Abstract
The impact of small RNA function has resonated throughout nearly every aspect of eukaryotic biology and captured the varied interests of researchers, whether they are endeavoring to understand the basis of development and disease or seeking novel therapeutic targets and tools. The genetic regulatory roles of microRNAs (miRNAs) are particularly interesting given that these often highly conserved factors post-transcriptionally silence many complementary target genes by inhibiting messenger RNA translation. In this regard, miRNAs can be considered as counterpart to transcription factors, the ensemble of which establish the set of expressed genes that define the characteristics of a specific cell type. In this review, evidence supporting a resounding role for small RNAs in development and maturation of sensory epithelia in the mouse inner ear will be considered with an emphasis on the contribution of one hair cell miRNA family (miR-183, miR-96, and miR-182). Although there is much yet to be explored in this fledgling aspect of ear biology, the breadth of miRNA expression and functional requirement for ear development are already sounding off.
Keywords: Hair cell, sensory epithelium, microRNA, Dicer, translational repression, genetic regulation
1. Introduction
Development of the mammalian inner ear requires coordinated transformation of a uniform sheet of cells to form an intricate labyrinthine structure that includes strategic positioning of vestibular and auditory sensory epithelia, and appropriate histological organization of epithelial supporting cells and mechanosensory hair cells. Many studies have revealed the importance of various regulatory proteins including morphogens and transcription factors on patterning, morphogenesis and histogenesis (reviewed in Fritzsch et al., 2007; Kelley, 2007), where coordinated expression and interaction contribute to precision in developmental transitions from precursor cells to differentiated cell types. Nevertheless, recent studies regarding the genetic regulatory roles of small RNAs (reviewed in Amaral et al., 2008) suggest that such developmental transitions in the inner ear are not exclusively orchestrated by the regulatory functions of proteins. Indeed, there is substantial evidence for the widespread importance of microRNAs (miRNAs) in post-transcriptional regulation of target gene expression affecting development, cell differentiation and maintenance, and disease (reviewed in Hobert, 2008; Makeyev and Maniatis, 2008). There are approximately 500 mammalian miRNAs representing about 2% of known genes and estimated to affect the expression of one-third of known protein-coding genes (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006). In this review, consideration will be given to the general function of miRNAs in post-transcriptional regulation of target gene expression and challenges to determining individual miRNA functions. Moreover, evidence for the expression and biological significance of miRNAs in development of the mouse inner ear are presented with a particular focus on specific miRNA families contributing to neurogenesis and innervation, epithelial development, and hair cell differentiation.
2. miRNA biogenesis and function
To best appreciate the role of miRNA-mediate gene regulation and the challenges to determining individual miRNA functions in development and maintenance of the inner ear, a brief review of miRNA biogenesis and function is warranted. The topic has been reviewed in detail from a number of interesting viewpoints including development and disease (Ambros, 2004; He and Hannon, 2004; Wienholds and Plasterk, 2005; Flynt and Lai, 2008; Stefani and Slack, 2008).
2.1. miRNA biogenesis
MicroRNA genes are expressed as capped and polyadenylated RNA polymerase II transcripts (Cai et al., 2004). Among human and mouse miRNA genes, approximately half reside within introns and are presumably co-expressed with known protein-coding genes, and approximately one-third reside in tandem with another miRNA gene(s) and exhibit coordinated expression (Saini et al, 2008). The production of functional miRNAs is mediated by the miRNA pathway (Fig. 1A). Unprocessed miRNA transcript, termed primary miRNA (pri-miR), contains the miRNA sequence within either side of a mostly base-paired stem-loop or hairpin structure. In the nucleus, DiGeorge syndrome critical region 8 (Dgcr8) protein is required to direct the activity of Drosha, a double-stranded RNA (dsRNA)-specific ribonuclease (RNase) III family member, to the pri-miR hairpin (Denli et al., 2004; Gregory et al., 2004; Landthaler et al., 2004; Wang et al., 2007; Yi et al., 2008). Drosha cleavage of the pri-miR in this ‘microprocessor’ complex produces an approximately 70-nucleotide (nt) hairpin called a precursor miRNA (pre-miR), which is exported from the nucleus through a Ran-GTP/Exportin-5-dependent process (Kim, 2004).
Fig. 1.
