Abstract
Human pigmentation is a polygenic quantitative trait with high heritability. Although a large number of single nucleotide polymorphisms (SNPs) have been identified in pigmentation genes, very few SNPs have been examined in relation to human pigmentary phenotypes and skin cancer risk. We evaluated the associations between fifteen SNPs in eight candidate pigmentation genes (TYR, TYRP1, OCA2, SLC24A5, SLC45A2, POMC, ASIP, and ATRN) and both pigmentary phenotypes (hair color, skin color, and tanning ability) and skin cancer risk in a nested case-control study of Caucasians within the Nurses’ Health Study (NHS) among 218 melanoma cases, 285 squamous cell carcinoma (SCC) cases, 300 basal cell carcinoma (BCC) cases, and 870 common controls. We found that the TYR Arg402Gln variant was significantly associated with skin color (p-value =7.7×10−4) and tanning ability (p-value =7.3×10−4); the SLC45A2 Phe374Leu variant was significantly associated with hair color (black to blonde) (p-value =2.4×10−7), skin color (p-value =1.1×10−7), and tanning ability (p-value =2.5×10−4). These associations remained significant after controlling for MC1R variants. No significant associations were found between these polymorphisms and the risk of skin cancer. We observed that the TYRP1 rs1408799 and SLC45A2 -1721 C>G were associated with melanoma risk (OR, 0.77; 95% CI, 0.60–0.98 and OR, 0.75; 95% CI, 0.60–0.95, respectively). The TYR Ser192Tyr was associated with SCC risk (OR, 1.23; 95% CI, 1.00–1.50). The TYR haplotype carrying only the Arg402Gln variant allele was significantly associated with SCC risk (OR, 1.35; 95% CI, 1.04–1.74). The OCA2 Arg419Gln and ASIP g.8818 A>G were associated with BCC risk (OR, 1.50; 95% CI, 1.06–2.13 and OR, 0.73; 95% CI, 0.53–1.00, respectively). The haplotype near ASIP (rs4911414[T] and rs1015362[G]) was significantly associated with fair skin color (OR, 2.28; 95% CI, 1.46–3.57) as well as the risks of melanoma (OR, 1.68; 95% CI, 1.18–2.39) and SCC (OR, 1.54; 95% CI, 1.08–2.19). These associations remained similar after adjusting for pigmentary phenotypes and MC1R variants. The statistical power of this study was modest and additional studies are warranted to confirm the associations observed in the present study. This study provides evidence for the contribution of pigmentation genetic variants, in addition to the MC1R variants, to variation in human pigmentary phenotypes and possibly the development of skin cancer.
Keywords: SNPs, pigmentation gene, pigmentary phenotypes, skin cancer
Introduction
Human pigmentation shows substantial variation both within and among human populations, with high heritability 1, 2. Ultraviolet (UV) exposure is one of the most important environmental variables partially influencing evolutionary selective pressure on human pigmentation 3. Melanin synthesized within melanosomes in melanocyte is the main contributor to human pigmentation. There are two main types of melanin: pheomelanin (red or yellow) and eumelanin (black or brown) 4.
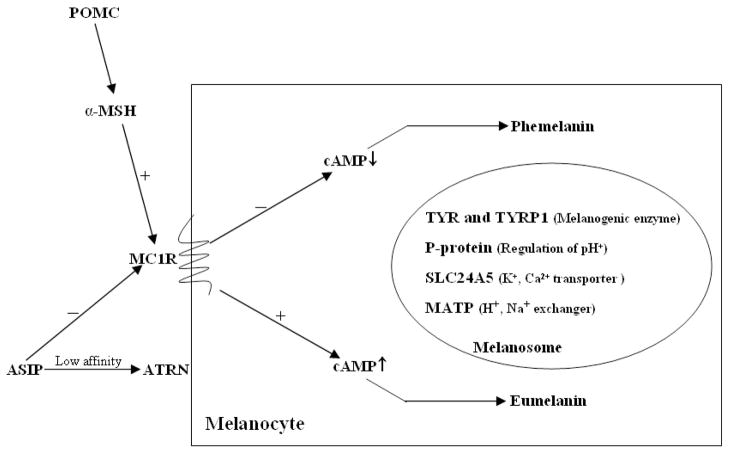
It has been hypothesized that human pigmentation is tightly regulated by multiple pigmentation genes harboring a handful of genetic variants (Figure 1). The genes involved in the process of pigmentation, such as formation, transport, and distribution of melanosome, have been identified through animal models 5. Melanin production is initiated by α-melanocyte-stimulating hormone (α-MSH), which is produced by proteolysis from a multicomponent precursor polypeptide encoded by the pro-opiomelanocortin (POMC) gene 6. A previously well-documented pigmentation gene, MC1R (melanocortin 1 receptor), encodes a 317-amino acid 7-pass transmembrane G-protein coupled receptor. As an agonist of MC1R, the induced POMC/α-MSH binds to MC1R, leading to elevated cAMP levels and resulting in eumelanin production 7, 8. Alternatively, the agouti signaling protein (ASIP) can also bind to the MC1R, blocking the MC1R-stimulated elevation of cAMP, and over-expression of ASIP produces a yellow coat color in mice 5, 9, 10. Attractin encoded by the ATRN gene is a low-affinity receptor for the ASIP protein product. A recessive color mutation mahogany (Atrnmg) was recognized as a modifier of agouti coat color in mice 11, 12. Tyrosinase (TYR) is required for melanization in both types of melanosome, whereas the tyrosinase-related protein 1 (TYRP1) is exclusive to the melanization of eumelanosome 13, 14. Hence, tyrosinase is a critical enzyme during melanosomal maturation and its high activity leads to the formation of eumelanosome 15, 16. The optimal activity of tyrosinase in human melanocytes requires an appropriate ionic environment, which is partially controlled by P-protein functioning as a pH exchange membrane channel 17, 18. The twelve transmembrane-spanning P-protein encoded by the OCA2 gene (human type II oculocutaneous albinism-related gene) was discovered in the “pink-eyed dilution” mouse mutant 19. In addition to P-protein, MATP, a membrane-associated transporter protein encoded by the SLC45A2 gene, has been considered as a sodium-hydrogen exchanger of melanosomes, regulating tyrosinase activity in human melanocyte 20. Another cation exchanger, SLC24A5, transports calcium or potassium ions into the melanosome and is involved in melanogenesis. It has been proposed that the human SLC24A5 gene is required for maturation of melanosome and has a role in skin pigmentation 21, 22.
Figure 1. The function of select pigmentation genes in the pigmentation pathway.
Induction of POMC/α-MSH activates the MC1R, inducing cAMP production in melanocyte. This elevated signaling leads to eumelanin production, resulting in the maturation of the pheomelanosome to the eumelanosome. Attractin (ATRN) is a low-affinity receptor for agouti signaling protein (ASIP), an antagonist of MC1R. Some proteins, such as TYR, TYRP1, P-protein, SLC24A5, and MATP, are involved in inducing melanization of pheomelanosome or eumelanosome, as described in the text.
Lighter pigmentation is the host susceptibility risk factor for skin cancer 23. Although a large number of single nucleotide polymorphisms (SNPs) were identified in pigmentation genes, very few SNPs have been examined in relation to human pigmentary phenotypes and skin cancer risk. Recent genome-wide association studies on pigmentary traits and skin cancer risks (melanoma and basal cell carcinoma) have generated additional information on both pigmentary phenotype related- and skin cancer related-genetic variants 24–28. In the present study, we evaluated the associations of fifteen SNPs in eight candidate pigmentation genes (TYR, TYRP1, OCA2, SLC24A5, SLC45A2, POMC, ASIP, and ATRN) with both pigmentary phenotypes (hair color, skin color, and tanning ability) and the risk of melanoma and non-melanoma skin cancer (squamous cell carcinoma (SCC) and basal cell carcinoma (BCC)) in a nested case-control study within the Nurses’ Health Study (NHS).
