Summary
Natural killer (NK) cells are important in the host resistance to viral infections. They are among the first cells to sense the release of proinflammatory cytokines, as well as the downregulation of surface MHC class I molecules and molecules induced by viral invasion of cells. Various viral functions have evolved to counter NK cell responses illustrating the evolutionary struggles between viruses and NK cells. Ligands for NK cell receptors are primary targets for viral immunoevasion. In order to counteract NK cell activation via the “missing self”-axis, viruses encode proteins which serve as ligands for inhibitory NK cell receptors. Viruses also downmodulate the ligands for the activating NK cell receptors and encode soluble ligands which block these receptors. In addition to viral immunoregulatory proteins, regulatory RNAs can also inhibit the expression of ligands for NK cell receptors. Improving our understanding of viral regulation of NK cell function could be essential for designing more efficient measures in the prophylaxis and treatment of virus-induced pathology.
Introduction
Natural killer (NK) cells control viral replication during the time preceding the induction of the adaptive immune response. They are activated through soluble mediators and by direct cell-to-cell contact. Also, by interacting with dendritic cells (DCs), NK cells are involved in regulation of the adaptive immunity [1,2]. NK cells control the infection by cytolysis and secreting proinflammatory cytokines [3]. The activation of NK cells is regulated through the integration of signals from a number of inhibitory and activating receptors [4], many of which employ MHC class I or class I-like proteins as their ligands. Furthermore, in order to become fully functional during ontogenesis, individual NK cells must engage self-reactive inhibitory receptors specific for appropriate MHC class I molecules [5,6]. NK cell receptors, which utilize MHC class I as their ligands, include the human killer cell Ig-like receptor (KIR) family, the mouse Ly49 family, human and mouse CD94/NKG2 heterodimers and leukocyte Ig-like receptors (LIR) [7]. Other activating NK cell receptors include natural cytotoxicity receptors (NCRs) and NKG2D, among others [8]. While normal cellular ligands for NCRs remain unknown, the NKG2D receptor recognizes several different ligands induced by cellular stress and infection [9].
Viruses have acquired numerous mechanisms aimed at subverting or evading the NK cell immune surveillance. Apart from interfering with NK cell receptors and their ligands, viruses affect NK cell responses by regulating apoptosis, modulating cytokines and chemokines, and by compromising DC functions [10,11]. Here we review the unique relationship between NK cells and viruses, with a focus on viral strategies for interfering with the expression of ligands for NK cell receptors (Table 1).
Table 1.
Common viral interference with NK cell receptor ligands.
| Virus | Viral immunoevasin | Mode of function | References |
|---|---|---|---|
| Interference with “missing self” axis | |||
| HCMV | UL18 | Binding to NK cell inhibitory receptor LIR-1 | [15] |
| MCMV | m144 | Unknown | [22] |
| MCMV | m157 | Binding to Ly49I | [62] |
| RCMV | r144 | Unknown | [76] |
| RCMV | RCTL | Binding to inhibitory receptor Nkrp1b | [27*] |
| Differential regulation of MHC class I | |||
| HCMV | US2, US11 | Downmodulate HLA-A but spare HLA-E; act differentially on other HLA molecules | [77–79] |
| HCMV | UL40 | UL40 leader peptide loads HLA-E and provides inhibitory signal to NK cells | [28,29] |
| MCMV | m04/gp34 | Binds MHC class I and rescues its expression on the cell surface, possible NK cell decoy | [80,81] |
| KSHV | K5 | Downmodulates HLA-A and HLA-B and weakly HLA-C, but shows no effect on HLA-E | [82] |
| HIV | Nef | Downmodulates HLA-A and -B | [31] |
| Interference with ligands for activating NK cell receptors | |||
| HCMV | UL16 | Downmodulates ULBP-1, -2 and MICB | [38,39] |
| HCMV | UL141 | Downmodulates CD155 | [83] |
| HCMV | UL142 | Downmodulates full length MICA | [44*] |
| HCMV | miR-UL112 | Prevents translation of MICB mRNA | [55**] |
| HCMV | UL83/pp65 | Abolishes activation via NKp30 by dissociating adaptor molecule CD3ζ | [74] |
| MCMV | m138 | Viral FcγR, downmodulates MULT-1 and H60, interferes with clathrin-dependent endocytosis | [49**] |
| MCMV | m145 | Downmodulates MULT-1 | [23] |
| MCMV | m152/gp40 | Downmodulates RAE-1 family members | [47] |
| MCMV | m155 | Downmodulates H60 | [48,50] |
| KSHV | K5 | Induces endocytosis of CD86 and CD54 | [32] |
| Zoonotic orthopoxvirus | OMCP | Competitively binds to NKG2D | [56**] |
| Other | |||
| MCMV | m157 | Binds to Ly49H and activates NK cells | [61,62] |
| MCMV | unknown | Activates Ly49P in context of H-2Dk | [64] |
| influenza virus | HA | Binds to NKp46 | [72] |
| HCV | E2 | Binds to CD81 | [84] |
| HTLV | p12I | Downmodulates ICAM-1 and -2 and reduces NK cell adherence to HTLV-infected CD4+ T cells | [85] |
Viral avoidance of NK cells activation via “missing self” axis
Viruses downregulate surface MHC class I molecules in order to avoid recognition by CD8+ T cells [12]. However, the loss of MHC class I molecules renders the cells susceptible to NK cells as described by the “missing-self” hypothesis [13]. To avoid NK cell-mediated control, viruses encode surrogate ligands for inhibitory NK cell receptors and/or are able to differentially regulate MHC class I molecules by downmodulating those that present viral peptides to CD8+ T cells, while sparing those that serve as ligands for inhibitory NK cell receptors [14].
