Abstract
Previous linkage studies have identified chromosome 8q24 as a promising positional candidate region to search for bipolar disorder (BP) susceptibility genes. We, therefore, sought to identify BP susceptibility genes on chromosome 8q24 using a family-based association study of a dense panel of SNPs selected to tag the known common variation across the region of interest. A total of 1,458 SNPs across 16 Mb of 8q24 were examined in 3,512 subjects, 1,954 of whom were affected with BP, from 737 multiplex families. Single-locus tests were carried out with FBAT and Geno-PDT, and multi-locus test were carried out with HBAT and multi-locus Geno-PDT. None of the SNPs were associated with BP in the single-locus tests at a level that exceeded our threshold for study-wide significance (p<3.00×10−5). However, there was consistent evidence at our threshold for the suggestive level (p<7.00×10−4) from both the single locus and multi-locus tests of associations with SNPs in the genes ADCY8, ST3GAL1 and NSE2. Multi-locus analyses suggested joint effects between ADCY8 and ST3GAL1 (p=3.00×10−4), with at least one copy of the “high risk” allele required at both genes for association with BP, consistent with a jointly dominant-dominant model of action. These findings with ADCY8 and ST3GAL1 warrant further investigation in order to confirm the observed associations and their functional significance for BP susceptibility.
Introduction
Linkage studies have implicated chromosome 8q24 as a promising positional candidate regions in bipolar disorder (BP). Two independent genome-wide scans of BP reported findings in the region that met stringent criteria for genome-wide significance established by Lander and Kruglyak [1995]. Cichon et al. [2001] studied 75 BP pedigrees of German, Israeli and Italian origin and reported a LOD of 3.6 at marker D8S514 (deCODE genetic map: 124.62 cM; Build 35 physical map: 123,811,284-123,811,642). We [McInnis et al., 2003; Avramopoulos et al., 2004] studied 65 pedigrees of European descent and reported a LOD of 3.32 at marker D8S256 (deCODE genetic map: 145.26 cM; Build 35 physical map: 134,513,446- 134,513,788). Moreover, McQueen et al. [2005] recently pooled the primary genotype data from 11 BP genome-wide linkage scans (including from our study) and carried out a non-parametric linkage analysis of the combined data. Two regions on 6q21 and 8q24 achieved genome-wide significance in the pooled data analysis.
In a previous study, we genotyped a panel of SNPs in a sub-sample of 583 affected offspring from 258 nuclear families with prior evidence of linkage to the region. The panel included 249 informative SNPs covering a narrowly defined region of interest of 3.4 Mb centered at D8S256, the peak marker in our genome-wide linkage scan. The findings from this association study have been reported elsewhere [Zandi et al., 2007].
In the current study, we expanded our search of the region to cover a larger territory spanning ∼16 Mb and including the marker identified in the genome-scan by Cichon et al. [2001]. In addition, we genotyped all 3,512 individuals from 737 extended pedigrees for which we had available DNA in order to extract maximal information for association testing. We report here the initial results of our primary analyses of these data, which we plan to make publicly available.
Methods
The Sample
This study combined two samples of families. The first sample was collected by our group in the Mood Disorders Research Program at Johns Hopkins University [McInnis et al., 2003]. The families were ascertained by opportunistic means through a treated bipolar I disorder (BPI) proband, and evaluated by trained interviewers using either the Schedule for Affective Disorders and Schizophrenia – Lifetime Version (SADS-L) [Endicott and Spitzer, 1978] or the Diagnostic Interview for Genetic Studies (DIGS) [Nurnberger, Jr. et al., 1994]. Family history information was collected and medical records also obtained when available. Diagnoses were made according to Research Diagnostic Criteria (RDC) [Spitzer et al., 1975] using best estimate procedures in which two non-interviewing psychiatrists who were blinded to each other assigned separate diagnoses and a third psychiatrist arbitrated a final decision if they disagreed. All subjects provided written informed consent prior to enrolling in the study, which was approved by the Johns Hopkins Institutional Review Board.
The second sample of families was collected as part of the NIMH Genetics Initiative on Bipolar Disorder [Nurnberger et al., 1997], a multi-site collaboration involving the Johns Hopkins University, Indiana University, Washington University in St. Louis, University of California, San Diego, University of Iowa, University of Pennsylvania, University of Chicago, Rush-Presbyterian Medical Center, University of California, Irvine, and the NIMH Intramural program. The goal of the collaboration was to ascertain and assess families using common procedures for genetic studies of BP. Families were ascertained by opportunistic methods through a BPI proband, and all participants were assessed by trained interviewers with the DIGS. Diagnoses were assigned by psychiatrists using similar best estimate procedures as described above based on either DSM-III-R or DSM-IV criteria. All subjects provided informed consent at the institutions where they were seen, and all protocols were approved by the local IRBs.