Materials and Methods
Study population
The NHS was established in 1976, when 121,700 female registered nurses between the ages of 30 and 55, residing in 11 larger U.S. states, completed and returned the initial self-administered questionnaire on their medical histories and baseline health-related exposures, forming the basis for the NHS cohort. Updated information has been obtained by questionnaires every two years. From May 1989 through September 1990, we collected blood samples from 32,826 participants in the NHS cohort. The distributions of risk factors for skin cancer were very similar in the subcohort of those who donated blood samples as in the overall cohort 29. Eligible cases in this study consisted of women with incident skin cancer from the subcohort who had given a blood specimen, including SCC and BCC cases with a diagnosis anytime after blood collection up to June 1, 1998 and melanoma cases up to June 1, 2000 that had no previously diagnosed skin cancer. All available pathologically confirmed melanoma and SCC cases and 300 self-reported BCC cases randomly selected from ~2,600 available self-reported BCC cases were included. The validity of self-report of BCC is high in this medically sophisticated population (90%) 30, 31. A common control series was randomly selected from participants who gave a blood sample and were free of diagnosed skin cancer up to and including the questionnaire cycle in which the case was diagnosed. One or two controls were matched to each case by year of birth (±1 year). All subjects were the U.S. non-Hispanic Caucasian women in this study. The nested case-control study consisted of 218 melanoma cases, 285 SCC cases, a sample of 300 BCC cases from the large number of incident cases, and 870 matched controls. The study protocol was approved by the Committee on Use of Human Subjects of the Brigham and Women’s Hospital, Boston, MA.
Exposure data
We obtained information regarding skin cancer risk factors from the prospective biennial questionnaires and a retrospective supplementary questionnaire. Information on natural hair color at age 20 and childhood and adolescent tanning tendency were collected in the 1982 prospective questionnaire into five categories (black, dark brown, light brown, blonde, and red) and four categories (practically none, light tan, average tan, and deep tan), respectively. Question on ethnic group was ascertained in the 1992 questionnaire. In the skin cancer nested case-control study, natural skin color and other sun exposure-related information were collected by the retrospective supplementary questionnaire in 2002. The response rate of cases and controls were 92% and 89%, respectively. Information on natural skin color was classified into three categories (fair, medium, and olive). In addition, the eleven states of residence of cohort members at baseline were grouped into three regions: Northeast (Connecticut, Massachusetts, Maryland, New Jersey, New York, and Pennsylvania), Northcentral (Michigan and Ohio), and West and South (California, Texas, and Florida).
Laboratory assays
We genotyped fifteen SNPs in eight candidate pigmentation genes (TYR, TYRP1, OCA2, SLC24A5, SLC45A2, POMC, ASIP, and ATRN) using the OpenArray™ SNP Genotyping System (BioTrove, Woburn, MA). Due to the assay failed we genotyped rs1393350 as a surrogate for the TYR Arg402Gln (rs1126809) (D′=1 and r2=0.86) (http://snp500cancer.nci.nih.gov). Laboratory personnel were blinded to the case-control status, and 42 blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes, and conditions for genotyping assays are available upon request. The genotyping method for the MC1R variants was described previously 29.
Statistical methods
We used the χ2 test to assess whether the genotypes for all fifteen SNPs were in Hardy-Weinberg equilibrium among the controls.
The MC1R gene has been strongly associated with human pigmentary phenotypes, especially with red hair color 32–34. We previously reported the frequency distribution of seven common MC1R variants among controls, including three “red hair color” (RHC) variants (Arg151Cys, Arg160Trp, and Asp294His) and four “non-red hair color” (NRHC) variants (Val60Leu, Val92Met, Ile155Thr, and Arg163Gln) 29. In order to compare the contribution of these fifteen SNPs to pigmentary phenotypes with that of the MC1R variants, we evaluated the associations between the MC1R variants and pigmentary phenotypes in parallel. We regressed an ordinal coding for skin color (1=fair; 2=medium; and 3=olive) or tanning ability (1=practically none; 2=light tan; 3=average tan; and 4=deep tan) on an ordinal coding for genotype (0, 1, or 2 copies of SNP minor allele). For hair color, we used two different statistical models: A) we tested the association between the ordinal genotype coding and an ordinal coding of hair color excluding the women with red hair (1=black; 2=dark brown; 3=light brown; and 4=blonde) using linear regression; and B) we used logistic regression to test the association between the ordinal genotype coding and a binary red hair phenotype (red hair vs. non-red hair color). For the SLC45A2 Gln272Lys and three MC1R NRHC variants (Val92Met, Ile155Thr, and Arg163Gln), we used Fisher’s exact test for “red vs. non-red hair color” analysis because none of the women with red hair color carried the variant allele.
We evaluated the association between each genotype and skin cancer risk using unconditional logistic regression. We compared each type of skin cancer with the common control series to increase the statistical power.
In the haplotype analysis, haplotype frequencies and expected haplotype counts for each individual were estimated using a simple expectation-maximization algorithm, as implemented in SAS PROC HAPLOTYPE. The association analyses between haplotypes and binary pigmentary phenotypes and skin cancer risk were performed using the expectation-substitution technique 35. All statistical analyses were two-sided and carried out using SAS V9.1 (SAS Institute, Cary, NC).
Results
Descriptive characteristics of cases and controls
At the beginning of the follow-up of this nested case-control study, the women were between 43 and 68 years old with a mean age of 58.7. The mean age at diagnosis of melanoma cases was 63.4 and that of SCC and BCC cases was 64.7 and 64.0, respectively. Basic characteristics of cases and controls in this study are presented in Table 1. Detailed description and statistical tests were published previously 23. Briefly, skin cancer cases were more likely to possess red hair color and fair skin color. The childhood tanning ability of cases was less than that of controls. Women in the West and South regions were more likely to be diagnosed with SCC or BCC compared with those in Northeast. A family history of skin cancer was a risk factor for the three types of skin cancer. Those with skin cancers were more likely to have used sunlamps or attended tanning salons. Those with skin cancers had higher cumulative sun exposure while wearing a bathing suit and more lifetime severe sunburns that blistered.
Table 1.
Characteristics of skin cancer cases and controls in the nested case-control study
| Characteristic | Controls (n=870) | Melanoma cases (n=218) | SCC cases (n=285) | BCC cases (n=300) |
|---|---|---|---|---|
| Age at diagnosis (mean, years) | 64.5 | 63.4 | 64.7 | 64.0 |
| Natural hair color at age 20 (%) | ||||
| Black or dark brown | 43.9 | 31.5 | 41.3 | 30.3 |
| Light brown | 40.0 | 42.5 | 34.6 | 45.7 |
| Blonde | 12.0 | 15.5 | 16.8 | 18.0 |
| Red | 2.9 | 10.5 | 5.2 | 4.7 |
| Natural skin color (%) | ||||
| Fair | 40.0 | 57.1 | 54.6 | 53.0 |
| Medium | 36.7 | 25.6 | 32.2 | 31.3 |
| Olive | 4.8 | 0.9 | 1.8 | 1.3 |
| Tanning ability (%) | ||||
| Practically none | 8.3 | 13.8 | 13.9 | 11.7 |
| Light tan | 21.1 | 29.5 | 24.1 | 25.5 |
| Average tan | 47.3 | 40.0 | 47.4 | 46.5 |
| Tan | 23.3 | 16.7 | 14.7 | 16.3 |
| Geographic region at baseline (%) | ||||
| Northeast | 55.2 | 58.0 | 51.7 | 49.3 |
| Northcentral | 23.4 | 16.9 | 17.1 | 20.3 |
| West and South | 21.4 | 25.1 | 31.1 | 30.3 |
| Family history of skin cancer (%) | 25.1 | 36.5 | 35.7 | 42.7 |
| Sunlamp use or tanning salon attendance (%) | 10.0 | 19.2 | 14.3 | 14.7 |
| Highest tertile of cumulative sun exposure with a bathing suit (%) | 33.4 | 53.3 | 46.1 | 42.6 |
| Number of lifetime severe sunburns (mean) | 5.4 | 9.6 | 7.8 | 8.2 |
The percentages may not sum to 100 due to rounding.
