeted by xenobiotic compounds.
Materials and Methods
Quantitative Real-Time PCR. Human tissue RNA was obtained from the FirstChoice Human total RNA survey panel (Ambion, Austin, TX). The total RNA was converted to cDNA using the High Capacity cDNA Archive Kit according to the manufacturer's instructions. Real-time PCR was performed using Custom Taqman Gene Expression Products (Applied Biosystems, Foster City, CA) following the manufacturer's standard protocol. The E1 variant was detected using the forward primer (5′-CTCCACAGCTCTCTTTCCCAA-3′), reverse primer (5′-CCACCAGGCTCCACGTT-3′), and probe (5′-TCACCCTCTGATTACTCC-3′). Likewise, the E1-b variant was detected using the forward primer (5′-GATCGCGCGCCTGC-3′), reverse primer (5′-GTGAGGAGGATTTCTAGCCACATG-3′), and probe (5′-CTCGCAGGCTCCGGC-3′). Real-time reverse transcription-PCR data were analyzed using methods described previously (Page et al., 2007). Plasmids containing cloned E1 or E1-b full-length sequences were diluted to create standard curves, ranging from 30 copies to 3 × 107 copies. DNA samples of known target sequence were used to verify the specificity of the assays.
Cell Culture. The hepatoma HepG2 and lung carcinoma A549 cell lines were grown in minimal essential medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, 0.1 mM nonessential amino acids, and 1.5 g/l sodium bicarbonate. The transformed human embryonic kidney lines 293A and 293T and breast adenocarcinoma MCF7 cell line were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. All cells were maintained at 37°C with 5% CO2.
Plasmid Construction and PCR Genotyping. A ∼2.8-kb DNA fragment upstream of E1-b was obtained by a first PCR amplification of genomic DNA by forward (5′-GGGTAGTAAACTGATTGGCCTC-3′) and reverse (5′-CGGGCGCTTACGGTCTCG-3′) primers followed by a second PCR with nested primers (5′-GACTGGTACCGGAATTGATCTACAATTTTTATCC-3′ and (5′-GAAGATCTCTC-TCCGGCTCCCTGGCTCTCCTC-3′) using UniPOL DNA polymerase (Gene-Choice, San Diego, CA). This 2.8-kb fragment was inserted into the KpnI and XhoI sites of the luciferase-reporter vector, pGL3-basic. The insert sequences were sequenced with forward (5′-CTAGCAAAATAGGCTGTCCC-3′) and reverse (5′-CTTTATGTTTTTGGCGTCTTCCA-3′) primers using a CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA). For the PCR genotyping of the Alu insertion, the primers (5′-ACCAAGTAGGAGGGTATTAG-3′) and (5′-ACCTGCAGTCTGGG-AGAGTTCTTT-3′) were used for PCR of 10 to 30 ng of genomic DNA (DNA Polymorphism Discovery Resource, Coriell Institute, Camden, NJ) with Taq DNA polymerase (GeneChoice).
Western Immunoblotting. Equal amounts (10 μg) of total cell lysate obtained from HepG2, A549, and 293A cells were loaded on a 10% SDS-polyacrylamide gel. After separation, proteins were transferred to a polyvinylidene fluoride membrane. Detection of EPHX1 proteins was performed after membranes were incubated with EPHX1 peptide-directed polyclonal antibody and mouse anti rabbit secondary antibodies as described previously (Hassett et al., 1998). Signals were visualized using the Lumi-Light Western blotting substrate (Roche Diagnostics, Indianapolis, IL). The glyceraldehyde-3-phosphate dehydrogenase antibody (Sigma-Aldrich, St. Louis, MO) was used as a loading control.
Cell Transfection and Luciferase Activity Assay. A549, HepG2, and MCF7 cells were transfected with Lipofectamine 2000 (Invitrogen). The 293 cells were transfected with FuGENE 6 (Roche Diagnostics). The transfection procedure and luciferase assay were performed as described previously (Liang et al., 2005).
Haplotype Analysis. The EPHX1 haplotype block structure was determined by the Haploview algorithm based on data provided from the HapMap Project (Barrett et al., 2005).
Statistical Analysis. All statistical analyses were performed using GraphPad Prism version 4.00 for Windows (GraphPad Software Inc., San Diego, CA). To assess the impact of the Alu repeat polymorphism on transcriptional activities in A549, HepG2, MCF7, and 293A cells, a two-tailed Student's t test was used to examine differences between each pair of haplotypes. One-way analysis of variance in combination with Tukey's post hoc test was used to determine significance of the difference in the E1-b EPHX1 transcript levels between 293T and either the A549 or HepG2 cell lines. Significant differences were designated in instances where p was <0.05.