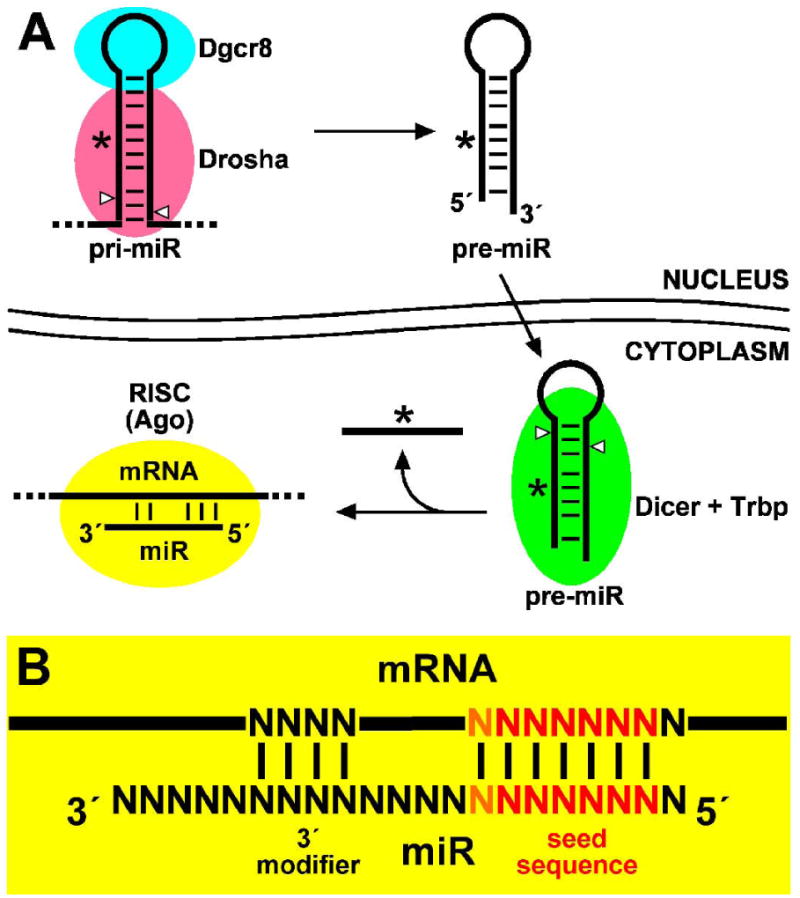
miRNA biogenesis and function. (A) The miRNA pathway. Primary miRNA (pri-miR) transcript processing in the nucleus requires Dgcr8 and the ribonuclease III family member, Drosha. The short hairpin precursor miRNA (pre-miR) is exported from the nucleus and subsequently processed by another ribonuclease III family member, Dicer, with Trbp. The mature miRNA strand is separated from the complementary strand (so-called “star” strand denoted by an asterisk) and is associated with an Argonaute (Ago) protein within the RNA induced silencing complex (RISC). RISC utilizes the miRNA as a guide sequence to discriminately bind target mRNAs and primarily effects translational repression. (B) miRNA-target mRNA interaction. Target mRNA is primarily discriminated through Watson-Crick base pairing to the miRNA “seed” sequence comprising nucleotides 2 through 7 (red) or 8 (orange). Other base-pairing interactions near the 3′ end of the miRNA can contribute further specificity and/or affinity.
In the cytoplasm, the pre-miR is further processed by Dicer, another dsRNA-specific RNase III family member, to produce a small dsRNA with approximately 21-nt strands that is similar to a small interfering RNA (siRNA) (Knight et al., 2001; Grishok et al., 2001; Harfe et al., 2005; Kanellopoulou et al., 2005). Indeed, both miRNA and siRNA biogenesis share Dicer and downstream components of the miRNA pathway (Tabara et al., 1999; Meister et al., 2004; Okamura et al., 2004; Chendrimada et al., 2005, Hasse et al., 2005). Selection of the mature miRNA strand from the complementary or so-called “star” strand is based on thermodynamic stability at each end of the dsRNA, where the miRNA strand is distinguished as that with its 5′ terminus at the lower stability dsRNA end (Schwarz et al., 2003; Tomari et al., 2004). Nevertheless, in some cases both strands of a pre-miR can represent mature functional miRNAs. The mature miRNA finally associates with an Argonaute (Ago) protein within the RNA-induced silencing complex (RISC), the cellular apparatus that exerts post-transcriptional silencing upon complementary target mRNAs using the miRNA as a guide sequence (Tabara et al., 1999; Meister et al., 2004; Okamura et al., 2004).
2.2. miRNA function
Post-transcriptional silencing of miRNA target genes can occur through two RISC-mediated mechanisms dependent upon the extent of complementarity between the miRNA and target mRNA sequence. Perfect complementarity between a miRNA and target mRNA sequence, like that of an siRNA, can elicit ribonuclease activity and mRNA cleavage in Ago2-containing RISC complexes (i.e. “Slicer” activity; Liu et al., 2004; Meister et al., 2004; Okamura et al., 2004). However, partial complementarity results primarily in translational repression through mechanisms not yet well understood (reviewed in Eulalio et al., 2008; Stefani et al., 2008). In fact, the vast majority of mammalian miRNAs lack perfect complementarity to any genomic sequence (other than their own) and thus function by the latter mechanism. The task of predicting target mRNAs is therefore made challenging by the lesser extent of base-pairing with the miRNA, which relies foremost on the miRNA “seed” sequence comprised of nucleotides 2 through 7 or 8 (Fig. 1B) (Lewis et al., 2003; Lewis et al., 2005). Nevertheless, several algorithms including TargetScan, miRanda, and PicTar (Lewis et al., 2003; John et al., 2004; Krek et al., 2005) have been devised to predict target mRNAs based upon the presence and phylogenetic conservation of seed matches within the 3′ untranslated regions (UTRs) of homologous genes.