Genotype Data
Genotyping was done by CIDR using Illumina technology on a BeadLab system with Golden Gate chemistries. A total of 1,536 SNPs from 123.1 to 139.1 Mb (Build 35) on chromosome 8q24 were selected for genotyping. The SNPs were selected using FESTA [Qin et al., 2006], an LD based tagSNP selection program. It uses the same criterion as LDSelect [Carlson et al., 2004], but uses a more efficient algorithm that selects a smaller number of tagSNPs. The tagSNPs were selected using HapMap Phase I data such that all the common SNPs (minor allele frequency [MAF]<0.05) in the region were either included or were in LD (pairwise r2>0.80) with at least one other SNP that was selected. Of the SNPs that were selected, 1,461 were successfully genotyped. The average median GenCall score across all 1,461 SNPs was 0.87. The GenCall scores, which were calculated using Illumina's proprietary BeadStudio software, range from 0 to 1 and reflect the proximity within a cluster plot of the intensities of that genotype to the centroid of the nearest cluster. The median GenCall score is a high level indicator of how well the DNA samples performed.
Quality control of the genotyping was monitored in two ways. First, we provided duplicate samples on each genotyping plate that were genotyped at all 1,461 SNPs. A total of 42 duplicates had sufficient data to estimate the genotyping error rate, which after removal of Mendelian inheritance errors (see below) was 0.0004 % (1 event / 239,348 genotypes). Second, CIDR included 4 controls, a parent-child trio plus one duplicate child, on each of the genotyping plates. Among these controls, the overall duplicate error rate was 0.0005% (1 event / 189,790 genotypes) and the parent-child Mendelian discordance rate was 0.00% (0 events / 124,110 genotypes).
Several steps were taken to clean the genotype data. First, we corrected problematic familial relationships that had been identified through previous analyses of microsatellite marker genotype data collected as part of genome-wide linkage scans with these samples. This included the removal of monozygotic twins and re-assignment of established incompatible paternities. If the problems could not be corrected, the offending subjects were removed from the dataset. Second, Mendelian inheritance errors were identified using PEDCHECK, and corrected by removing the problematic genotypes according to a pre-specified algorithm established by CIDR. We then re-checked the data for Mendelian errors using Merlin's PEDSTATS. Finally, we used Merlin's error routine to examine the data for evidence of unlikely double recombinations. Genotypes associated with an unlikely double recombination at p<0.005 were removed. We chose this threshold by examining the distribution of p-values from the analysis and selecting a cut-off at the point of inflection away from the expected uniform distribution under the null hypothesis. After the above data cleaning steps, we removed SNPs with more than 2% missing data (n=3), leaving a total of 1,458 SNPs for the analysis. We did not exclude any SNPs solely based on deviation from Hardy-Weinberg equilibrium (HWE), because of considerations that a disease associated SNP may deviate from HWE.
Statistical Analyses
We used Haploview to calculate H-W statistics for each marker using the founders, as well as to characterize the linkage disequilibrium across the region of interest. We also used the Tagger function of Haploview to estimate the proportion of common variation (MAF >0.05) identified in the HapMap Phase II data that was tagged at an r2>0.80 by the SNPs genotyped in the current experiment.
For our primary analysis, we carried out single-locus tests of association with each individual SNP using two different methods. First, we conducted an allelic test of association using FBAT [Horvath et al., 2001]. FBAT is a flexible program that is appropriate for analyses of family data. Under certain conditions, the FBAT reduces to the commonly used transmission/diseqiulibrium test (TDT). However, FBAT is more general and allows tests of associations that are robust to population confounds in the case where parental data is missing and/or other offspring are included in the analysis [Laird and Lange, 2006]. We used the biallelic mode and examined the additive model for counting alleles. We specified the option to calculate the variance empirically in order to provide valid tests of association in the presence of linkage [Lake et al., 2000]. Second, we conducted genotypic tests of association using the program Geno-PDT (Genotype-Pedigree Disequilibrium Tests) [Martin et al., 2003]. Geno-PDT provides valid tests of association in the presence of linkage for genotypes in extended pedigrees. We obtained global 2 degree of freedom significance values for each test.