Results
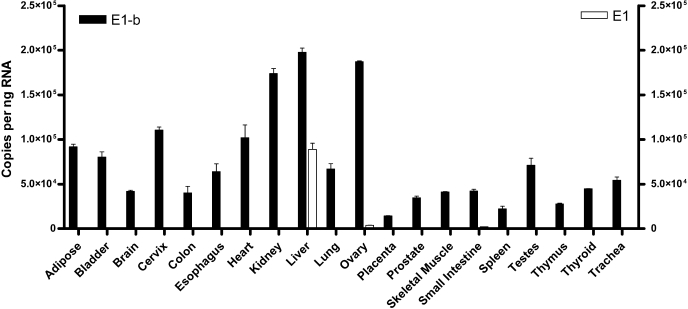
The E1-b Variant Is the Primary Transcript in Human Tissues. We designed specific, quantitative real-time PCR assays to assess the expression of the E1 and E1-b alternative EPHX1 transcripts across a panel of 20 human tissues. Each tissue sample is a pooled sample, obtained from at least three different adult individuals. As shown in Fig. 1, both the E1 and the E1-b EPHX1 transcripts were detected at high levels in the liver, although E1-b was the major hepatic transcript, accounting for ∼70% of the total EPHX1 RNA expressed in this tissue. Although the expression of the E1 transcript was largely liver-selective, very low levels of E1 were also detected in the ovary, small intestine, and testes. In contrast, the E1-b transcript was detected as the predominant EPHX1 transcript in all tissues examined.
Fig. 1.
Quantification of the EPHX1 E1 and E1-b transcripts expression in different tissues. Real-time PCR was performed with RNA obtained from 20 human tissues; each sample represents a pool of at least three individuals. E1-b (black bar) and E1 (white bar) transcript copy numbers were quantified by absolute quantification based on standard curves determined using plasmid DNA templates.
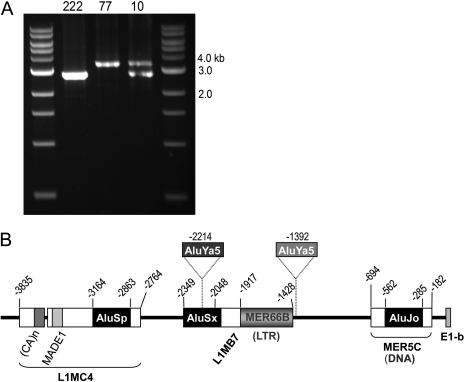
Identification of Alu Insertion Polymorphisms in the Proximal Upstream Region of the E1-b mEH Variant. In efforts to further characterize the structure of the E1-b promoter region, we cloned and analyzed a 2.8-kb DNA fragment from various individuals within the Polymorphism Discovery Resource genomic panel (DNA Polymorphism Discovery Resource, Coriell Institute). During these studies, we observed different genomic patterns occurring in different individuals. Overall, three types of PCR profiles were encountered: 1) a homozygous, wide-type fragment profile; 2) a profile consisting of a wild-type fragment plus a larger fragment; and 3) a profile containing a homozygous, larger molecular weight fragment (Fig. 2A). All of the genomic fragments were subjected to DNA sequence analysis. The larger genomic fragment was characterized as containing two Alu insertions (2xAlu) at nucleotide positions -2214 and -1392 (Fig. 2B). It is interesting these two Alu insertions that occurred at separate positions were always linked together, i.e., in no instance have we detected only single Alu polymorphic insertions within a given haplotype. By searching the RepeatMasker database (http://www.repeatmasker.org), we determined that both the -2214 and -1392 Alu insertional elements belong to the Ya5 Alu family, evolutionarily a relatively young Alu insertion. Although largely conserved in their sequence, the Alu insertions did possess several single base differences within their nucleotide content (data not shown).
Fig. 2.
Identification of Alu-insertion polymorphisms in the 5′-flanking region of E1-b. A, PCR analysis of polymorphic DNA fragments ∼2.8 kb upstream of E1-b. The genomic DNA (20 ng) of three individuals (nos. 10, 77, and 222 of DNA Polymorphism Discovery Resource, Coriell Institute) were subsequently amplified using two primer sets as described under Materials and Methods. B, schematic representation of the polymorphic Alu (Ya5) insertions upstream of E1-b. Also indicated are the TE cassettes formed through combinations of different types of TEs that belong to SINE (AluSp, AluSx, AluJo), long interspersed element (L1MC4, L1MB7), long terminal repeat (MER66B), tandem repeat ((Ca)n), or DNA transposon (MADE1, MER5C) classifications.