Predicted miRNA target genes must be experimentally validated on a case-by-case basis or elucidated by other experimental approaches. The incorporation of miRNA target sequences within the 3′ UTR of reporter gene constructs provides validation by demonstrating miRNA-mediated reporter gene repression in cell culture and in vivo (Doench et al., 2003; Zeng et al., 2003; Mansfield et al., 2004; Vella et al., 2004), where silencing is typically enhanced by the presence of multiple miRNA-binding sites (Johnson et al., 2005; Mayr et al., 2007). Moreover, microarray analysis of gene expression demonstrates that target mRNA levels can be reduced upon miRNA transfection in cell culture (Lewis et al., 2005; Lim et al., 2005). This effect is likely mediated by miRNA-directed mRNA deadenylation independent of translational repression (Giraldez et al., 2006; Wu et al., 2006), although it is not an absolute feature of miRNA regulation. Nevertheless, microarray analysis provides a relatively accessible means of determining miRNA target genes and assessing miRNA functions. Importantly, recent advances have been made toward biochemical isolation and identification miRNA-target transcripts by immunoprecipitation of silencing complexes from C. elegans (Zhang et al., 2007; Hammell et al., 2008), Drosophila (Easow et al., 2007), and human cell culture (Karginov et al., 2007; Landthaler et al., 2008). In conclusion, miRNAs can post-transcriptionally silence target genes through immediate effects upon mRNA stability and/or translation, and such interactions can be determined through the combination of bioinformatic prediction and experimental methodologies.
2.3. Challenges to determining function
The determination of individual miRNA functions faces many challenges. Although predicted target mRNAs can be ranked with regard to the likelihood of miRNA-mediated repression based on the number, type, and context of interactions (Hon et al., 2007; Lewis et al., 2005; Grimson et al., 2007; Nielsen et al., 2007), individual miRNAs might have as many as hundreds of predicted targets. Microarray analysis of predicted target gene expression is dependent upon the transcriptome of cell culture models suitable to assessment of a particular miRNA. Moreover, the effect of miRNAs on target mRNA levels does not reflect translational inhibition and does not necessarily coincide with miRNA regulation. Whereas proteomic analysis is more suitable to the task, it is considerably more laborious and technically challenging. Nevertheless, recent studies applying both transcriptome and proteome analyses have revealed that miRNAs indeed impact the expression of hundreds of genes enriched for predicted targets, but that miRNA-mediated regulation rarely accounts for effects on target gene expression exceeding 50% reduction (Baek et al., 2008; Selbach et al., 2008). Individual miRNAs thus appear to function as “rheostats” that fine-tune the output of many expressed target genes more so than absolute genetic “switches” affecting one or a few target genes. Therefore, it becomes considerably more challenging to attribute a phenotypic outcome resulting from an individual miRNA's contribution to one or few determinable genetic mechanisms.
3. miRNA expression in the inner ear
As is the case with most regulatory genes of interest, considerable insight regarding their prospective functions can be gained by examining their tissue and cell-specific expression. Among the first studies of miRNA expression, cloning and sequencing revealed the existence of a large number of unique miRNAs, many of which are differentially expressed in various cell types or tissues (Lagos-Quintana et al., 2002; Houbaviy et al., 2003, Landgraf et al., 2007). With the further development of techniques and sequence-specific probes appropriate for microarray analysis (Krichevsky et al., 2003; Liu et al., 2004; Babak et al., 2004), quantitative RT-PCR (Schmittgen et al., 2004; Chen et al., 2005), and in situ hybridization (Weinholds et al., 2005; Kloosterman et al., 2006), studies have been facilely extended to examine changes in miRNA expression coincident with temporospatial aspects of development, disease, and cellular responses to biological factors and stresses. Moreover, the generation of animal models wherein small RNA biogenesis and function is disrupted by deletion of Dicer has facilitated the analysis of miRNA effects in organisms including zebrafish and mouse (Giraldez et al., 2005; Harfe et al., 2005).
Microarray analysis of miRNA expression in the postnatal mouse inner ear has revealed that at least 100 or approximately one-fourth of known mouse miRNAs are present (Weston et al., 2006). Expression profiles are not substantially altered from the newborn mouse inner ear, through maturation and functional onset of hearing, and into adulthood, suggesting that miRNA expression is largely established in embryonic development. Nevertheless, the abundance of miRNAs implies a considerable contribution to the regulation of genetic programs amongst the various cell types that are important to inner ear development, maturation, and function. Moreover, the cell-specific expression of certain miRNAs demonstrated by in situ hybridization in this work and others predicts the relevance of such miRNAs to attaining mature cell characteristics in neurosensory epithelia. The following sections explore just a handful of miRNAs that appear to be important to the establishment of neurons, epithelial cells, and hair cells in the mouse inner ear.
3.1. Neuronal miRNA expression
Many miRNAs have been demonstrated to contribute to neuronal development, plasticity, and disease (reviewed by Fiore et al., 2008). With regard to neurosensory development, miRNAs and their target genes demonstrably constitute genetic mechanisms that dictate sensory neuron cell fates and properties in C. elegans, Drosophila, and mouse. The establishment of left/right asymmetry in C. elegans taste receptor neurons is enforced by a double-negative feedback loop involving cell-specific miRNAs and transcription factor targets (Johnston and Hobert, 2003; Johnston et al., 2005). Whereas the left neuron expresses die-1 transcription factor and lys-6 miRNA which represses cog-1 transcription factor expression, the right neuron expresses cog-1 transcription factor and miR-273 which represses die-1 transcription factor expression. In Drosophila, miR-279 expression in olfactory neurons prevents carbon dioxide (CO2) neuron development by repressing nerfin-1 expression among other genes (Cayirlioglu et al., 2008). In mouse, neuronal miR-124 expression has been shown to represses RE1 silencing transcription factor (REST) and phosphatase SCP1, stifling the anti-neural function of the REST/SCP1 pathway (Conaco et al., 2006; Visvanathan et al., 2007), and to repress polypyrimidine tract binding protein 1 (PTBP1), a global repressor of alternative splicing in non-neuronal cells (Makeyev et al., 2007).