Association between the fifteen SNPs in pigmentation genes and pigmentary phenotypes
Information on the fifteen SNPs in pigmentation genes is presented in Table 2. We selected putative functional SNPs in the 6 pigmentation genes (TYR, OCA2, SLC45A2, POMC, ASIP, and ATRN), including non-synonymous SNPs and those in the promoter and UTR regions. For the SLC24A5 gene, the Ala111Thr (rs1426654) is monomorphic in HapMap CEU samples. The SNP rs17426596 is the only one with minor allele frequency >1% in the HapMap CEU samples. Recently, an eye-color variant in TYRP1 (rs1408799) was reported to be associated with melanoma risk 25, 27. A haplotype near ASIP carrying rs4911414 variant allele[T] and rs1015362 major allele[G] (hereafter called ASIP AH) was associated with pigmentary phenotypes (skin sensitivity to sun, burn, and freckle) as well as the risks of melanoma and BCC 25, 27. These three SNPs were evaluated for the association with pigmentary phenotypes and the risk of skin cancer as well in this study. The distributions of genotypes for these fifteen SNPs were in Hardy-Weinberg equilibrium among controls. The participants of this study were from 11 states, which were grouped into three regions (Northeast, Northcentral, and West and South). The minor allele frequencies of the fifteen genetic variants according to the 11 individual states are presented in Supplementary Table 1. There were no significant differences in minor allele frequencies of these fifteen SNPs across the 11 states (all p-values>0.05).
Table 2.
Fifteen SNPs in the selected pigmentation genes
| SNP | rs# | Gene | Protein | MAF - controls (%)b | MAF - CEU (%)c | MAF - CHB/JPT (%)d | MAF - YRI (%)e |
|---|---|---|---|---|---|---|---|
| TYR Ser192Tyr | rs1042602 | TYR | Tyrosinase | 35 | 42 | 0 | 0 |
| TYR Arg402Gln | rs1126809a | TYR | Tyrosinase | - | 22 | 0 | 0 |
| TYR -6895 G>A | rs1393350a | TYR | Tyrosinase | 28 | 19 | 0 | 0 |
| TYRP1 C>T | rs1408799 | TYRP1 | Tyrosinase-related protein 1 | 32 | 30 | 98 | 78 |
| OCA2 Arg305Trp | rs1800401 | OCA2 | P-protein | 6 | - | - | - |
| OCA2 Arg419Gln | rs1800407 | OCA2 | P-protein | 6 | 7 | 0 | 0 |
| SLC45A2 -1721 C>G | rs13289 | SLC45A2 | MATP | 40 | 32 | 34 | 73 |
| SLC45A2 Glu272Lys | rs26722 | SLC45A2 | MATP | 2 | 0 | 40 | 5 |
| SLC45A2 Phe374Leu | rs16891982 | SLC45A2 | MATP | 4 | 2 | 99 | 100 |
| SLC24A5 intron2 T>C | rs17426596 | SLC24A5 | SLC24A5 | 4 | 3 | 0 | 0 |
| POMC3′ UTR C>T | rs1042571 | POMC | POMC, MSH, ACTH | 19 | - | - | - |
| ASIPG>T | rs4911414 | ASIP | Agouti signaling protein | 31 | 28 | 19 | 13 |
| ASIPG>A | rs1015362 | ASIP | Agouti signaling protein | 27 | 23 | 19 | 83 |
| ASIPg.8818 A>G | rs6058017 | ASIP | Agouti signaling protein | 13 | - | - | - |
| ATRN Ile426Thr | rs17782078 | ATRN | Attractin | 5 | 4 | 0 | 0 |
| ATRN Arg1152Lys | rs3886999 | ATRN | Attractin | 6 | 4 | 0 | 0 |
The SNP rs1126809 failed the assay and the rs1393350 was genotyped instead (D′=1 and r2=0.86).
Minor allele frequency (MAF) was calculated among controls in this study.
MAF was based on the HapMap CEU (Utah residents with ancestry from northern and western Europe) samples.
MAF was based on the HapMap CHB and JPT (CHB: Han Chinese in Beijing, China; JPT: Japanese in Tokyo, Japan ) samples.
MAF was based on the HapMap YRI (Yoruba in Ibadan, Nigeria) samples.
We evaluated the associations between the fifteen SNPs and pigmentary phenotypes including hair color, skin color, and tanning ability among controls (Table 3). We observed significant associations between SLC45A2 Phe374Leu (p-value =2.4×10−7) and SLC45A2 Glu272Lys (p-value =6.0×10−5), and hair color (black to blonde). We then mutually adjusted these two significant SLC45A2 SNPs and found that only the SLC45A2 Phe374Leu remained significant. The p-value for SLC45A2 Phe374Leu and SLC45A2 Glu272Lys was 8.0×10−4 and 0.15, respectively. The SLC45A2 Phe374Leu remained significantly associated with hair color after adjusting for MC1R variants (p-value =3.3×10−6) (Supplementary Table 2a and 2b). However, in the analysis of “red hair vs. non-red hair color”, there was no significant association between any of the fifteen polymorphisms and red hair color (Table 3). In contrast, the three MC1R RHC variants were significantly associated with red hair color phenotype.
Table 3.
Associations between the fifteen SNPs in the selected pigmentation genes and pigmentary phenotypes among controls
| SNP | Hair color (black to blonde) |
Hair color (red vs. nonred) |
Skin color |
Tanning ability |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | SE | p-value | β | SE | p-value | β | SE | p-value | β | SE | p-value | |
| TYR Ser192Tyr | 0.08 | 0.04 | 0.06 | −0.26 | 0.33 | 0.42 | 0.06 | 0.04 | 0.10 | 0.00 | 0.05 | 0.97 |
| TYR Arg402Gln** | 0.00 | 0.04 | 0.92 | −0.31 | 0.35 | 0.38 | −0.12 | 0.04 | 7.7E-04 | −0.16 | 0.05 | 7.3E-04 |
| TYRP1 C>T (rs1408799) | 0.02 | 0.04 | 0.60 | 0.32 | 0.30 | 0.28 | 0.02 | 0.04 | 0.65 | 0.04 | 0.05 | 0.46 |
| OCA2 Arg305Trp | −0.08 | 0.08 | 0.31 | 0.11 | 0.62 | 0.85 | 0.15 | 0.07 | 0.04 | 0.02 | 0.10 | 0.84 |
| OCA2 Arg419Gln | −0.09 | 0.08 | 0.23 | −1.05 | 1.01 | 0.30 | −0.10 | 0.07 | 0.18 | −0.05 | 0.09 | 0.61 |
| SLC45A2 -1721 C>G | 0.03 | 0.04 | 0.49 | −0.11 | 0.30 | 0.71 | 0.02 | 0.03 | 0.64 | 0.04 | 0.04 | 0.33 |
| SLC45A2 Glu272Lys | −0.58 | 0.14 | 6.0E-05 | - | - | 1.00* | 0.44 | 0.13 | 1.1E-03 | 0.48 | 0.18 | 7.0E-03 |
| SLC45A2 Phe374Leu | −0.49 | 0.10 | 2.4E-07 | 0.51 | 0.60 | 0.39 | 0.48 | 0.09 | 1.1E-07 | 0.42 | 0.11 | 2.5E-04 |
| SLC24A5 intron2 T>C | 0.11 | 0.10 | 0.26 | 0.97 | 0.54 | 0.08 | −0.08 | 0.08 | 0.34 | −0.03 | 0.12 | 0.78 |
| POMC 3′ UTR C>T | −0.06 | 0.05 | 0.23 | 0.09 | 0.36 | 0.79 | −0.03 | 0.04 | 0.52 | −0.01 | 0.06 | 0.86 |
| ASIP G>T (rs4911414) | 0.02 | 0.04 | 0.68 | 0.33 | 0.30 | 0.27 | −0.11 | 0.03 | 1.4E-03 | −0.11 | 0.05 | 0.02 |
| ASIP G>A (rs1015362) | −0.06 | 0.04 | 0.15 | −0.37 | 0.35 | 0.29 | −0.01 | 0.04 | 0.76 | 0.00 | 0.05 | 0.96 |
| ASIP g.8818 A>G | −0.01 | 0.06 | 0.81 | 0.07 | 0.44 | 0.88 | 0.00 | 0.05 | 0.98 | 0.08 | 0.07 | 0.25 |
| ATRN Ile426Thr | 0.04 | 0.08 | 0.65 | 0.50 | 0.54 | 0.35 | −0.07 | 0.07 | 0.31 | −0.02 | 0.10 | 0.87 |