A PCR genotyping protocol was used to determine the frequency of these Alu insertions in the human population. In this analysis, the E1-b upstream DNA region (-2428 to -1305) was analyzed across 450 individuals of the Coriell Institute's human genomic DNA collection, containing samples from 450 U.S. residents with ancestry from all the major regions of the world (Collins et al., 1998). The results revealed that ∼60% of the population had no Alu insertion [Alu(-/-)], whereas ∼36% of the individuals were heterozygous for the 2xAlu insertion elements [Alu(+/-)], and 4% of the population were homozygous for the 2xAlu insertion (Table 1).
TABLE 1.
Frequency of the Alu insertion polymorphisms within the 5′-flanking region of the EPXH1 E1-b promoter region in a sample of the United States population
| Genotype | Number of Samples | Percentage |
|---|---|---|
| % | ||
| Alu(–/–) | 269 | 59.9 |
| Alu(+/–) | 162 | 36.1 |
| Alu(+/+) | 18 | 4.0 |
Multispecies Sequence Alignments of the E1-b Upstream Promoter Region. In analyses not shown, we used the UCSC Genome Browser (http://www.genome.ucsc.edu) to assess evolutionary conservation within the distal 3-kb 5′-flanking sequence of the EPHX1 E1-b promoter region among 28 vertebrates, including mammalian, amphibian, bird, and fish species. To reduce potential errors in the browser alignment algorithms, segments of EPHX1 sequences were also submitted to the BLASTN program to search directly for sequence conservation against all genomic National Center for Biotechnology Information databases. The results of the analysis revealed that the E1-b 5′-flanking region is highly conserved among human, chimp, and rhesus but is not identified in any species outside the primate clade (data not shown). The region comprising the immediate 300 base pairs 5′ of the human E1-b transcriptional start site was conserved in both chimp and rhesus; however, the Alu insertion polymorphic elements identified in the human EPXH1 gene locus (Fig. 2B; chromosome 1, 224062300–224063000) and the repetitive element structure surrounding this insertion region were present only in the human and chimp genomes. In contrast, within the structural regions of the gene in the human and other vertebrate species, the protein coding sequences of EPHX1 are relatively highly conserved. It is interesting that in the human and chimp genomes, EPHX1 was localized to the chromosome 1 “plus” strand, whereas for rhesus, EPHX1 sequences were localized in the chromosome 1 “reverse” strand (data not shown).
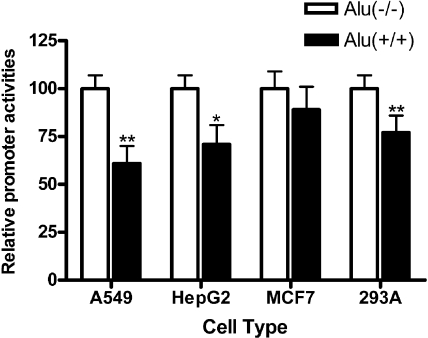
Analyses of Promoter Activities of the Polymorphic E1-b 5′-Flanking Region. We reasoned that the existence of multiple polymorphisms within the proximal upstream region of the E1-b promoter region may affect the transcriptional activity of the gene, perhaps leading to interindividual differences in EPHX1 expression. To assess the functional significance of these sequence variations, two polymorphic haplotypes were cloned by PCR amplification and ligated into the luciferase reporter vector, pGL3-basic. The promoter activities of the resulting 2xAlu and wild-type constructs were analyzed in vitro using transfection assays conducted in four different human cell lines, 293A, A549, HepG2, and MCF7. The presence of the 2xAlu double insertion resulted in lower associated transcriptional activities, although these activities varied among the cell types tested. A statistically significant reduction in promoter activity was associated with the insertion assayed in A549, HepG2, and 293A cells (Fig. 3), with the maximal difference (40% reduction) occurring in A549 cells. Also trending lower was the Alu-associated transcriptional activity level assessed in the MCF7 breast cancer cell line; however, the lower trend observed in the MCF7 cells was not statistically significant. Overall, these data suggest that the presence of the genetically polymorphic Alu repeat elements within the 5′-flanking region of the EPHX1 E1-b promoter down-regulates resultant EPHX1 gene transcriptional activity.