We then carried out multi-locus tests of association again using two different methods. First, we tested haplotypes of adjacent SNPs using HBAT [Horvath et al., 2004]. It has been shown that in some situations (such as when r2=1 between the risk variant and a particular multi-SNP haplotype) haplotypes may provide more information for association than corresponding single-locus tests [Clayton et al., 2004]. HBAT is an elaboration of FBAT that allows for family-based association tests of haplotypes, even when the phasing of the haplotypes is ambiguous. We used a sliding window approach to test haplotypes of 2, 3 and 4 adjacent SNPs across the region of interest. Significance values were obtained for tests of each specific haplotype. Second, we tested for associations with combinations of genotypes between pairs of SNPs using multi-locus Geno-PDT [van der Walt et al., 2004]. This program does not require phase information and, is therefore useful for dissecting joint effects of SNPs. It provides significance values for individual as well as global tests of the different genotype combinations. To limit the number of tests, we only considered pair-wise combinations of SNPs that provided at least suggestive evidence (defined as below) in the single locus tests.
For all tests of association described above, subjects diagnosed with BPI, schizo-affective disorder, bipolar type (SABP), or bipolar II disorder (BPII) were included as affected. This model of affection was chosen because the best findings on chromosome 8q24 from the previous linkage studies cited above were all obtained under such a model. Subjects who were determined to be never mentally ill were included as unaffected. All other subjects were included with unknown status.
We set the threshold for declaring a finding significant to p<3.00×10−5, reflecting a nominal level of p<0.05 and correcting for 1,458 tests. We used the probability of identifying one false positive out of 1,458 tests (p<7.00×10−4) as the threshold for declaring a finding suggestive. These thresholds do not account for the dependence of the 1,458 SNPs tested or for the multiple methods that were applied to the data.
Results
The final dataset consisted of 3,512 genotyped subjects, including 1,383 males and 2,129 females. Of these, 1,954 were affected (94 SABP, 1,546 BPI, and 314 BPII), 513 were unaffected (i.e., never mentally ill), and 1,045 had some other diagnosis and were included in the analyses as unknown phenotype. There were a total of 737 families, consisting of 1,142 nuclear families. The nuclear families broke down into 1,840 parent affected offspring triads (573 with zero genotyped parents, 641 with one genotyped parent, and 626 with two genotyped parents) and 333 discordant sib-pairs.
A total of 1,458 SNPs met our quality control standards and were examined across the ∼16Mb region of interest at a mean density of 1 SNP per 11 Kb with 5 gaps of greater than 100 Kb. The mean minor allele frequency (MAF) was 28.6%. In the CEPH cohort of the Phase II data of the HapMap project, this collection of SNPs tagged ∼54% of the known common (MAF>0.05) variation across the region at a threshold of r2>0.80.
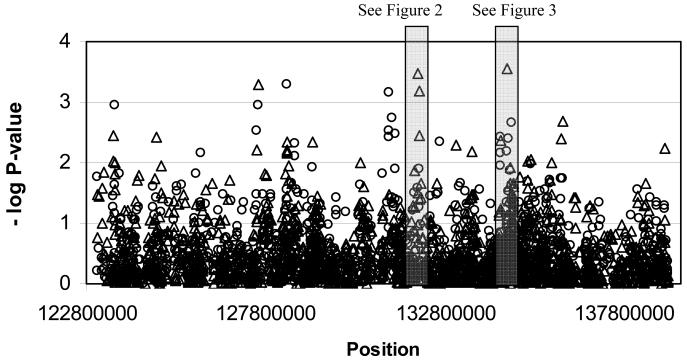
The results of the single locus tests of association between each SNP and BP using FBAT and Geno-PDT are shown in Figure 1, with greater detail at two sites of particular interest shown in Figure 2a and b. None of the SNPs that met our quality control standards were associated at a level that exceeded our threshold for study-wide significance (p<3.00×10−5). Data for SNPs that were associated at the suggestive threshold of p<7.00×10−4 on either of the single locus tests are provided in Table 1. There were six such SNPs, none of which significantly deviated from HWE. These included rs6986303 (FBAT, p=2.82×10−4) in the fourth intron of ST3GAL1; rs1411187 (FBAT, 6.40×10−4) and rs3750889 (FBAT, p=3.40×10−4) in the first and second introns, respectively, of ADCY8; and rs7462286 (Geno-PDT, p=7.00×10−4) in the eleventh intron of DDEF1. The other two SNPs were not located in genes and included rs10101440 (FBAT, p=4.99×10−4) 29 Kb 3′ (centromeric) of the nearest gene, NSE2, and rs1668875 (Geno-PDT, p=5.00×10−4) 407 Kb 5′ (centromeric) of MYC.