In the mouse inner ear, miR-124 is strongly expressed in sensory neurons derived from the otocyst (Weston et al., 2006). In addition to promoting neuronal cell differentiation and expression profiles (Lim et al., 2005; Krichevsky et al., 2006), miR-124 has been demonstrated to affect neurite outgrowth in differentiating mouse cells (Yu et al., 2008) and branching in Drosophila dentritic arborization sensory neurons through association with fragile X mental retardation protein 1 (dFMRP) and components of the miRNA pathway (Xu et al., 2008). Taken together, these studies suggest that miR-124 among other neuronal miRNAs figure prominently in neurogenesis and innervation of the inner ear.
3.2. Epithelial miRNA expression
The miR-200 family is comprised of five homologous miRNAs (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) that are coordinately expressed from two different genetic loci in epithelia including those of the inner ear in zebrafish (Wienholds et al., 2005), chick (Darnell et al., 2006), and mouse (Kloosterman et al., 2006; Weston et al., 2006). The miRNAs are key regulators of mesenchymal to epithelial transition (MET; reviewed by Gregory et al., 2008a), where they enforce epithelial cell fate by repressing expression of the E-cadherin transcriptional repressors zinc finger E-box binding homeobox 1 (Zeb1) and Smad-interacting protein 1 (Sip1/Zeb2) (Hurteau et al., 2006; Christoffersen et al., 2007; Gregory et al., 2008b; Korpal et al., 2008; Park et al., 2008). The miRNAs and their target genes constitute a double-negative feedback loop wherein Zeb1/Sip1 expression induced by the transforming growth factor beta 1 (Tgfb1) and bone morphogenic protein (Bmp) signaling pathway also represses miR-200 family transcription to promote epithelial to mesenchymal transition (EMT) (Bracken et al., 2008; Burk et al., 2008). Given that Bmp signaling and Smad proteins affect formation of the bony labyrinth and sensory epithelial cell differentiation (Chang et al., 2002; Pujades et al., 2006; Liu et al., 2007) and that hair cell specification requires the basic helix-loop-helix (bHLH) transcription factor Atonal 1 (Atoh1/Math1) which binds E-box motifs (Bermingham et al., 1999; Woods et al., 2004; Krizhanovsky et al., 2006), it is likely that the miR-200 family plays a key role in establishing prosensory epithelial domains in which E-box motifs are receptive to bHLH transcription factors.
3.3. Hair cell miRNA expression
The miR-183 family is comprised of three homologous miRNAs (miR-183, miR-96, and miR-182) that are coordinately expressed from a single genetic locus in vertebrates (Fig. 2A). Moreover, homologous miRNAs are found in Drosophila (miR-263b) and C. elegans (miR-228; Fig. 2A). Importantly, this highly conserved family of miRNAs demonstrates expression in ciliated neurosensory organs across phyla (Pierce et al., 2008). In C. elegans, miR-228 appears to be specifically expressed in various chemosensory and mechanosensory sensilla. In Drosophila, miR-263b is expressed in sensory organ precursors (SOPs) in the embryo (Aboobaker et al., 2005), and in chemosensory and mechanosensory segments of the adult antenna and the haltere (Pierce et al., 2008). In vertebrates, miR-183 family members are expressed in sensory cells of various organs including the olfactory epithelium, eye, neuromast, and ear of zebrafish (Wienholds et al., 2005), and in sensory neurons (cranial and spinal ganglia) and sensory cells of eye and ear in chicken and mouse (Darnell et al., 2006; Kloosterman et al., 2006; Weston et al., 2006). In the mouse eye, the miRNAs are expressed in photoreceptor, retinal bipolar, and amacrine cells (Xu et al., 2007), and their expression is perturbed in a mutant rhodopsin (Rho) model of retinitis pigmentosa (Loscher et al., 2007). In the mouse inner ear, the miRNAs are expressed in sensory neurons and hair cells throughout the vestibular end organs and organ of Corti (Fig. 2B; Weston et al., 2006). Although little is known regarding target genes affected by miR-183 family members, which will be explored in a following section, the high degree of sequence conservation and expression in neurosensory cells suggest these miRNAs play important if not varied roles in the development of functional sensory cells including inner ear hair cells.
Fig. 2.
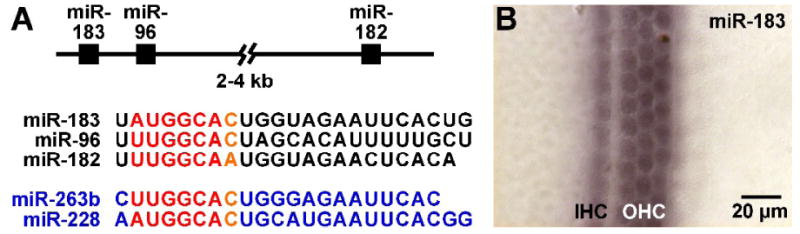
Neurosensory miR-183 family. (A) miR-183 family member sequence and homology. miR-183, miR-96, and miR-182 are clustered within 2-4 kb in vertebrate genomes and co-expressed from a single pri-miR transcript. The vertebrate miRNAs (black) share substantial sequence homology, particularly with the seed sequence (red/orange). Other members of the neurosensory miRNA family include homologous miR-263b and miR-228 (blue) respectively found in D. melanogaster and C. elegans. (B) miR-183 expression in newborn mouse cochlea. miR-183 is detected specifically in inner and outer hair cells (IHC and OHC, respectively) in the organ of Corti by in situ hybridization using a digoxigenin-labeled LNA probe.