Fig. 3.
Analysis of promoter activities of the polymorphic E1-b 5′-flanking region. Luciferase-based promoter activities were analyzed by cloning the E1-b 5′-flanking DNA region (-2763/+6) for different haplotypes into the pGL3-basic vector. The data shown depict means and S.D. values derived from five separate experiments, each performed in duplicate. Inclusion of the 2xAlu element results in a statistically significant decreased promoter activity in A549, HepG2, and 293A cells (*, p < 0.05; **, p < 0.01).
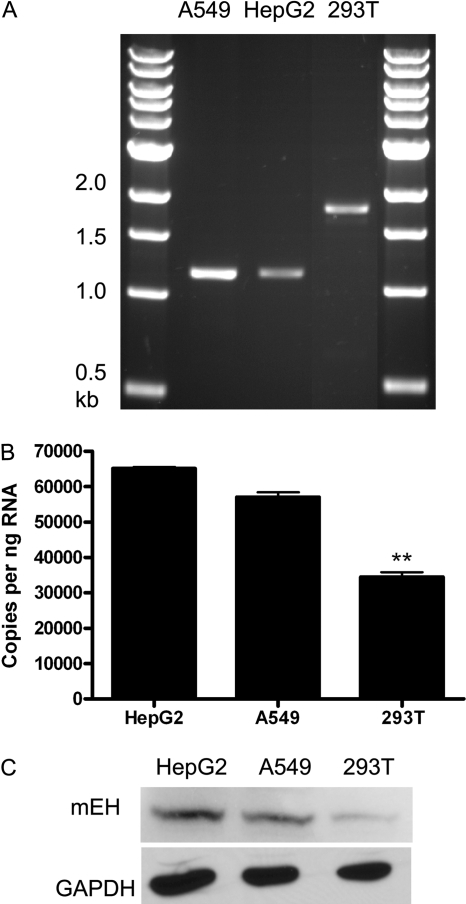
We further analyzed the Alu insertion region within selected cell lines by the specific PCR amplification of a DNA fragment between -2428 and -1305 of E1-b. Of interest, 293A (data not shown) and 293T kidney cells possessed the homozygous 2xAlu insertion genotype, whereas the 2xAlu elements were not present in A549 lung cells and HepG2 hepatoma cells (Fig. 4A). We used our quantitative reverse transcription-PCR assays to measure the E1-b transcript levels in these respective cell lines and determined that the 293T cells expressed significantly less transcript than the either of the other cell lines (Fig. 4B). We further assessed the EPHX1 protein levels in these cells using immunoblotting methods and observed that the 293T cells were similarly reduced in their corresponding EPHX1 protein content (Fig. 4C).
Fig. 4.
Characterization of the E1-b promoter region and EPHX1 expression in human cell lines. A, PCR genotyping of Alu insertion polymorphism in A549, HepG2, and 293T cells. The DNA fragment between -2428 and -1305 of E1-b was specifically amplified from genomic DNA demonstrating the presence of the 2xAlu insertion in 293T cells but not A549 or HepG2 cells. B, real-time PCR was performed with RNA obtained from the respective cell lines. The data shown depict means and S.D. values derived from two separate experiments, each performed in triplicate. The 293T cells exhibited a significantly lower level of EPHX1 E1-b transcript expression compared either with HepG2 or A549 cells (**, p < 0.01). C, Western immunoblot blot analysis assessing corresponding EPHX1 protein levels.
HapMap Structure of the EPHX1 Coding Region and the 5′ Upstream Region. The EPHX1 gene plays an important role in the activation of carcinogenic compounds, such as polycyclic aromatic hydrocarbons in cigarette smoke. Results from several epidemiological investigations have now suggested that EPHX1 coding region polymorphisms may alter the risk of developing lung cancer, as summarized by Kiyohara et al. (2006), and other human diseases, such as chronic obstructive pulmonary disease (Park et al., 2005; Brøgger et al., 2006) and emphysema (Smith and Harrison, 1997). Thus, it was of interest to examine the genetic linkage of the coding polymorphisms with that of the E1-b promoter region Aluinsertion polymorphism.