Figure 1.
Results from the FBAT and Geno-PDT tests for all 1,458 SNPs across the region of interest are shown. Open triangles show the negative log p-values for the FBAT test under the additive model with the empirical estimator of the variance to appropriately account for association in the presence of linkage. Open circles show the negative log p-values for the Geno-PDT global tests with two degrees of freedom. Highlighted are two regions of interest that are shown at greater resolution in Figures 2 and 3.
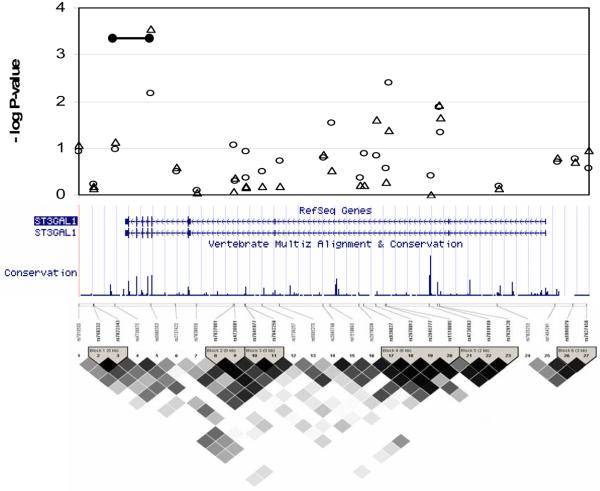
Figure 2.
Detailed findings from the targeted region around ST3GAL1 including: the results from the single-locus tests with FBAT (open triangles) and Geno-PDT (open circles) of all SNPs in the more narrowly defined region; any findings from the multi-locus haplotype analyses with suggestive evidence (p < 7.00×10−4 of association (black bar); a snapshot of the region from UCSC Golden Path showing tracks of known genes and a measure of conservation across 17 species;28 and a picture of the linkage disequilibrium (LD) structure from Haploview (stronger LD in darker shades of black) with blocks defined by the algorithm of Gabriel et al.29
Table 1.
Results from Single SNP Tests with Suggestive Evidence of Association using FBAT and GenoPDTa
| FBAT | GenoPDT | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positionb | SNP | Allelec | S | E(S) | Var(S) | p-value | Genotype | Trans | NotTrans | Aff | Unaff | p-valued |
| 127604613 | rs10101440 | G | 234 | 201.08 | 89.40 | 4.99×10−4 | T,T | 469 | 506 | 179 | 129 | 1.10×10−3 |
| T,G | 147 | 113 | 75 | 36 | ||||||||
| G,G | 10 | 7 | 0 | 1 | ||||||||
| 128410285 | rs1668875 | G | 499 | 534.63 | 173.02 | 6.75×10−3 | G,G | 98 | 99 | 37 | 21 | 5.00×10−4 |
| G,C | 280 | 325 | 107 | 87 | ||||||||
| C,C | 246 | 200 | 110 | 58 | ||||||||
| 131236496 | rs7462286 | C | 274 | 289.28 | 126.02 | 1.73×10−1 | T,T | 440 | 403 | 186 | 119 | 7.00×10−4 |
| T,C | 152 | 200 | 59 | 43 | ||||||||
| C,C | 34 | 23 | 8 | 4 | ||||||||
| 132071516 | rs3750889 | G | 525 | 576.37 | 205.59 | 3.40×10−4 | A,A | 229 | 193 | 93 | 56 | 2.63×10−2 |
| A,G | 302 | 315 | 128 | 80 | ||||||||
| G,G | 95 | 118 | 33 | 30 | ||||||||
| 132091145 | rs1411187 | G | 777 | 725.74 | 227.26 | 6.70×10−4 | A,A | 167 | 142 | 71 | 41 | 1.30×10−2 |
| A,G | 323 | 307 | 139 | 85 | ||||||||
| G,G | 136 | 177 | 44 | 40 | ||||||||
| 134547711 | rs6986303 | A | 514 | 460.23 | 219.30 | 2.82×10−4 | G,G | 57 | 38 | 26 | 14 | 6.60×10−3 |
| G,A | 272 | 249 | 110 | 63 | ||||||||
| A,A | 297 | 339 | 118 | 89 | ||||||||
Results are reported for SNPs that are associated with BP at p < 7.00 × 10-4 with either FBAT or Geno-PDT; The FBAT tests are conducted under the additive model with the empirical variance estimator to appropriately account for association in the presence of linkage, and the Geno-PDT results are shown for the 2-degree of freedom global test
Physical position is based on the May 2004 human reference sequence (Build 34)
The minor allele is shown
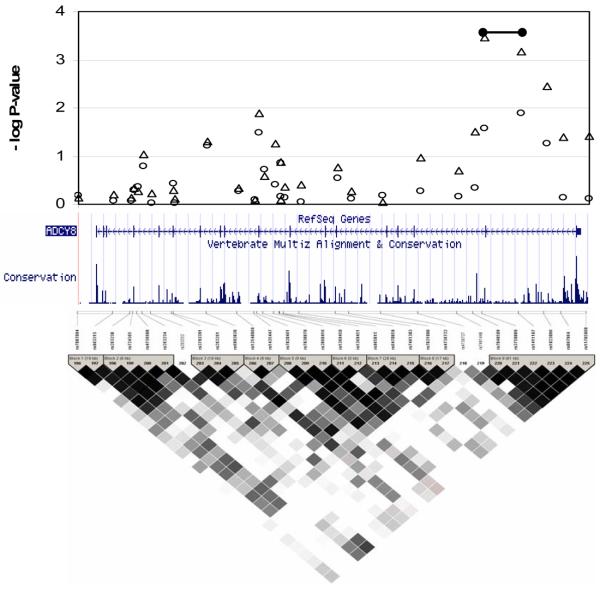