4. miRNA function in inner ear development
MicroRNA function is typically assessed using genetic models for loss-of-function (LOF) or gain-of-function (GOF), although recent studies have employed antisense oligonucleotides termed antigomirs to abrogate specific miRNA function in vivo (Krützfeldt et al., 2005; Carè et al., 2007). In Drosophila, LOF and GOF studies have facilitated determination of miR-9a repression of senseless expression in regulating the formation of SOPs (Li et al., 2006), and miR-7 repression of yan expression in photoreceptor differentiation (Li and Carthew, 2005). In mouse, targeted deletions of certain miRNAs have recently revealed their specific roles in cardiac and immune functions (Rodriguez et al., 2007; Thai et al., 2007; van Rooij et al., 2007; Zhao et al., 2007; Ventura et al., 2008), but no specific miRNA deletions with relevance to neurosensory organs have yet been generated. Nevertheless, miRNA function as a whole can be assessed through the analysis of Dicer knockout (KO) models. Whereas Dicer KO in zebrafish has a demonstrated effect on ear development and otoconia formation (Giraldez et al., 2005), Dicer KO mice exhibit early embryonic lethality preceding otocyst development (Bernstein et al., 2003). However, the roles of miRNAs in development of specific mouse tissues can be assessed by conditional KO (CKO) of floxed Dicer alleles through Cre recombinase-mediated deletion (Harfe et al., 2005; Andl et al., 2006; Yi et al., 2006).
4.1 Effect of Dicer CKO on mouse inner ear
The significance of small RNAs to inner ear development has recently been examined in Pax2-Cre;Dicer CKO mice (Soukup et al., 2009). The Pax2-Cre transgene is transiently expressed in the otic placode and results in floxed allele deletion in most cells of the inner ear including sensory neurons, and supporting cell and hair cells in all sensory epithelia (Fig. 3A; Ohyama and Groves, 2004). The transgene is also expressed in the mid-hindbrain, olfactory bulb, and kidneys. Analysis of Pax2-Cre;Dicer CKO mouse inner ear demonstrates major defects in neurogenesis and innervation, morphogenesis, and sensory epithelial histogenesis (summarized in Fig. 3B). The effect of small RNA depletion in this model is similar in severity to LOF models for key regulators of prosensory development such as the SRY-box containing gene 2 protein (Sox2; Kiernan et al., 2005), suggesting a prominent role for miRNAs in the regulation of genetic programs that contribute to ear development.
Fig. 3.
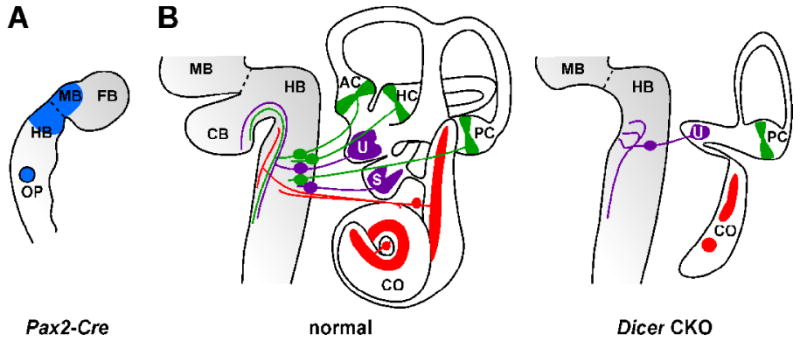
Dicer conditional knockout (CKO) and affect of miRNA depletion on inner ear development. (A) Tissue-specific Cre recombination in transgenic Pax2-Cre mouse embryo (∼8.5 days post coitus). Cre-mediated recombination supports conditional reporter gene expression (blue) in the midbrain (BM), anterior hindbrain (HB), and otic placode (OP). Also indicated are the mid-hindbrain boundary (dashed line) and forebrain (FB). (B) Overview of developmental defects in Pax2-Cre Dicer CKO inner ear (∼17.5 days post coitus). Schematically depicted is normal morphology, histology and innervation of the inner ear and brain (left) versus that of the Dicer CKO (right). Dicer CKO and loss of miRNAs results in a substantial loss of mid-hindbrain structure including the cerebellum (CB), auditory and vestibular neurons, and certain sensory epithelia of the inner ear (colored) are variously affected. Remaining fibers innervating the utricular macula fail to project properly to the brain. AC, anterior crista; HC, horizontal crista; PC, posterior crista; U, utricular macula; S, saccular macula; CO, cochlea.
In Pax2-Cre;Dicer CKO mouse inner ear, sensory neurons initially form but are rapidily depleted of miRNAs including miR-124 and undergo apoptosis. The few surviving neurons fail to properly project to targets in the hindbrain and inner ear, with the exception of some innervation of the utricular macula. The effect of miRNA depletion on surviving neurons is consistent with the apparent roles of miRNAs including miR-124 in neuronal differentiation and neurite outgrowth (Krichevsky et al., 2006; Yu et al., 2008). Morphogenesis and sensory epithelial development are variously affected by small RNA depletion, where there is a complete loss of anterior and horizontal cristae, loss or severe reduction of the saccular macula, reduction of the utricular macula, and severe truncation of the cochlea which typically exhibits two Sox2-positive sensory patches. Interestingly, the posterior crista is least affected in the model, where it was shown to retain some residual miR-183 expression despite Dicer deletion (Soukup et al., 2009), suggesting that Dicer mRNA and/or Dicer protein are relatively more stable in the development of this tissue.