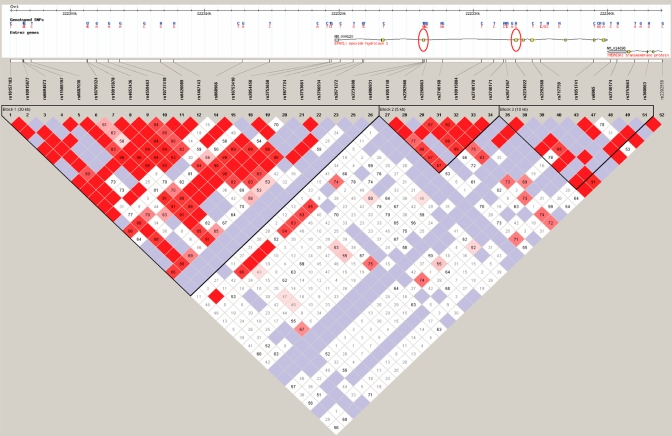
We used Haploview software, the algorithm derived for analysis of HapMap project data, to determine the EPHX1 gene locus linkage disequilibrium (LD) structure. LD describes the situation where combinations of alleles or genetic markers occur more or less frequently in a population than would otherwise be predicted as a random combination of allelic haplotypes based on their known frequencies of occurrence. Thus, nonrandom associations between genetic polymorphisms at different loci are measured by the degree of LD. The map of these projections is presented in Fig. 5. The structural region of the gene together with its extended 5′ sequence, including the distal E1-b promoter region (human chromosome 1, 222304957–222301977), is indicated in the upper portion of the figure, with the exon 3 and exon 4 polymorphism positions marked with red ovals. Across this genetic region, three distinct haplotype blocks are apparent, delineated by the black triangular borders that are filled in with predominantly red squares. What is clear from the HapMap is that the distal E1-b promoter region is not in linkage disequilibrium with the coding region of the gene, which itself is divided into two haplotype blocks, with one block harboring the exon 3 113 SNP and another harboring the exon 4 139 SNP. Therefore, in the EPHX1 coding-region, the exon 3 and exon 4 nonsynonomous SNPs were not in linkage disequilibrium with each other.
Fig. 5.
LD structure of a 52-kb region spanning EPHX1 and extended promoter in European Americans. EPHX1 genotypes from HapMap (http://www.hapmap.org/) were downloaded, and the LD was determined using Haploview software. D′ values are displayed in the squares (empty squares have a pairwise D′ 1.00). Red squares show high pairwise LD, gradually coloring down to white squares of low pairwise LD. The black triangles indicate the haplotype blocks. There are three haplotype blocks in this region (one block in the large promoter region that spans into the first two exons of the gene, one block that spans exon 3 and nearby intronic regions, and one block that spans exons 4–9). At the upper portion gene structure region, red ovals indicate the coding polymorphisms in exons 3 (rs1051740) and 4 (rs2234922), which are not in LD with each other, nor with the promoter region.
Discussion
The primary coding sequences of the human, rodent, and rabbit EPHX1 genes are highly conserved, with alignments possessing >75% similarity (Fretland and Omiecinski, 2000). These observations alone may suggest that the EPHX1 gene product has important biological function. EPHX1 is an important contributor to the metabolism of many xenobiotic epoxides, including those derived from the metabolism of certain pharmaceuticals and a host of environmental toxins such as polyaromatic hydrocarbons, butadiene, and acrylamide (Fretland and Omiecinski, 2000; Morisseau and Hammock, 2005). The data presented in this study demonstrate conclusively that EPHX1 expression in most human tissues is directed predominantly through the use of a far upstream alternative gene promoter, termed the E1-b promoter. Our analyses of the E1-b 5′-flanking promoter region further identified a large concentration of transposable elements (TEs) (Fig. 2B). Nearly half of the human genome is derived from ancient TEs, including short interspersed elements (SINEs) and long interspersed elements, long terminal repeats, and DNA transposons (Jasinska and Krzyzosiak, 2004). TEs can promote genetic diversification and regulatory variation by serving as recombination hot spots or acquiring specific cellular functions such as adopting portions of protein coding regions or by affecting gene transcription (Lorenc and Makałowski, 2003; van de Lagemaat et al., 2003). Furthermore, TEs have been identified to function as alternative promoters of many genes, including aromatase, carbonic anhydrase 1, and bile acid CoA/amino acid N-transferase, resulting in tissue-specific regulation of gene expression.