To perform multilocus tests of association, we derived haplotypes by sliding windows of 2, 3 and 4 adjacent SNPs across the region of interest. We observed 7 clusters of haplotypes with suggestive evidence of association exceeding the threshold of p<7.00×10−4. Three of these included SNPs that were highlighted in the single locus tests described above. The first set of haplotypes included both rs3750889 and rs141187 and straddled the second exon of ADCY8 (p=3.40×10−4). The second included rs10101440 and spanned the entire gene of NSE2, which only consists of two exons (p=3.80×10−4). The third included rs6986303 and extended from the fourth intron to 2.3 Kb past the 3′ end of ST3GAL1 (p=6.20×10−4). The other four clusters of haplotypes included SNPs that were not highlighted in the single locus tests. The most significant of these (p=3.90×10−5) was approximately 657 kb away from ST3GAL1 in a relative gene desert that was implicated in our previous, more limited study of the region [Zandi et al., 2007]. The other three were 27 Kb 5′ (telomeric) of KCNQ3 (p=4.30×10−4), 1.6 Mb 3′ (telomeric) of MYC (p=5.90×10−4), and 246 Kb 5′ (centromeric) of MYC (p=3.50×10−4).
We next carried out an additional multi-locus test of associations with Geno-PDT. There was only one combination of SNPs in which the pairwise test exceeded the threshold of suggestive evidence (p<7.00×10−4). This was between rs6986303 in ST3GAL1 and rs1411187 in ADCY8 (Table 2). The p-value for the global pairwise test was 0.0003. Examination of the results suggested that over-transmission to the affected cases (as evidenced by a positive Z-score>1.0) occurred whenever there was at least one copy of the “high-risk” (T) allele at both SNPs.
Table 2.
Results from multilocus test using GenoPDT with rs1411187 and rs6986303
| Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| rs1411187 | rs6986303 | Trans | NotTrans | Aff | Unaff | Z-score | p-valuea | Global Testa |
| T,T | T,T | 12 | 8 | 6 | 3 | 1.58 | 1.14×10−1 | χ2=28.90, p=3.00×10−4 |
| T,T | T,C | 81 | 43 | 29 | 14 | 3.21 | 1.00×10−3 | |
| T,T | C,C | 74 | 91 | 36 | 24 | −1.17 | 2.42×10−1 | |
| T,C | T,T | 30 | 17 | 14 | 9 | 1.37 | 1.72×10−1 | |
| T,C | T,C | 136 | 123 | 66 | 33 | 1.88 | 6.00×10−2 | |
| T,C | C,C | 157 | 167 | 59 | 43 | −1.17 | 2.42×10−1 | |
| C,C | T,T | 15 | 13 | 6 | 2 | 0.79 | 4.31×10−1 | |
| C,C | T,C | 55 | 83 | 15 | 16 | −2.79 | 5.00×10−3 | |
| C,C | C,C | 66 | 81 | 23 | 22 | −1.79 | 7.30×10−2 | |
Trans=transmitted; NotTrans=not transmitted; Aff=affected sib; Unaff=unaffected sib
There is 1 degree of freedom for the genotype specific tests, and 8 degrees of freedom for the global test
Discussion
Previous linkage studies indicate chromosome 8q24 is one of the more promising regions to search for BP susceptibility genes [Cichon et al., 2001; McInnis et al., 2003; Avramopoulos et al., 2004; McQueen et al., 2005]. We carried out a large, family-based association study with a panel of SNPs spanning ∼16 Mb across the locus. We report here the results of our primary analyses of these data. There was consistent evidence at the suggestive level from both the single locus and multi-locus tests of associations with several SNPs in genes across the region, including ADCY8, ST3GAL1 and NSE2. It is possible there are multiple genes in the region that contribute to BP susceptibility. Indeed, it may have been the clustering of such genes that led to a detectable linkage signal which originally drew attention to the region. The associations with ADCY8 and ST3GAL1 are of particular interest, because of evidence suggesting joint effects between the two genes.