4.2 Importance of hair cell miRNAs
The unexpected differential and residual miRNA expression in Pax2-Cre;Dicer CKO mouse inner ear sensory epithelia provides an interesting perspective on the role of hair cell miRNAs in differentiation. Whereas the production of mature miR-183 is evident in posterior crista hair cells, expression in the utricular macula and cochlea is weak or absent. Examination of hair cell apical specializations by scanning electron microscopy (SEM) revealed that hair cell differentiation in this regard correlates with residual miRNA expression (Fig. 4), where hair cells in the posterior crista show well developed kinocilia and those in the utricular macula are notably bulbous and microvillous.
Fig. 4.
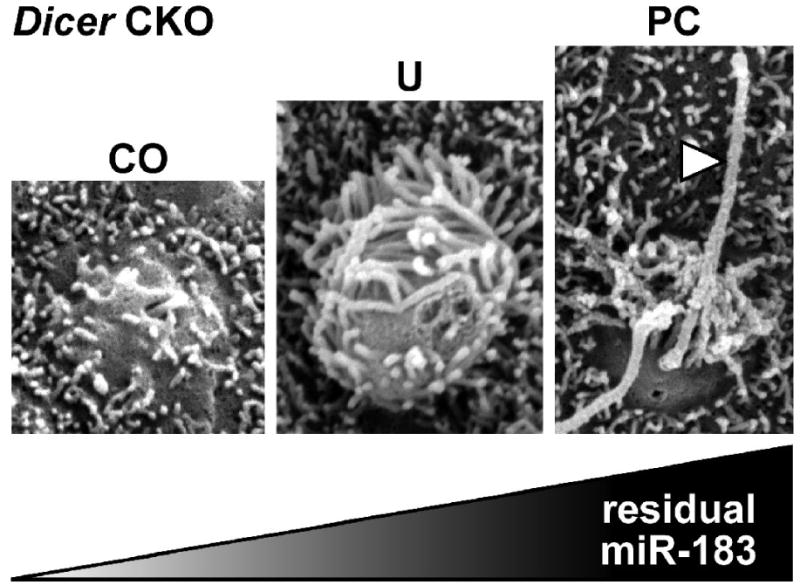
Residual miRNA expression correlates with extent of hair cell differentiation in Dicer CKO inner ear sensory epithelia (∼17.5 days post coitus). Absence or presence of apical specializations for presumptive hair cells in the cochlea (CO), utricular macula (U), and posterior crista (PC) correlate with observed loss or retention of residual miR-183. Hair cells in the utricular macula appear as bulbous microvillus cells whereas those in the posterior crista exhibit well-developed kinocilia (arrowhead).
The range of effects in the Pax2-Cre;Dicer CKO mouse thus demonstrate the importance of small RNAs in inner ear development and cell differentiation. Loss of mid-hindbrain structures and sensory neurons are consistent with effects on proliferation and apoptosis observed in other Dicer CKO models (Davis et al., 2008), and might in part be attributable to loss of natural siRNA functions in Dicer null cells (Kanellopoulou et al., 2005). However, it is interesting to note that the specific depletion of miRNAs in Dgcr8 CKO stem cells reveals similar defects in cell differentiation with little effect on proliferation (Wang et al.,2007), and that a recent model of Dgcr8 CKO in mouse skin is indistinguishable from that of Dicer CKO (Yi et al., 2006; Yi et al., 2009). These studies suggest that the bulk of developmental effects observed in the Pax2-Cre;Dicer CKO inner ear are attributable to the depletion of miRNAs. Ongoing studies seek to examine the post-developmental effects of miRNA depletion on degeneration of sensory epithelia in the ear, as has recently been described for the mouse eye (Damiani et al., 2008).
5. Potential hair cell miRNA mechanisms
Among the many hundreds of predicted target genes for neurosensory/hair cell miR-183 family members reside several of particular interest to the development of hair cells from prosensory epithelia. The mutual exclusion model for miRNA and target gene expression (Flynt and Lai, 2008) predicts that miRNAs contribute to differentiating cell transitions by targeting genes expressed in precursor cells and/or alternative cell fates. Therefore, it is of interest to note the cadre of genes whose interactions contribute to supporting and hair cell fates that possess putative miR-183, miR-96, and/or miR-182 target sites (Fig. 5). These include genes encoding Sox2, which is required to establish prosensory domains and is downregulated in hair cells (Kiernan et al., 2005); Notch1, which through interactions with its ligands Jagged 2 (Jag2) and delta-like 1 (Dll1) expressed in hair cells is key to establishing supporting cell versus hair cell fate (Lanford et al., 1999; Kiernan et al., 2005; Brooker et al., 2006); other prosensory Notch ligands such as Jag1 (Brooker et al., 2006; Kiernan et al., 2006); and Notch1-regulated factors such as Hes1, which antagonizes the function of Atoh1 in specifying hair cell fate (Zheng et al., 2000; Zine and de Ribaupierre, 2002). Importantly, the 3′ UTR of Sox2 mRNA appears to be a bona fide target of miR-182, which mildly albeit significantly downregulates expression in a reporter assay (Weston and Soukup, unpublished data). Considering the rheostat-like function of miRNAs, the whole of these putative interactions suggest that miR-183 family members serve to reinforce hair cell fate and differentiation through a network of integrated effects.