We used comparative sequence analysis to discover that the EPHX1 E1-b promoter region is polymorphic among individuals, specifically with respect to the presence of dual TEs of the Alu-class. As one important component of TEs, Alu elements are primate-specific and represent the most abundant of the SINE group; their 1.1 million copies occupy over 10% of the human genome (Mighell et al., 1997). Alu repeats are unevenly distributed, with transcriptionally active regions of the genome especially densely populated, and probably play roles in human gene regulation (Korenberg and Rykowski, 1988; Moyzis et al., 1989; Grover et al., 2004). Alu elements are divided into several subfamilies according to their insertion age (Kapitonov and Jurka, 1996; Mighell et al., 1997). Almost all of the insertions occurring specifically in the human genome belong to four closely related subfamilies, Alu Y, Ya5, Ya8, and Yb8 (Deininger and Batzer, 1999), with the majority of human Alu elements belonging to old or intermediate subfamilies. Although present in relatively low copy numbers, the younger Alus are considered the most active and those most likely to possess biological function. These more recent Alu insertions also result in genetic variation in human populations by generating polymorphic loci. In this study, we report the new finding of Alu insertion polymorphisms of the youngest Alu class, the Ya5 class, in the E1-b 5′-flanking region of the EPHX1 gene.
Frequency analysis in a panel of 450 individuals demonstrated that the combination of Alu insertion/deletion polymorphisms results in three different EPHX1 E1-b promoter haplotypes: Alu(-/-) genotype (59.9%), Alu(+/-) genotype (36.1%), and Alu(+/+) genotype (4.0%) (Table 1). Furthermore, data obtained from in vitro promoter activity assays indicate that the presence of the 2xAlu insertion down-regulates the transcriptional level of the associated E1-b EPHX1 transcription in several cells types, suggesting that individuals with the Alu(+/+) genotype may possess lower EPXH1 activities in certain tissues. Comparative species assessments using the UCSC genome browser indicated that the EPHX1 E1-b promoter region context was a specific attribute of primate species (data not shown). The EPHX1 Alu element polymorphism may serve as a future marker for the study of human population genetics, disease associations, and genomic diversity and evolution.
These results, together with our previous findings that genetic polymorphisms exist both in the coding region and the proximal 5′-flanking region of the EPHX1 gene (Hassett et al., 1994a; Raaka et al., 1998), may constitute determinants of interindividual and tissue-specific differences in capacities to biotransform xenobiotic-derived epoxides. The upstream promoter region Alu element polymorphism identified in this study is of particular interest in the respect that E1-b is the primary EPHX1 transcript expressed in human tissues. EPHX1 transcript level analyses within a panel of human tissues demonstrated the largely liver-specific expression pattern of the E1 proximal promoter-derived transcript; in contrast, the far distal E1-b promoter ubiquitously and predominantly drives EPHX1 expression in all tissues examined. Human/primate genetic evolution apparently has selected the alternative E1-b promoter as the primary regulator of EPXH1 expression.
The association between genetic polymorphisms of the biotransformation enzymes and the risk of cancer has been of particular interest for the study of carcinogenesis. One major group of strong carcinogenic materials in tobacco smoking is the polycyclic aromatic hydrocarbon with bay or fjord regions (Hecht et al., 1994). The ultimate carcinogenic polycyclic aromatic hydrocarbon metabolites are generated through the concerted action of multiple enzymes, such as the cytochrome P450s, glutathione S-transferases, and EPHX1. EPHX1-null mice are completely resistant to the tumorigenic effects of dimethylbenz[a]anthracene in a complete carcinogenesis assay (Miyata et al., 1999), and results of several human epidemiologic studies suggest a reduced risk of lung cancer in individuals with a H113 genotype, as summarized by Kiyohara et al. (2006). The pair of ∼300-base pair Alu insertion elements described in the current investigation provides several potential binding sites for transcription factors and hormone receptors (Shankar et al., 2004). Additional epidemiological studies are required to investigate any potential associations of the polymorphic Alu insertions within the upstream promoter region of the EPHX1 gene with human cancers or with the incidence of other disease states previously implicated with EPHX1 genetic polymorphisms (e.g., Smith and Harrison, 1997; Fretland and Omiecinski, 2000; Brøgger et al., 2006).
When considering the association of genetic polymorphisms with the risk of human disease, to better predict complex phenotypes or disease processes, one clearly needs to consider pairwise LD or haplotype blocks (several linked polymorphic alleles), rather than reliance on a single polymorphic site (Crawford and Nickerson, 2005). The HapMap structure of the EPHX1 gene (Fig. 5) indicated that the E1-b Alu polymorphisms in the regulatory region were not in the same haplotype block with either the Tyr113/His or Arg139/His coding region polymorphisms previously identified by our laboratory (Hassett et al., 1994a) and that the respective coding region polymorphisms also are predicted to reside in separate haplotype blocks, indicating that there is not a strong linkage association among these three polymorphisms and that variations in each polymorphism can affect the phenotype independently.