| ATRN Arg1152Lys | 0.05 | 0.08 | 0.50 | 0.45 | 0.53 | 0.40 | −0.08 | 0.07 | 0.24 | −0.02 | 0.10 | 0.82 |
|
| ||||||||||||
| MC1R variants | ||||||||||||
| MC1R Val60Leu | −0.03 | 0.05 | 0.63 | −1.91 | 1.01 | 0.06 | −0.06 | 0.05 | 0.23 | −0.13 | 0.06 | 0.05 |
| MC1R Val92Met | −0.13 | 0.06 | 0.04 | - | - | 0.01* | 0.00 | 0.06 | 1.00 | −0.10 | 0.07 | 0.18 |
| MC1R Arg151Cys | 0.17 | 0.08 | 0.03 | 2.21 | 0.40 | 2.6E-08 | −0.24 | 0.06 | 1.4E-04 | −0.46 | 0.09 | 1.4E-07 |
| MC1R Ile155Thr | 0.09 | 0.16 | 0.58 | - | - | 1.00* | −0.05 | 0.13 | 0.71 | −0.18 | 0.19 | 0.34 |
| MC1R Arg160Trp | 0.20 | 0.07 | 5.9E-03 | 2.14 | 0.34 | 5.0E-10 | −0.19 | 0.06 | 9.4E-04 | −0.28 | 0.08 | 4.1E-04 |
| MC1R Arg163Gln | 0.05 | 0.09 | 0.58 | - | - | 0.25* | 0.01 | 0.09 | 0.92 | −0.07 | 0.11 | 0.54 |
| MC1R Asp294His | 0.27 | 0.16 | 0.08 | 1.94 | 0.59 | 9.6E-04 | −0.37 | 0.13 | 4.2E-03 | −0.67 | 0.18 | 2.1E-04 |
The regression parameter beta refers to the mean change in scoring in hair color (black to blonde), skin color, and tanning ability (or change in log odds of red hair for red hair analyses) per copy of the SNP minor allele.
Fisher’s exact test was used. The β value was not calculated because none of the women with red hair color carried the variant allele.
TYR -6895 G>A (rs1393350) was genotyped instead.
We found that the TYR Arg402Gln and SLC45A2 Phe374Leu were significantly associated with skin color (p-value =7.7×10−4 and 1.1×10−7, respectively) and tanning ability (p-value =7.3×10−4 and 2.5×10−4, respectively). The OCA2 Arg305Trp was associated with skin color (p-value=0.04). These associations remained significant after controlling for the MC1R variants (Table 3, Supplementary Table 2a and 2b). The significant associations between the SLC45A2 Glu272Lys and skin color and tanning ability were eliminated after controlling for the SLC45A2 Phe374Leu (p-value =0.97 and 0.49, respectively).
We found a significant association of ASIP G>T (rs4911414) polymorphism with skin color and tanning ability (p-value =1.4×10−3 and 0.02, respectively). However, the association with tanning ability was eliminated after adjusting for the MC1R variants (p-value =0.19), while the association with skin color remained significant (p-value =0.01) (Table 3, Supplementary Table 2a and 2b). We also performed a global test to evaluate whether the haplotype frequencies were different between various pigmentary phenotypes (Supplementary Table 3). Consistent with the single SNP analysis of ASIP G>T (rs4911414), the haplotype ASIP AH was significantly associated with fair skin color (OR, 2.28; 95% CI, 1.46–3.57) (p-value for global test, 0.003). This association remained significant after adjusting for the ASIP g.8818 A>G (OR, 2.54; 95% CI, 1.58–4.07).
All the significant associations described above with pigmentary phenotypes remained significant after controlling for the geographic regions (either by 11 states or 3 combined groups) (data not shown).
Association between the fifteen SNPs in pigmentation genes and skin cancer risk
The main effect of each polymorphism was evaluated across the three types of skin cancer (Table 4). In the analyses controlling for age, we observed that the TYRP1 rs1408799 and SLC45A2 -1721 C>G were associated with melanoma risk (OR, 0.77; 95% CI, 0.60–0.98 and OR, 0.75; 95% CI, 0.60–0.95, respectively); the TYR Ser192Tyr and ASIP rs4911414 were associated with SCC risk (OR, 1.23; 95% CI, 1.00–1.50 and OR, 1.29; 95% CI, 1.05–1.59, respectively); the OCA2 Arg419Gln and ASIP g.8818 A>G were associated with BCC risk (OR, 1.50; 95% CI, 1.06–2.13 and OR, 0.73; 95% CI, 0.53–1.00, respectively). These associations remained similar after adjusting for either pigmentary phenotypes (hair color, skin color, and tanning ability) (Supplementary Table 4) or MC1R variants (Supplementary Table 5a and 5b) or skin cancer risk factors including constitutional susceptibility score (tertiles), family history of skin cancer, the number of lifetime severe sunburns that blistered, sunlamp use or tanning salon attendance, cumulative sun exposure while wearing a bathing suit, and geographic regions (either by 11 states or 3 combined groups) (data not shown). None of those significant associations with skin cancer risk remained significant after the Bonferroni correction (all p-values >0.05/45 (15 SNPs and 3 types of skin cancer) =0.001).
Table 4.
Associations between the fifteen SNPs in the selected pigmentation genes and skin cancer risk
| SNP | Melanoma |
SCC |
BCC |
|||
|---|---|---|---|---|---|---|
| Additive OR* | p for trend | Additive OR* | p for trend | Additive OR* | p trend | |
| TYR Ser192Tyr | 1.18 (0.94–1.48) | 0.15 | 1.23 (1.00–1.50) | 0.05 | 1.08 (0.89–1.33) | 0.43 |
| TYR Arg402Gln** | 1.05 (0.83–1.32) | 0.71 | 1.07 (0.87–1.32) | 0.52 | 1.04 (0.84–1.27) | 0.74 |
| TYRP1 C>T (rs1408799) | 0.77 (0.60–0.98) | 0.03 | 0.96 (0.78–1.18) | 0.71 | 0.95 (0.77–1.16) | 0.60 |
| OCA2 Arg305Trp | 0.92 (0.56–1.52) | 0.76 | 0.87 (0.56–1.36) | 0.55 | 0.97 (0.63–1.50) | 0.91 |
| OCA2 Arg419Gln | 1.33 (0.89–2.01) | 0.17 | 1.39 (0.97–2.01) | 0.07 | 1.50 (1.06–2.13) | 0.02 |
| SLC45A2-1721 C>G | 0.75 (0.60–0.95) | 0.01 | 1.08 (0.89–1.31) | 0.42 | 0.91 (0.75–1.11) | 0.36 |
| SLC45A2 Glu272Lys | 1.19 (0.53–2.67) | 0.68 | 0.55 (0.21–1.45) | 0.23 | 1.04 (0.49–2.17) | 0.93 |
| SLC45A2 Phe374Leu | 0.66 (0.34–1.29) | 0.22 | 0.76 (0.43–1.34) | 0.34 | 0.61 (0.33–1.11) | 0.10 |
| SLC24A5 intron2 T>C | 0.75 (0.39–1.43) | 0.38 | 0.86 (0.50–1.48) | 0.58 | 0.80 (0.47–1.38) | 0.43 |
| POMC 3′ UTR C>T | 0.99 (0.74–1.31) | 0.92 | 0.95 (0.74–1.22) | 0.68 | 0.95 (0.75–1.22) | 0.71 |
| ASIP G>T (rs4911414) | 1.21 (0.96–1.51) | 0.10 | 1.29 (1.05–1.59) | 0.01 | 1.16 (0.95–1.42) | 0.14 |
| ASIP G>A (rs1015362) | 0.89 (0.69–1.13) | 0.34 | 1.14 (0.92–1.41) | 0.23 | 1.06 (0.86–1.31) | 0.59 |
| ASIP g.8818 A>G | 0.89 (0.64–1.24) | 0.50 | 0.79 (0.58–1.09) | 0.15 | 0.73 (0.53–1.00) | 0.05 |
| ATRN Ile426Thr | 0.87 (0.52–1.45) | 0.59 | 1.14 (0.75–1.72) | 0.54 | 1.08 (0.72–1.62) | 0.72 |
| ATRN Arg1152Lys | 0.86 (0.53–1.42) | 0.56 | 1.09 (0.72–1.63) | 0.69 | 1.09 (0.73–1.61) | 0.67 |
Additive OR was calculated based on the unconditional logistic regression adjusted for the age.