Fig. 5.
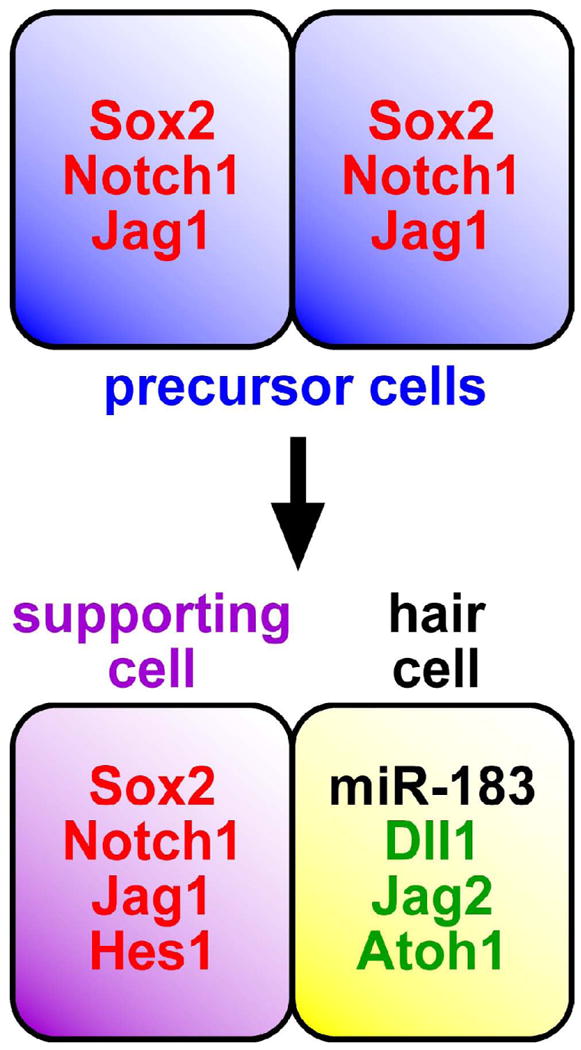
Mutually exclusive expression of miR-183 family members and predicted target genes. Precursor and/or supporting cells express predicted target genes (red) including Sox2, Notch1, Jag1, and Hes1, whereas hair cells expressing miR-183 family members (black) exclude such factors and otherwise express Dll1, Jag2, and Atoh1 (green). Genetic and/or biochemical interactions among these factors are key to establishing supporting versus hair cell fates, suggesting that miR-183 family members help tip the balance in favor of hair cell differentiation.
Other insights to miR-183 family member function can be derived from analyses of their dysregulation in cancer cells. The upregulation of miR-183 family members in leukemia, hepatic, and colorectal cancer cells (Bandrés et al., 2006; Agirre et al., 2008; Ladeiro et al., 2008) has been postulated to downregulate anti-apoptotic factors including forkhead transcription factor (Fox) proteins (Bandrés et al., 2006; Pineles et al., 2007), thus suggesting a pro-apoptotic function for the miRNAs. In other hematopoetic disorders, upregulation of the miRNAs correlates with downregulation of heparin-binding epidermal growth factor (Hbegf; Guglielmelli et al., 2007; Bruchova et al., 2008), which among other mitogens effects the formation of supernumerary hair cells (Zheng et al., 1999). Other studies of leukemia and lymphoma cells (Pal et al., 2007; Wang et al., 2008a) demonstrate that miR-183 family members downregulate expression of protein arginine methyltransferase 5 (Prmt5), reducing histone methylation and upregulating expression of suppressor of tumorigenicity 7 (St7) and retinoblastoma (Rb) pocket proteins, the latter of which maintain post-mitotic differentiation of hair cells (Rocha-Sanchez and Beisel, 2007). Moreover, Prmt5 methylation of p53 modulates responses to DNA damage (Jansson et al., 2008), where depletion of Prmt5 triggers p53-dependent apoptosis rather arrest. Lastly, miR-183 family members have been demonstrated in lung cancer to repress ezrin (Ezr; Wang et al., 2008b), a member of the ezrin/radixin/moesin (ERM) family of proteins that mediate cytoskeletal-membrane interactions and cell signaling (Niggli and Rossy, 2008), where radixin is crucial to hair cell maintenance (Kitajiri et al., 2004). Taken together, these studies suggest that miR-183 family members reinforce the post-mitotic differentiated state of hair cells, but might otherwise predispose hair cells to apoptosis particularly in response to oxidative stresses caused by ototoxic agents such as cisplatin (Rybak et al., 2007). Nevertheless, these putative roles for miR-183 family members in hair cell development and maintenance will require a substantial effort toward validation.