In summary, the results from our current study serve to further characterize the tissue-selective expression context of human EPHX1 and have identified the polymorphic nature of the far upstream E1-b promoter region of EPHX1, the promoter that serves as the predominant transcriptional driver of EPHX1 in human cells. These studies have also elucidated the haplotype structure of the extended EPHX1 genomic locus, thereby contributing a context for further investigations examining the associations of EPHX1 genetic polymorphisms with human diseases arising from environmental or pharmaceutical exposures that are subject to metabolism through the EPHX1 enzymatic pathway.
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES016358].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.150870.
ABBREVIATIONS: EPHX1, mEH, human microsomal epoxide hydrolase; E1-b, EPHX1 exon 1-b; kb, kilobase pair(s); E1, EPHX1 exon 1; PCR, polymerase chain reaction; LD, linkage disequilibrium; SNP, single nucleotide polymorphism; TE, transposable element; SINE, short interspersed element.
References
- Barrett JC, Fry B, Maller J, and Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 263-265. [DOI] [PubMed] [Google Scholar]
- Brøgger J, Steen VM, Eiken HG, Gulsvik A, and Bakke P (2006) Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur Respir J 27 682-688. [DOI] [PubMed] [Google Scholar]
- Collins AR, Gedik CM, Olmedilla B, Southon S, and Bellizzi M (1998) Oxidative DNA damage measured in human lymphocytes: large differences between sexes and between countries, and correlations with heart disease mortality rates. FASEB J 12 1397-1400. [PubMed] [Google Scholar]
- Crawford DC and Nickerson DA (2005) Definition and clinical importance of haplotypes. Ann Rev Med 56 303-320. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, and Huang TH (2008) The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24 167-177. [DOI] [PubMed] [Google Scholar]
- Deininger PL and Batzer MA (1999) Alu repeats and human disease. Mol Genet Metab 67 183-193. [DOI] [PubMed] [Google Scholar]
- Fretland AJ and Omiecinski CJ (2000) Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact 129 41-59. [DOI] [PubMed] [Google Scholar]
- Grover D, Mukerji M, Bhatnagar P, Kannan K, and Brahmachari SK (2004) Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics 20 813-817. [DOI] [PubMed] [Google Scholar]
- Hassett C, Aicher L, Sidhu JS, and Omiecinski CJ (1994a) Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet 3 421-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett C, Laurenzana EM, Sidhu JS, and Omiecinski CJ (1998) Effects of chemical inducers on human microsomal epoxide hydrolase in primary hepatocyte cultures. Biochem Pharmacol 55 1059-1069. [DOI] [PubMed] [Google Scholar]
- Hassett C, Lin J, Carty CL, Laurenzana EM, and Omiecinski CJ (1997) Human hepatic microsomal epoxide hydrolase: comparative analysis of polymorphic expression. Arch Biochem Biophys 337 275-283. [DOI] [PubMed] [Google Scholar]
- Hassett C, Robinson KB, Beck NB, and Omiecinski CJ (1994b) The human microsomal epoxide hydrolase gene (EPHX1): complete nucleotide sequence and structural characterization. Genomics 23 433-442. [DOI] [PubMed] [Google Scholar]
- Hattori N, Fujiwara H, Maeda M, Fujii S, and Ueda M (2000) Epoxide hydrolase affects estrogen production in the human ovary. Endocrinology 141 3353-3365. [DOI] [PubMed] [Google Scholar]
- Hecht SS, el-Bayoumy K, Rivenson A, and Amin S (1994) Potent mammary carcinogenicity in female Cd rats of a fjord region diol-epoxide of benzo[C]phenanthrene compared to a bay-region diol-epoxide of benzo[A]pyrene. Cancer Res 54 21-24. [PubMed] [Google Scholar]
- Jasinska A and Krzyzosiak WJ (2004) Repetitive sequences that shape the human transcriptome. FEBS Lett 567 136-141. [DOI] [PubMed] [Google Scholar]
- Kapitonov V and Jurka J (1996) The age of Alu subfamilies. J Mol Evol 42 59-65. [DOI] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. (2006) Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16 55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara C, Yoshimasu K, Takayama K, and Nakanishi Y (2006) EPHX1 polymorphisms and the risk of lung cancer: a HuGE review. Epidemiology 17 89-99. [DOI] [PubMed] [Google Scholar]