TYR -6895 G>A (rs1393350) was genotyped instead.
Haplotypes for the TYR, OCA2, SLC45A2, and ASIP genes and skin cancer risk
We performed the global test to evaluate the difference in haplotype frequencies between cases and controls (Table 5). We found significant differences in TYR haplotype frequency for SCC (p-value =0.007), OCA2 haplotype frequency for BCC (p-value =0.03), and ASIP haplotype frequencies for melanoma and SCC (p-value =0.008 and 0.004, respectively). For the TYR gene, the haplotypes that carried only one variant allele at the two sites were significantly associated with an increased risk of SCC. The adjusted ORs (95% CI) for the haplotype carrying only the Ser192Tyr or only the Arg402Gln variant alleles were 1.48 (1.16–1.89) and 1.35 (1.04–1.74), respectively. The Arg402Gln variant was not significantly associated with risk of SCC in the single SNP analysis. We observed that the haplotype carrying OCA2 Arg419Gln variant allele and the OCA2 Arg305Trp major allele was significantly associated with an increased risk of BCC (adjusted OR, 1.62; 95% CI, 1.13–2.32). For the ASIP gene, the haplotype AH was significantly associated with an increased risk of melanoma (OR, 1.68; 95% CI, 1.18–2.39) and SCC (OR, 1.54; 95% CI, 1.08–2.19). The ASIP rs4911414 variant allele was not significantly associated with melanoma risk in the single SNP analysis.
Table 5.
Haplotypes for SNPs in the TYR, OCA2, SLC45A2, and ASIP genes and skin cancer risk
| TYR | Melanoma | SCC | BCC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Cases | Cases | |||||||
| A | B | n | % | n | % | n | % | n | % | |
| 0 | 0 | 618 | 38.0 | 136 | 32.5 | 164 | 31.2 | 206 | 37.0 | |
| Multivariate OR | 1.00 | 1.00 | 1.00 | |||||||
| 1 | 0 | 551 | 33.9 | 157 | 37.6 | 206 | 39.4 | 190 | 34.2 | |
| Multivariate OR | 1.34 (1.03–1.75) | 1.48 (1.16–1.89) | 1.05 (0.83–1.34) | |||||||
| 0 | 1 | 438 | 26.9 | 122 | 29.2 | 150 | 28.7 | 153 | 27.6 | |
| Multivariate OR | 1.29 (0.97–1.71) | 1.35 (1.04–1.74) | 1.06 (0.83–1.35) | |||||||
| 1 | 1 | 19 | 1.2 | 3 | 0.7 | 4 | 0.7 | 7 | 1.2 | |
| Multivariate OR | 0.43 (0.07–2.79) | 0.34 (0.05–2.30) | 0.89 (0.32–2.51) | |||||||
|
| ||||||||||
| A: Ser192Tyr; B: Arg402Glna | ||||||||||
| OCA2 | Melanoma | SCC | BCCc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Cases | Cases | |||||||
| A | B | n | % | n | % | n | % | n | % | |
| 0 | 0 | 1417 | 88.3 | 351 | 87.3 | 468 | 86.7 | 465 | 85.2 | |
| Multivariate OR | 1.00 | 1.00 | 1.00 | |||||||
| 0 | 1 | 94 | 5.9 | 31 | 7.7 | 44 | 8.1 | 51 | 9.3 | |
| Multivariate OR | 1.33 (0.88–2.03) | 1.40 (0.97–2.03) | 1.62 (1.13–2.32) | |||||||
| 1 | 0 | 93 | 5.8 | 20 | 5.0 | 28 | 5.2 | 30 | 5.5 | |
| Multivariate OR | 0.91 (0.54–1.51) | 0.91 (0.58–1.41) | 0.99 (0.64–1.53) | |||||||
|
| ||||||||||
| A: Arg305Trp; B: Arg419Gln | ||||||||||
| SLC45A2 | Melanoma | SCC | BCC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Cases | Cases | |||||||
| A | B | C | n | % | n | % | n | % | n | % |
| 0 | 0 | 0 | 966 | 59.4 | 268 | 65.5 | 312 | 57.4 | 357 | 62.1 |
| Multivariate OR | 1.00 | 1.00 | 1.00 | |||||||
| 1 | 0 | 0 | 601 | 37.0 | 130 | 31.6 | 216 | 39.7 | 203 | 35.4 |
| Multivariate OR | 0.77 (0.61–0.97) | 1.11 (0.91–1.35) | 0.93 (0.76–1.13) | |||||||
| 1 | 0 | 1 | 23 | 1.4 | 4 | 0.9 | 8 | 1.5 | 4 | 0.7 |
| Multivariate OR | 0.54 (0.16–1.84) | 1.17 (0.48–2.84) | 0.48 (0.16–1.47) | |||||||
| Rare < 1% | 36 | 2.2 | 8 | 2.0 | 8 | 1.5 | 10 | 1.7 | ||
| Multivariate OR | 0.84 (0.38–1.86) | 0.66 (0.29–1.50) | 0.75 (0.36–1.57) | |||||||
|
| ||||||||||
| A: -1721C>G; B: Glu272Lys; C: Phe374Leu | ||||||||||
| ASIP | Melanomad | SCCe | BCC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Cases | Cases | |||||||
| A | B | n | % | n | % | n | % | n | % | |
| 0 | 0 | 1020 | 65.7 | 253 | 63 | 294 | 60.8 | 337 | 63.1 | |
| Multivariate OR | 1.00 | 1.00 | 1.00 | |||||||
| 1 | 1 | 366 | 23.6 | 91 | 22.6 | 132 | 27.3 | 134 | 25.1 | |
| Multivariate OR | 0.99 (0.76–1.29) | 1.25 (0.99–1.58) | 1.09 (0.87–1.38) | |||||||
| AHf | 1 | 0 | 120 | 7.7 | 52 | 13.0 | 52 | 10.7 | 48 | 9.0 |
| Multivariate OR | 1.68 (1.18–2.39) | 1.54 (1.08–2.19) | 1.22 (0.86–1.74) | |||||||
| 0 | 0 | 48 | 3.1 | 6 | 1.6 | 6 | 1.2 | 15 | 2.8 | |
| Multivariate OR | 0.52 (0.21–1.29) | 0.43 (0.17–1.08) | 1.00 (0.54–1.83) | |||||||
| A: rs4911414; B: rs1015362 | ||||||||||
0, common allele; 1, rare allele; Logistic regression adjusted for the age.
TYR -6895 G>A (rs1393350) was genotyped instead;
p-value for global test for SCC is 0.007;
p-value for global test for BCC is 0.03;
p-value for global test for melanoma is 0.008;
p-value for global test for SCC is 0.004;
AH means the ASIP haplotype carrying the rs4911414 variant allele [T] and the rs1015362 major allele [G].