ADCY8 codes for adenyl-cyclase in the brain. This enzyme catalyzes the formation of cyclic AMP (cAMP), an important second messenger that has been implicated in BP and is a target for lithium and other mood stabilizing agents [Mork, 1993; Stewart et al., 2001; Perez et al., 2000]. Thus, it is a promising functional candidate gene for BP. Interestingly, a recent study of another candidate region in BP on chromosome 18p reported an association with a SNP that is 1 Mb away from ADCYAP [Mulle et al., 2007], adenyl cylcase activating polypeptide 1, which stimulates adenyl cyclase in pituitary cells and cAMP levels in target cells.
ST3GAL1 codes for a membrane-bound protein that catalyzes the transfer of sialic acid from CMP-sialic acid to galactose containing substrates. It belongs to a family of glycosyltransferase proteins, several of which have been found to be highly expressed in the developing brain and to play a role in neurogenesis [Angata et al., 2004]. Recently, a member of this protein family, SIAT8B on chromosome 15q26, has been associated with schizophrenia in two separate studies in Japanese [Arai et al., 2006] and Chinese [Tao et al., 2006] populations, providing support for the potential relevance of polysaccharide synthesizing genes in major psychiatric disorders. In our previous, more limited association study of the region [Zandi et al., 2007], the best findings in that effort pointed to SNPs in the promoter region of ST3GAL1. In the current study, the most significant findings were at the 3′ end of the gene. It is possible that multiple variants in the gene are associated with disease (i.e., allelic heterogeneity).
The associations with ADCY8 and ST3GAL1 are particularly noteworthy because of evidence suggesting joint effects between the two genes. Our multi-locus analyses suggested that at least one copy of the “high risk” allele is required at both genes for association with BP. This is consistent with a jointly dominant-dominant model of action. Such two-locus models have been described in detail previously [Li and Reich, 2000]. The potential molecular basis for the joint effects between these two genes needs to be investigated further.
Two whole genome association studies (GWAS) of BP have recently been reported that tested SNPs in our region of interest. The first GWAS utilized a pooling approach with two separate samples [Baum et al., 2007]. One sample consisted of a subset of subjects from the NIMH Genetics Initiative on Bipolar Disorder that were included in the current study. The other sample included 772 cases and 876 matched controls from Germany, and in it there were 5 SNPs across ADCY8 and ST3GAL1 that were nominally (p<0.05) associated with BP. The second GWAS, which was completed as part of the Wellcome Trust Case Control Cosortium initiative (WTCCC), included 2000 cases and 3000 controls that were individually genotyped [Wellcome Trust Case Control Consortium, 2007] In this sample there were 13 different SNPs across ADCY8 and ST3GAL1 that were nominally (p<0.05) associated with BP. In neither the pooled German nor the WTCCC samples, however, were the observed associations significant after correcting for multiple tests. It is possible there is genetic heterogeneity across the samples. Alternatively, it is interesting to consider whether the findings across these two genes would have been more remarkable if their joint effects were considered as done in the current study.
The current study has a number of important strengths. It examined one of the largest samples of BP families available for genetic studies. Moreover, all members of the families with available DNA were genotyped, yielding maximal information from the data. We conservatively estimated, assuming one affected trio per family and a disease allele frequency of 0.30 under a multiplicative model, that at the level of study wide significance specified above this sample had 80% power to detect a susceptibility locus with a genotypic relative risk as small as 1.50. Finally, the family-based tests of association provided protection against spurious findings due to population stratification. A limitation of the study is that it included families collected under two different protocols, which could have introduced heterogeneity into the combined sample and biased the results towards the null. However, our group was involved in the design and conduct of both collection efforts, and the ascertainment and assessment procedures were comparable. Another limitation of the study is that it was originally designed based on HapMap Phase I data, and as a result, the coverage of the currently known common variation is incomplete.