6. Conclusion
The astounding impact of miRNA function in development has been widely examined in model organisms, and the roles of many conserved miRNAs in specific cells, organs, and biological processes including disease are well appreciated. Although miRNA function in development and maintenance of the inner ear is just beginning to be explored, studies already suggest that their genetic regulatory properties are essential to the proper establishment of sensory neurons and sensory epithelial supporting and hair cells. Moreover, a study of miRNA expression during hair cell regeneration in newt sensory epithelia suggest a role for let-7 miRNAs in the process (Tsonis et al., 2007), and miR-9 function has been suggested in the regulation of a COL9A1 expression in human cochlea (Sivakumaran et al., 2006). Many questions remain regarding the functional contributions and mechanistic actions of individual miRNAs amidst the complexity of genetic and biochemical interactions that contribute to development, differentiation, and maintenance of specific cell types throughout the inner ear. Nevertheless, determination of individual miRNA functions is sure to enrich our understanding of pathways and factors important to such processes. Moreover, one can envisage miRNAs among other factors as targets or agents to affect cellular properties or cell differentiation in various therapeutic strategies including stem cell approaches to hair cell regeneration (Beisel et al., 2008).
Acknowledgments
Supported by NIH grant P20RR018788 and Nebraska State Fund LB692 (GAS). Thanks to the many researchers in the miRNA and ear fields whose valued contributions could not been cited here, and to Mike Weston, Marsha Pierce, Bernd Fritzsch, Kirk Beisel, and Sonia Rocha-Sanchez for the many discussions contributing to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X, Jiménez-Velasco A, San José-Enériz E, Garate L, Bandrés E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, Pérez-Roger I, García-Foncillas J, Torres A, Heiniger A, Calasanz MJ, Fortes P, Román-Gómez J, Prósper F. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome is an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel K, Hansen L, Soukup G, Fritzsch B. Regenerating cochlear hair cells: quo vadis stem cell. Cell Tissue Res. 2008;333:373–379. doi: 10.1007/s00441-008-0639-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93:1009–1016. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chang W, ten Dijke P, Wu DK. BMP pathways are involved in otic capsule formation and epithelial-mesenchymal signaling in the developing chicken inner ear. Dev Biol. 2002;251:380–394. doi: 10.1006/dbio.2002.0822. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O'Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008b;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008a;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–44. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guglielmelli P, Tozzi L, Pancrazzi A, Bogani C, Antonioli E, Ponziani V, Poli G, Zini R, Ferrari S, Manfredini R, Bosi A, Vannucchi AM, MPD Research Consortium MicroRNA expression profile in granulocytes from primary myelofibrosis patients. Exp Hematol. 2007;35:1708–1718. doi: 10.1016/j.exphem.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Hurteau GJ, Spivack SD, Brock GJ. Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT-PCR. Cell Cycle. 2006;5:1951–1956. doi: 10.4161/cc.5.17.3133. [DOI] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Cellular commitment and differentiation in the organ of Corti. Int J Dev Biol. 2007;51:571–583. doi: 10.1387/ijdb.072388mk. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, Nakagawa T, Ito J, Tsukita S, Tsukita S. Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol. 2004;166:559–570. doi: 10.1083/jcb.200402007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Soreq L, Kliminski V, Ben-Arie N. Math1 target genes are enriched with evolutionarily conserved clustered E-box binding sites. J Mol Neurosci. 2006;28:211–229. doi: 10.1385/JMN:28:2:211. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1807–1809. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;10:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu W, Butts S, Kim H, Frenz DA. Negative regulation of otic capsule chondrogenesis: it can make you Smad. Ann N Y Acad Sci. 2007;1116:141–8. doi: 10.1196/annals.1402.005. 2007 Nov. [DOI] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, Hassan SS, Kim CJ. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261.e1–e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Pujades C, Kamaid A, Alsina B, Giraldez F. BMP-signaling regulates the generation of hair-cells. Dev Biol. 2006;292:55–67. doi: 10.1016/j.ydbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Rocha-Sanchez SM, Beisel KW. Pocket proteins and cell cycle regulation in inner ear development. Int J Dev Biol. 2007;51:585–595. doi: 10.1387/ijdb.072387sr. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak LP, Whitworth CA, Mukherjea D, Ramkumar V. Mechanisms of cisplatin-induced ototoxicity and prevention. Hear Res. 2007;226:157–167. doi: 10.1016/j.heares.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Saini HK, Enright AJ, Griffiths-Jones S. Annotation of Mammalian Primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sivakumaran TA, Resendes BL, Robertson NG, Giersch AB, Morton CC. Characterization of an abundant COL9A1 transcript in the cochlea with a novel 3′ UTR: Expression studies and detection of miRNA target sequence. J Assoc Res Otolaryngol. 2006;7:160–172. doi: 10.1007/s10162-006-0032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Call MK, Grogg MW, Sartor MA, Taylor RR, Forge A, Fyffe R, Goldenberg R, Cowper-Sal-lari R, Tomlinson CR. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem Biophys Res Commun. 2007;362:940–945. doi: 10.1016/j.bbrc.2007.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008b;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008a;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Xu XL, Li Y, Wang F, Gao FB. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28:11883–11999. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Frantz G, Lewis AK, Sliwkowski M, Gao WQ. Heregulin enhances regenerative proliferation in postnatal rat utricular sensory epithelium after ototoxic damage. J Neurocytol. 1999;28:901–912. doi: 10.1023/a:1007078307638. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- Zine A, de Ribaupierre F. Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear Res. 2002;170:22–31. doi: 10.1016/s0378-5955(02)00449-5. [DOI] [PubMed] [Google Scholar]