Power calculation
The Quanto statistical software version 1.2.3 was used for power calculation 36. We calculated the power to detect the specified ORs at various allele frequencies of variant allele in additive models. The calculations were based on a two-sided alpha of 0.05, and the particular sizes of different phenotypic population groups presented in the Table 1 of this study. For melanoma (SCC or BCC), we have 80% power to detect an OR of 1.80 (1.72 or 1.70), 1.48 (1.42 or 1.41), and 1.35 (1.32 or 1.31) if the minor allele frequency is 5%, 15%, and 40%, respectively. For pigmentary phenotypes, we calculated the power to detect the difference between dark and light pigmentation: black/brown and blonde hair color, medium/olive and fair skin color, and average/deep tan and practically none/light tan. For hair color (skin color or tanning ability), we have 80% power to detect an OR of 2.26 (1.99 or 1.90), 1.71 (1.52 or 1.50), and 1.52 (1.36 or 1.35) if the minor allele frequency is 5%, 15%, and 40%, respectively.
Discussion
Hair color and skin color show striking variations between human subgroups. The dark pigmentation and tanning response protect the skin from UV 37. To date, although more than 100 candidate pigmentation genes containing common genetic variants have been identified, only the variants in the MC1R gene have been consistently implicated in the variation of pigmentary phenotypes as well as skin cancer risk 5, 29, 34, 38–40. In our study only the three MC1R RHC variants were significantly associated with red hair color, supporting the major contribution of the MC1R gene to the red hair color phenotype, an autosomal recessive trait 28, 29, 33, 41. In addition, some genetic variants in the other pigmentation genes showed significant associations with non-red hair color (black to blonde) in this study, suggesting distinct mechanisms in the formation of non-red hair color and red hair color. Previous studies reported possible associations between some genetic variants evaluated in this study and pigmentary phenotypes. We summarized these studies in Supplementary Table 6.
TYR Arg402Gln, a common polymorphism of tyrosinase, was correlated with reduced pigmentation of the retina and iris resulting from low tyrosinase activity 42. In addition to the associations of this SNP with skin color and tanning ability observed in this study, Sulem et al. reported that this SNP was associated with eye color and possibly with blond hair color 28. Mutations in murine SLC45A2 gene lead to hypopigmentation of the eyes and fur 43. Two non-synonymous SNPs in this gene, SLC45A2 Phe374Leu and SLC45A2 Glu272Lys, were associated with darker pigmentary phenotypes in our study, which is consistent with a previous report 44. However, our multivariate analysis mutually adjusting for these two SNPs showed that the effect of SLC45A2 Glu272Lys on pigmentary phenotypes was explained by the variant SLC45A2 Phe374Leu. For the promoter polymorphism SLC45A2 -1721 C>G, we did not detect any significant associations with pigmentary traits, while a previous study reported an association between this SNP and olive skin color 45.
The deletion of OCA2 gene has been linked to reduced pigmentation of skin, hair, and eyes in Prader-Willi Syndrome 46. Two genetic variants in this gene, Arg305Trp and Arg419Gln, have been correlated with dark eye color 47. However, these two SNPs were not associated with pigmentary phenotypes measured in our study, such as hair color, skin color, and tanning ability except that the Arg305Trp was marginally associated with skin color. A polymorphism in the 3’ untranslated region of ASIP gene (g.8818 A>G) has been previously reported to be associated with dark pigmentary phenotypes among populations of African Americans or European-ancestry 48–51. Although we did not find a significant association of this SNP with any of the pigmentary phenotypes, our haplotype analysis showed that the haplotype ASIP AH was significantly associated with fair skin color and this association remained significant after adjusting for ASIP g.8818 A>G. Similarly, Sulem et al. reported that the ASIP AH haplotype remained significant for the pigmentary traits, such as burning and freckling, after adjusting for ASIP g.8818 A>G, while the association of ASIP g.8818 A>G with pigmentary traits were eliminated after adjusting for the haplotype 27.
We evaluated the contributions of genetic variants in the pigmentation genes not only to pigmentary phenotypes but also to the risks of three types of skin cancers among U.S. Caucasians, whereas most previous studies only evaluated the relation of those genetic variants to the pigmentary traits. Ours is the first report evaluating the association between genetic variants in the pigmentation genes and the three types of skin cancer simultaneously. We summarized the results from previous studies assessing the associations of the SNPs evaluated in this study with melanoma risk in Supplementary Table 7. Only one previous study examined pigmentation genes with BCC risk 25. Overall, in this study, the associations observed with an altered risk of at least one skin cancer in the single SNP analysis remained similar after adjusting for either pigmentary phenotypes or MC1R variants, suggesting that these genetic variants play a role in development of skin cancer beyond their influence on pigmentary phenotypes. Furthermore, most of these genetic variants were not associated with the pigmentary phenotypes. The genes involved in pigmentation process may also contribute to other cellular responses to UV exposure. For example, the immune and inflammatory responses to UV exposure are at least partially mediated by the MC1R gene 52–54. Also, the OCA2 gene increases cellular sensitivity to toxic compounds in addition to its role in controlling melanosome biogenesis 55. In addition, tyrosinase is recognized as melanoma-associated antigen by cytotoxic T lymphocytes 56. It is therefore plausible that these genetic variants associated with skin cancer risks may influence other cellular responses leading to skin cancer development. However, we cannot rule out the possibility that the associations with skin cancer risk could be due to chance considering the number of tests performed. Therefore, we should be cautious when interpreting the results on skin cancer risks.
Recently, Gudbjartsson et al. reported a significant association of the TYR 402Gln variant and the ASIP AH haplotype with the increased risks of melanoma and BCC 25. The haplotype analysis performed in our study showed that the risk estimate associated with the TYR 402Gln was elevated for melanoma (OR, 1.29; 95% CI, 0.97–1.71) and SCC (OR, 1.35; 95% CI, 1.04–1.74), which was not observed in the single SNP analysis. For the ASIP AH haplotype, in addition to the association with an increased risk of melanoma, we also found significant association with an increased risk of SCC. The inverse association between the OCA2 Arg305Trp polymorphism and melanoma risk among French Caucasians reported by Jannot et al. is inconsistent with the result of our study 57. We did not find any significant associations between this SNP and three types of skin cancer risks in this study.
Two of the four regions that we found to be associated with variation in pigmentary phenotypes among Europeans (SLC45A2 and OCA2) show strong evidence of recent positive selection, based on a comparison of allele frequencies across samples from three continental populations (Africa, Asia, and Europe) 58. Allele frequencies for the SLC45A2 Phe374Leu polymorphism have been shown to vary greatly across continental populations 58, and less drastically within Europe 59. The TYR SNP rs1126809 Arg402Gln also shows significant differences in allele frequency across the HapMap CEU, CHB, JPT, and YRI panels. This SNP and its surrogate SNP rs1393350 were monomorphic in CHB, JPT, and YRI panels (the minor allele was absent from these samples).
Eight SNPs out of the 15 SNPs were either genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) breast cancer genome-wide association scan or could be imputed with high confidence using the observed genotypes and the HapMap phased data 60. The CGEMS breast cancer scan consists of 1200 cases of breast cancer and 1200 controls from the Nurses’ Health Study of European ancestry genotyped using the Illumina 500k HumanHap platform 61. In order to assess the potential for within-Europe population stratification bias, we examined the association between these 8 SNPs and the top principal components of genetic variation inferred from the CGEMS genome-wide scan data 62, 63. For example, the TYR rs1393350 SNP, which was strongly associated with skin color and tanning ability in the skin cancer controls, was also significantly associated with two of the top 10 principal components of genetic variation (Supplementary Table 8), suggesting that allele frequency for this marker also vary among European populations. This SNP was strongly associated with tanning ability (information on the skin color is not recorded in the CGEMS data) in the CGEMS genome-wide association study samples (p-value =5.8×10−11), and this association remained significant after adjusting for the top four principal components (p-value =8.0×10−8). In fact, only 3 of the top ten principal components were associated with tanning ability, and together these explained 4.5% of the residual variation in tanning ability, while the TYR rs1126809 (a surrogate for Arg402Gln) polymorphism explained 1.5% of the residual variation in tanning ability beyond the effect of top 3 principal components. We believe it is unlikely that the strong associations we see between these markers and pigmentary phenotypes are solely due to population stratification bias. Rather it is likely that differences in the distribution of pigmentary phenotypes across Europe are due in part to differences in allele frequencies at these loci and other as-yet-unknown loci.