The findings from the current study suggest several loci on chromosome 8q24 may be associated with BP and warrant closer investigation. We intend to follow-up these findings by genotyping a denser panel of SNPs around the identified loci, especially in the ST3GAL1 and ADCY8 genes, and to fill in the current gaps in coverage. Our goal is to confirm which of the observed associations are real, and ultimately to determine their functional relevance for BP susceptibility. We plan to make the data from this study publicly available in order to encourage the development of new methods to analyze complex datasets if this kind and to facilitate continued research on chromosome 8q24 in BP.
Figure 3.
Detailed findings from the targeted region around ADCY8 including: the results from the single-locus tests with FBAT (open triangles) and Geno-PDT (open circles) of all SNPs in the more narrowly defined region; any findings from the multi-locus haplotype analyses with suggestive evidence (p < 7.00x10−4) of association (black bar); a snapshot of the region from UCSC Golden Path showing tracks of known genes and a measure of conservation across 17 species;28 and a picture of the linkage disequilibrium (LD) structure from Haploview (stronger LD in darker shades of black) with blocks defined by the algorithm of Gabriel et al.29
Acknowledgements
This work was supported by National Institutes of Health grants R01 MH70775, K01 MH72866 and U54-DA021519. The families were collected with support from the National Institute of Mental Health and the Charles A. Dana Foundation. We are grateful to the families for their participation. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403.
References
- Angata K, Long JM, Bukalo O, Lee W, Dityatev A, Wynshaw-Boris A, Schachner M, Fukuda M, Marth JD. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J.Biol.Chem. 2004;279(31):32603–32613. doi: 10.1074/jbc.M403429200. [DOI] [PubMed] [Google Scholar]
- Arai M, Yamada K, Toyota T, Obata N, Haga S, Yoshida Y, Nakamura K, Minabe Y, Ujike H, Sora I, Ikeda K, Mori N, Yoshikawa T, Itokawa M. Association between polymorphisms in the promoter region of the sialyltransferase 8B (SIAT8B) gene and schizophrenia. Biol.Psychiatry. 2006;59(7):652–659. doi: 10.1016/j.biopsych.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D, Willour VL, Zandi PP, Huo Y, MacKinnon DF, Potash JB, DePaulo JR, Jr., McInnis MG. Linkage of bipolar affective disorder on chromosome 8q24: follow-up and parametric analysis. Mol.Psychiatry. 2004;9(2):191–196. doi: 10.1038/sj.mp.4001388. [DOI] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nothen MM, Georgi A, Schumacher J, Schwarz M, Abou JR, Hofels S, Propping P, Satagopan J, tera-Wadleigh SD, Hardy J, McMahon FJ. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol.Psychiatry. 2007 doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am.J.Hum.Genet. 2004;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Schumacher J, Muller DJ, Hurter M, Windemuth C, Strauch K, Hemmer S, Schulze TG, Schmidt-Wolf G, Albus M, Borrmann-Hassenbach M, Franzek E, Lanczik M, Fritze J, Kreiner R, Reuner U, Weigelt B, Minges J, Lichtermann D, Lerer B, Kanyas K, Baur MP, Wienker TF, Maier W, Rietschel M, Propping P, Nothen MM. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum.Mol.Genet. 2001;10(25):2933–2944. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet.Epidemiol. 2004;27(4):415–428. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch.Gen.Psychiatry. 1978;35(7):837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur.J.Hum.Genet. 2001;9(4):301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet.Epidemiol. 2004;26(1):61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat.Rev.Genet. 2006;7(5):385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- Lake SL, Blacker D, Laird NM. Family-based tests of association in the presence of linkage. Am.J.Hum.Genet. 2000;67(6):1515–1525. doi: 10.1086/316895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat.Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Li W, Reich J. A complete enumeration and classification of two-locus disease models. Hum.Hered. 2000;50(6):334–349. doi: 10.1159/000022939. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet.Epidemiol. 2003;25(3):203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, MacKinnon DF, Potash JB, Mahoney AT, Chellis J, Huo Y, Swift-Scanlan T, Chen H, Koskela R, Stine OC, Jamison KR, Holmans P, Folstein SE, Ranade K, Friddle C, Botstein D, Marr T, Beaty TH, Zandi P, DePaulo JR. Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol.Psychiatry. 2003;8(3):288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, Abou JR, Albus M, Bacanu SA, Baron M, Barrett TB, Berrettini W, Blacker D, Byerley W, Cichon S, Coryell W, Craddock N, Daly MJ, DePaulo JR, Edenberg HJ, Foroud T, Gill M, Gilliam TC, Hamshere M, Jones I, Jones L, Juo SH, Kelsoe JR, Lambert D, Lange C, Lerer B, Liu J, Maier W, Mackinnon JD, McInnis MG, McMahon FJ, Murphy DL, Nothen MM, Nurnberger JI, Pato CN, Pato MT, Potash JB, Propping P, Pulver AE, Rice JP, Rietschel M, Scheftner W, Schumacher J, Segurado R, Van SK, Xie W, Zandi PP, Laird NM. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am.J.Hum.Genet. 2005;77(4):582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork A. Actions of lithium on the cyclic AMP signalling system in various regions of the brain--possible relations to its psychotropic actions. A study on the adenylate cyclase in rat cerebral cortex, corpus striatum and hippocampus. Pharmacol.Toxicol. 1993;73(Suppl 3):1–47. doi: 10.1111/j.1600-0773.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Fallin MD, Lasseter VK, McGrath JA, Wolyniec PS, Pulver AE. Dense SNP association study for bipolar I disorder on chromosome 18p11 suggests two loci with excess paternal transmission. Mol.Psychiatry. 2007;12(4):367–375. doi: 10.1038/sj.mp.4001916. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch.Gen.Psychiatry. 1994;51(11):849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, DePaulo JR, Gershon ES, Reich T, Blehar MC, Edenberg HJ, Foroud T, Miller M, Bowman E, Mayeda A, Rau NL, Smiley C, Conneally PM, McMahon FJ, Meyers D, Simpson S, McInnis MG, Stine OC, Detera-Wadleigh S, Goldin L, Guroff J, Maxwell E, Kazuba D, Gejman PV, Badner J, Sanders A, Rice J, Bierut L, Goate A. Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: a preliminary report. Am.J.Med.Genet. 1997;74(3):227–237. doi: 10.1002/(sici)1096-8628(19970531)74:3<227::aid-ajmg1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Perez J, Tardito D, Mori S, Racagni G, Smeraldi E, Zanardi R. Abnormalities of cAMP signaling in affective disorders: implication for pathophysiology and treatment. Bipolar.Disord. 2000;2(1):27–36. doi: 10.1034/j.1399-5618.2000.020104.x. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Gopalakrishnan S, Abecasis GR. An efficient comprehensive search algorithm for tagSNP selection using linkage disequilibrium criteria. Bioinformatics. 2006;22(2):220–225. doi: 10.1093/bioinformatics/bti762. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Endicott J, Robins E. Research Diagnostic Criteria for a Selected Group of Functional Disorders. Biometrics Research; New York: 1975. [Google Scholar]
- Stewart RJ, Chen B, Dowlatshahi D, MacQueen GM, Young LT. Abnormalities in the cAMP signaling pathway in post-mortem brain tissue from the Stanley Neuropathology Consortium. Brain Res.Bull. 2001;55(5):625–629. doi: 10.1016/s0361-9230(01)00524-x. [DOI] [PubMed] [Google Scholar]
- Tao R, Li C, Zheng Y, Qin W, Zhang J, Li X, Xu Y, Shi YY, Feng G, He L. Positive association between SIAT8B and schizophrenia in the Chinese Han population. Schizophr.Res. 2006 doi: 10.1016/j.schres.2006.09.029. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Noureddine MA, Kittappa R, Hauser MA, Scott WK, McKay R, Zhang F, Stajich JM, Fujiwara K, Scott BL, Pericak-Vance MA, Vance JM, Martin ER. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am.J.Hum.Genet. 2004;74(6):1121–1127. doi: 10.1086/421052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi PP, Avramopoulos D, Willour VL, Huo Y, Miao K, MacKinnon DF, McInnis MG, Potash JB, DePaulo JR., Jr SNP Fine Mapping of Chromosome 8q24 in Bipolar Disorder. Am.J.Med.Genet.B Neuropsychiatr.Genet. 2007 doi: 10.1002/ajmg.b.30486. [DOI] [PubMed] [Google Scholar]