- Korenberg JR and Rykowski MC (1988) Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53 391-400. [DOI] [PubMed] [Google Scholar]
- Liang SH, Hassett C, and Omiecinski CJ (2005) Alternative promoters determine tissue-specific expression profiles of the human microsomal epoxide hydrolase gene (EPHX1). Mol Pharmacol 67 220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebstein R, Vecsler M, Kurnik D, Austerweil N, Gak E, Halkin H, and Almog S (2005) Common genetic variants of microsomal epoxide hydrolase affect warfarin dose requirements beyond the effect of cytochrome P450 2C9. Clin Pharmacol Ther 77 365-372. [DOI] [PubMed] [Google Scholar]
- Lorenc A and Makałowski W (2003) Transposable elements and vertebrate protein diversity. Genetica 118 183-191. [PubMed] [Google Scholar]
- Mertes I, Fleischmann R, Glatt HR, and Oesch F (1985) Interindividual variations in the activities of cytosolic and microsomal epoxide hydrolase in human liver. Carcinogenesis 6 219-223. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Markham AF, and Robinson PA (1997) Alu sequences. FEBS Lett 417 1-5. [DOI] [PubMed] [Google Scholar]
- Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, and Gonzalez FJ (1999) Targeted disruption of the microsomal epoxide hydrolase gene: microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem 274 23963-23968. [DOI] [PubMed] [Google Scholar]
- Morisseau C and Hammock BD (2005) Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol 45 311-333. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Torney DC, Meyne J, Buckingham JM, Wu JR, Burks C, Sirotkin KM, and Goad WB (1989) The distribution of interspersed repetitive DNA sequences in the human genome. Genomics 4 273-289. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Aicher L, Holubkov R, and Checkoway H (1993) Human peripheral lymphocytes as indicators of microsomal epoxide hydrolase activity in liver and lung. Pharmacogenetics 3 150-158. [DOI] [PubMed] [Google Scholar]
- Page JL, Johnson MC, Olsavsky KM, Strom SC, Zarbl H, and Omiecinski CJ (2007) Gene expression profiling of extracellular matrix as an effector of human hepatocyte phenotype in primary cell culture. Toxicol Sci 97 384-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Chen L, Wadhwa N, and Tockman MS (2005) Polymorphisms for microsomal epoxide hydrolase and genetic susceptibility to COPD. Int J Mol Med 15 443-448. [PMC free article] [PubMed] [Google Scholar]
- Raaka S, Hassett C, and Omiencinski CJ (1998) Human microsomal epoxide hydrolase: 5′-flanking region genetic polymorphisms. Carcinogenesis 19 387-393. [DOI] [PubMed] [Google Scholar]
- Robbins DK, Wedlund PJ, Kuhn R, Baumann RJ, Levy RH, and Chang SL (1990) Inhibition of epoxide hydrolase by valproic acid in epileptic patients receiving carbamazepine. Br J Clin Pharmacol 29 759-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer JM, Yagi H, van Bladeren PJ, Levin W, and Jerina DM (1985) Stereoselectivity of microsomal epoxide hydrolase toward diol epoxides and tetrahydroepoxides derived from benz[a]anthracene. J Biol Chem 260 1630-1640. [PubMed] [Google Scholar]
- Shankar R, Grover D, Brahmachari SK, and Mukerji M (2004) Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol Biol 4 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoda RC, Demierre A, McBride OW, Gonzalez FJ, and Meyer UA (1988) Human microsomal xenobiotic epoxide hydrolase: complementary DNA sequence, complementary DNA-directed expression in COS-1 cells, and chromosomal localization. J Biol Chem 263 1549-1554. [PubMed] [Google Scholar]
- Smith CA and Harrison DJ (1997) Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet 350 630-633. [DOI] [PubMed] [Google Scholar]
- Thakker DR, Levin W, Yagi H, Yeh HJ, Ryan DE, Thomas PE, Conney AH, and Jerina DM (1986) Stereoselective metabolism of the (+)-(S,S)- and (-)-(R,R)-enantiomers of trans-3,4-dihydroxy-3,4-dihydrobenzo[c]- phenanthrene by rat and mouse liver microsomes and by a purified and reconstituted cytochrome P-450 system. J Biol Chem 261 5404-5413. [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, and Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19 530-536. [DOI] [PubMed] [Google Scholar]