Population stratification may be a particularly important issue in assessment of the association of the pigmentation genetic polymorphism with skin cancer risk because of the variation of the allele frequency and the predominance of the disease among light pigmented people, even among European populations. However, the ancestry informative variants mentioned above were not associated with any type of skin cancer, suggesting that the associations of other variants with skin cancer risk is not due to bias rooted in stratification, a possibility that is also made less likely by the fact that these associations remained significant after controlling for the geographic regions (either by 11 states or 3 combined groups). However, these modest associations require further replication in other populations.
One limitation of this study would be the modest statistical power. It is possible that the modest effects of some genetic variants cannot be detected due to insufficient statistical power. Another limitation of our study was that we used self-reported pigmentary phenotypes. Such assessment may miss certain aspects of pigmentary phenotypes influenced by these genetic variants, such as melanin content and composition 64, 65. In addition, misclassification is always a concern in epidemiologic studies. The high education level and interest in health of cohort members allows high quality information to be collected. Test-retest reliability of collecting phenotypic factors from questionnaires is moderate to substantial, including skin color, tanning/burning tendency, and sunburn history 66–68.
In conclusion, our study evaluated the associations between genetic variants in the pigmentation genes and pigmentary phenotypes and skin cancer risk. As this reported is one of the very few studies examining such associations, additional studies are warranted to confirm these associations. This information may be useful in understanding the involvement of different pigmentation genes in Caucasian pigmentary phenotypes and skin cancer risk.
Acknowledgments
We thank Dr. Hardeep Ranu and Ms. Pati Soule for their laboratory assistance, and Ms. Carolyn Guo and Ms. Constance Chen for their programming support. We are indebted to the participants in the Nurses’ Health Study for their dedication and commitment.
Grant sponsor: NIH; Grant numbers: CA128080 and CA132175.
Abbreviations
- BCC
basal cell carcinoma
- SCC
squamous cell carcinoma
- CI
confidence interval
- OR
odds ratio
- UV
ultraviolet
References
- 1.Frisancho AR, Wainwright R, Way A. Heritability and components of phenotypic expression in skin reflectance of Mestizos from the Peruvian lowlands. Am J Phys Anthropol. 1981;55:203–8. doi: 10.1002/ajpa.1330550207. [DOI] [PubMed] [Google Scholar]
- 2.Harrison GA, Owen JJ. Studies on the Inheritance of Human Skin Colour. Ann Hum Genet. 1964;28:27–37. doi: 10.1111/j.1469-1809.1964.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 3.Jablonski NG, Chaplin G. The evolution of human skin coloration. J Hum Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 4.Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne JP. The pigmentary system - Physiology and pathophysiology. Oxford University Press; Oxford: 1998. [Google Scholar]
- 5.Jackson IJ. Molecular and developmental genetics of mouse coat color. Annu Rev Genet. 1994;28:189–217. doi: 10.1146/annurev.ge.28.120194.001201. [DOI] [PubMed] [Google Scholar]
- 6.Schauer E, Trautinger F, Kock A, Schwarz A, Bhardwaj R, Simon M, Ansel JC, Schwarz T, Luger TA. Proopiomelanocortin-derived peptides are synthesized and released by human keratinocytes. J Clin Invest. 1994;93:2258–62. doi: 10.1172/JCI117224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barsh G, Gunn T, He L, Schlossman S, Duke-Cohan J. Biochemical and genetic studies of pigment-type switching. Pigment Cell Res. 2000;13(Suppl 8):48–53. doi: 10.1034/j.1600-0749.13.s8.10.x. [DOI] [PubMed] [Google Scholar]
- 8.D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–4. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 9.Furumura M, Sakai C, Abdel-Malek Z, Barsh GS, Hearing VJ. The interaction of agouti signal protein and melanocyte stimulating hormone to regulate melanin formation in mammals. Pigment Cell Res. 1996;9:191–203. doi: 10.1111/j.1600-0749.1996.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 10.Wilson BD, Ollmann MM, Kang L, Stoffel M, Bell GI, Barsh GS. Structure and function of ASP, the human homolog of the mouse agouti gene. Hum Mol Genet. 1995;4:223–30. doi: 10.1093/hmg/4.2.223. [DOI] [PubMed] [Google Scholar]
- 11.Gunn TM, Miller KA, He L, Hyman RW, Davis RW, Azarani A, Schlossman SF, Duke-Cohan JS, Barsh GS. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature. 1999;398:152–6. doi: 10.1038/18217. [DOI] [PubMed] [Google Scholar]
- 12.He L, Gunn TM, Bouley DM, Lu XY, Watson SJ, Schlossman SF, Duke-Cohan JS, Barsh GS. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat Genet. 2001;27:40–7. doi: 10.1038/83741. [DOI] [PubMed] [Google Scholar]
- 13.Sturm RA, O’Sullivan BJ, Box NF, Smith AG, Smit SE, Puttick ER, Parsons PG, Dunn IS. Chromosomal structure of the human TYRP1 and TYRP2 loci and comparison of the tyrosinase-related protein gene family. Genomics. 1995;29:24–34. doi: 10.1006/geno.1995.1211. [DOI] [PubMed] [Google Scholar]
- 14.Sturm RA, Teasdale RD, Box NF. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene. 2001;277:49–62. doi: 10.1016/s0378-1119(01)00694-1. [DOI] [PubMed] [Google Scholar]
- 15.Jimbow K, Hua C, Gomez PF, Hirosaki K, Shinoda K, Salopek TG, Matsusaka H, Jin HY, Yamashita T. Intracellular vesicular trafficking of tyrosinase gene family protein in eu- and pheomelanosome biogenesis. Pigment Cell Res. 2000;13(Suppl 8):110–7. doi: 10.1034/j.1600-0749.13.s8.20.x. [DOI] [PubMed] [Google Scholar]
- 16.Spritz RA. Molecular genetics of oculocutaneous albinism. Hum Mol Genet. 1994;3:1469–75. doi: 10.1093/hmg/3.suppl_1.1469. Spec No. [DOI] [PubMed] [Google Scholar]
- 17.Fuller BB, Spaulding DT, Smith DR. Regulation of the catalytic activity of preexisting tyrosinase in black and Caucasian human melanocyte cell cultures. Exp Cell Res. 2001;262:197–208. doi: 10.1006/excr.2000.5092. [DOI] [PubMed] [Google Scholar]
- 18.Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol. 2000;115:607–13. doi: 10.1046/j.1523-1747.2000.00108.x. [DOI] [PubMed] [Google Scholar]
- 19.Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–6. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 20.Smith DR, Spaulding DT, Glenn HM, Fuller BB. The relationship between Na(+)/H(+) exchanger expression and tyrosinase activity in human melanocytes. Exp Cell Res. 2004;298:521–34. doi: 10.1016/j.yexcr.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, Sinha S, Moore JL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 22.Schnetkamp PP. The SLC24 Na+/Ca2+-K+ exchanger family: vision and beyond. Pflugers Arch. 2004;447:683–8. doi: 10.1007/s00424-003-1069-0. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Colditz GA, Hunter DJ. Risk factors for skin cancers: a nested case-control study within the Nurses’ Health Study. Int J Epidemiol. 2006;35:1514–21. doi: 10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- 24.Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–40. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Sveinsdottir SG, Magnusson V, Lindblom A, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 26.Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, Martin NG, Montgomery GW, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, Palsson S, Sigurgeirsson B, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–7. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 28.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, Jakobsdottir M, Steinberg S, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 29.Han J, Kraft P, Colditz GA, Wong J, Hunter DJ. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer. 2006;119:1976–84. doi: 10.1002/ijc.22074. [DOI] [PubMed] [Google Scholar]
- 30.Colditz GA, Martin P, Stampfer MJ, Willett WC, Sampson L, Rosner B, Hennekens CH, Speizer FE. Validation of questionnaire information on risk factors and disease outcomes in a prospective cohort study of women. Am J Epidemiol. 1986;123:894–900. doi: 10.1093/oxfordjournals.aje.a114319. [DOI] [PubMed] [Google Scholar]
- 31.Hunter DJ, Colditz GA, Stampfer MJ, Rosner B, Willett WC, Speizer FE. Risk factors for basal cell carcinoma in a prospective cohort of women. Ann Epidemiol. 1990;1:13–23. doi: 10.1016/1047-2797(90)90015-k. [DOI] [PubMed] [Google Scholar]
- 32.Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, Birch-Machin MA, Rees JL. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000;9:2531–7. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 33.Healy E, Jordan SA, Budd PS, Suffolk R, Rees JL, Jackson IJ. Functional variation of MC1R alleles from red-haired individuals. Hum Mol Genet. 2001;10:2397–402. doi: 10.1093/hmg/10.21.2397. [DOI] [PubMed] [Google Scholar]
- 34.Rees JL. Genetics of hair and skin color. Annu Rev Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- 35.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol. 2005;28:261–72. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 36.Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra.usc.edu/gxe.
- 37.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–50. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 38.Bastiaens MT, ter Huurne JA, Kielich C, Gruis NA, Westendorp RG, Vermeer BJ, Bavinck JN. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am J Hum Genet. 2001;68:884–94. doi: 10.1086/319500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117:294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 40.Palmer JS, Duffy DL, Box NF, Aitken JF, O’Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66:176–86. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–30. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 42.Fukai K, Holmes SA, Lucchese NJ, Siu VM, Weleber RG, Schnur RE, Spritz RA. Autosomal recessive ocular albinism associated with a functionally significant tyrosinase gene polymorphism. Nat Genet. 1995;9:92–5. doi: 10.1038/ng0195-92. [DOI] [PubMed] [Google Scholar]
- 43.Sweet HO, Brilliant MH, Cook SA, Johnson KR, Davisson MT. A new allelic series for the underwhite gene on mouse chromosome 15. J Hered. 1998;89:546–51. doi: 10.1093/jhered/89.6.546. [DOI] [PubMed] [Google Scholar]
- 44.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–84. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 45.Graf J, Voisey J, Hughes I, van Daal A. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat. 2007;28:710–7. doi: 10.1002/humu.20504. [DOI] [PubMed] [Google Scholar]
- 46.Spritz RA, Bailin T, Nicholls RD, Lee ST, Park SK, Mascari MJ, Butler MG. Hypopigmentation in the Prader-Willi syndrome correlates with P gene deletion but not with haplotype of the hemizygous P allele. Am J Med Genet. 1997;71:57–62. doi: 10.1002/(sici)1096-8628(19970711)71:1<57::aid-ajmg11>3.0.co;2-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebbeck TR, Kanetsky PA, Walker AH, Holmes R, Halpern AC, Schuchter LM, Elder DE, Guerry D. P gene as an inherited biomarker of human eye color. Cancer Epidemiol Biomarkers Prev. 2002;11:782–4. [PubMed] [Google Scholar]
- 48.Bonilla C, Boxill LA, Donald SA, Williams T, Sylvester N, Parra EJ, Dios S, Norton HL, Shriver MD, Kittles RA. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet. 2005;116:402–6. doi: 10.1007/s00439-004-1251-2. [DOI] [PubMed] [Google Scholar]
- 49.Kanetsky PA, Swoyer J, Panossian S, Holmes R, Guerry D, Rebbeck TR. A polymorphism in the agouti signaling protein gene is associated with human pigmentation. Am J Hum Genet. 2002;70:770–5. doi: 10.1086/339076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meziani R, Descamps V, Gerard B, Matichard E, Bertrand G, Archimbaud A, Ollivaud L, Saiag P, Lebbe C, Basset-Seguin N, Alberti C, Crickx B, et al. Association study of the g.8818A>G polymorphism of the human agouti gene with melanoma risk and pigmentary characteristics in a French population. J Dermatol Sci. 2005;40:133–6. doi: 10.1016/j.jdermsci.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Voisey J, Gomez-Cabrera Mdel C, Smit DJ, Leonard JH, Sturm RA, van Daal A. A polymorphism in the agouti signalling protein (ASIP) is associated with decreased levels of mRNA. Pigment Cell Res. 2006;19:226–31. doi: 10.1111/j.1600-0749.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- 52.Kalden DH, Scholzen T, Brzoska T, Luger TA. Mechanisms of the antiinflammatory effects of alpha-MSH. Role of transcription factor NF-kappa B and adhesion molecule expression. Ann N Y Acad Sci. 1999;885:254–61. doi: 10.1111/j.1749-6632.1999.tb08682.x. [DOI] [PubMed] [Google Scholar]
- 53.Luger TA, Scholzen TE, Brzoska T, Bohm M. New insights into the functions of alpha-MSH and related peptides in the immune system. Ann N Y Acad Sci. 2003;994:133–40. doi: 10.1111/j.1749-6632.2003.tb03172.x. [DOI] [PubMed] [Google Scholar]
- 54.Roberts DW, Newton RA, Beaumont KA, Helen Leonard J, Sturm RA. Quantitative analysis of MC1R gene expression in human skin cell cultures. Pigment Cell Res. 2006;19:76–89. doi: 10.1111/j.1600-0749.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 55.Staleva L, Manga P, Orlow SJ. Pink-eyed dilution protein modulates arsenic sensitivity and intracellular glutathione metabolism. Mol Biol Cell. 2002;13:4206–20. doi: 10.1091/mbc.E02-05-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirkin AF, Dzhandzhugazyan K, Zeuthen J. The immunogenic properties of melanoma-associated antigens recognized by cytotoxic T lymphocytes. Exp Clin Immunogenet. 1998;15:19–32. doi: 10.1159/000019050. [DOI] [PubMed] [Google Scholar]
- 57.Jannot AS, Meziani R, Bertrand G, Gerard B, Descamps V, Archimbaud A, Picard C, Ollivaud L, Basset-Seguin N, Kerob D, Lanternier G, Lebbe C, et al. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur J Hum Genet. 2005;13:913–20. doi: 10.1038/sj.ejhg.5201415. [DOI] [PubMed] [Google Scholar]
- 58.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–5. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 59.Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, Dashnyam B, Jin F, Lucotte G, Chattopadhyay PK, Henke L, Henke J. Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet. 2006;70:802–11. doi: 10.1111/j.1469-1809.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- 60.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat Genet. 2008;40:491–2. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 63.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, Chanock S, Thomas G. Population substructure and control selection in genome-wide association studies. PLoS ONE. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alaluf S, Heath A, Carter N, Atkins D, Mahalingam H, Barrett K, Kolb R, Smit N. Variation in melanin content and composition in type V and VI photoexposed and photoprotected human skin: the dominant role of DHI. Pigment Cell Res. 2001;14:337–47. doi: 10.1034/j.1600-0749.2001.140505.x. [DOI] [PubMed] [Google Scholar]
- 65.Rees JL. The genetics of sun sensitivity in humans. Am J Hum Genet. 2004;75:739–51. doi: 10.1086/425285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Branstrom R, Kristjansson S, Ullen H, Brandberg Y. Stability of questionnaire items measuring behaviours, attitudes and stages of change related to sun exposure. Melanoma Res. 2002;12:513–9. doi: 10.1097/00008390-200209000-00014. [DOI] [PubMed] [Google Scholar]
- 67.Glanz K, Schoenfeld E, Weinstock MA, Layi G, Kidd J, Shigaki DM. Development and reliability of a brief skin cancer risk assessment tool. Cancer Detect Prev. 2003;27:311–5. doi: 10.1016/s0361-090x(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 68.Westerdahl J, Anderson H, Olsson H, Ingvar C. Reproducibility of a self-administered questionnaire for assessment of melanoma risk. Int J Epidemiol. 1996;25:245–51. doi: 10.1093/ije/25.2.245. [DOI] [PubMed] [Google Scholar]