The human cytomegalovirus (HCMV) MHC class I homolog, UL18, was originally described as a decoy ligand for the inhibitory NK cell receptor [15]. The fact that its receptor, LIR-1, was found only on a minor subset of NK cells makes this finding controversial [16]. Later on, it was reported that UL18 could both activate and inhibit NK cell response [16,17]. Its inhibitory function is dependent on the interaction with the LIR-1 molecule [18*]. The mode of activation by UL18 still remains unknown, although it is LIR-1 independent [19*]. Of note, the UL18 sequence and its affinity for LIR-1 vary among clinical HCMV isolates [20,21]. The cellular receptor for mouse cytomegalovirus (MCMV) MHC class I homolog, m144, remains unknown, although mutant viruses lacking this gene show enhanced NK cell sensitivity in vivo [22]. However, in our recent study the virus lacking m144 acted more like the wild type (wt) MCMV strain [23]. The basis for this discrepancy is unclear, but it could be attributed to the use of different viral strains.
Although the “missing self” hypothesis assumes that NK cells can sense the altered expression of MHC class I molecules [13], recent studies showed that the “missing self” axis can also be activated via non-classical MHC molecules [24]. The Nkrp1b and d receptors mediate inhibition via the recognition of Ocil/Clr molecules [25,26]. It has been postulated that the Nkrp1-Ocil/Clr receptor-ligand recognition system may represent an alternative mechanism for discriminating between normal and damaged cells [24]. Recently, it has been shown that cellular Ocil/Clr-b are subject to downmodulation by rat CMV (RCMV) [27*]. This should make the RCMV-infected cells susceptible to the control by NK cell killing due to the lack of inhibition. To avoid this, RCMV encodes its own homolog of Ocil/Clr-b, RCMV C-type lectin-like (RCTL), which serves as a viral ligand for Nkrp1b [27*]. While downmodulation of MHC class I molecules is aimed at protecting the infected host cells against CD8+ T cells, the rationale for cells undergoing RCMV infection to selectively downmodulate the ligand for the inhibitory receptor that could sensitize them to NK cell killing, remains elusive. Since NK cells possess no stimulatory receptors for Ocil/Clr-b, the selective pressure for the virus to downmodulate the ligand is unlikely [25,26]. The explanation for the loss of Ocil/Clr-b could be a default reaction of cells to stress induced by infection.
Differential modulation of MHC class I expression by viruses
HLA-E and mouse Qa-1 are MHC class Ib molecules that are loaded with a peptide derived from a conserved signal sequence of MHC class Ia molecules and are recognized by CD94/NKG2 receptors. Only cells that synthesize MHC class Ia molecules generate a functional ligand for the CD94/NKG2 receptors. While KIRs and Ly49 NK receptors directly survey surface MHC class Ia, the HLA-E-CD94/NKG2A ligand-receptor interaction is therefore an indirect monitoring system for the MHC class Ia expression. Thus, viral functions that spare the expression of HLA-E/Qa-1 should be an optimal strategy to evade both NK and CD8+ T cells.
The HCMV UL40 derived peptide, identical to nonameric HLA-C leader sequence, binds to HLA-E and upregulates its surface expression [28,29]. Although the published data on the efficacy of HCMV US2 and US11 in downmodulation of individual HLA class I molecules are not consistent, it is well established that these proteins spare the inhibitory receptor ligand HLA-E, while targeting HLA-A for degradation [30]. A similar function was attributed to the HIV Nef, causing internalization and lysosomal degradation of HLA-A and HLA-B, but having little or no effect on HLA-C and HLA-E [31]. Similar selectivity was described for Kaposi’s sarcoma herpes virus (KSHV) K5 protein [32].
The functional effect on HLA-E expression has some caveats, however. Although the ability of viruses to preserve HLA-E seems to be an attractive mechanism to avoid NK cells via the inhibitory CD94/NKG2A receptor, its functional impact is questionable since the activating receptor CD94/NKG2C also binds HLA-E [33]. In HCMV-infected humans, either healthy or aviremic HIV-1 infected, a substantial increase of CD94/NKG2C+ over CD94/NKG2A+ cells was observed [34,35]. Furthermore, co-cultivation of peripheral blood mononuclear cells from HCMV-positive donors with virus-infected fibroblasts resulted in the expansion of CD94/NKG2C+ cells [36*]. This function was independent of HCMV UL16, UL18 and UL40, but could be impaired if the virus is lacking MHC class I inhibitors, suggesting that the engagement of inhibitory receptors via MHC class I prevail over activation via CD94/NKG2C.
Viral interference with NK cell activation via NKG2D
The ligand engagement by some activating NK cell receptors, such as NKG2D, can cause activation of NK cells even if target cells express normal levels of MHC class I. NKG2D serves as an activating and co-stimulatory receptor on NK cells and CD8+ T lymphocytes, respectively [9]. NKG2D ligands are usually not constitutively expressed on normal cells but can be induced upon infection or stress [9,37].
NKG2D serves as a target for viral subversion. ULBP-1 and -2, as well as MICB, members of NKG2D-ligand family in humans, can bind to HCMV protein UL16 [38,39] resulting in the retention and sequestration of these ligands [40,41] (Figure 1b). MICA is not exempt from regulation by HCMV [42]. However, only expression of full-length MICA is affected, whereas the truncated form is resistant. The truncated allele is the most common allele in human population, suggesting that polymorphism in the MICA represent an advantage in the ability of the host to control virus infection [43]. Subsequent studies demonstrated that the product of the HCMV UL142 gene downregulates full-length MICA [44*]. Of note is that the laboratory strain AD169, which lacks the UL/b’ region, containing 14 genes among which is UL142, also downregulates MICA [42]. Therefore, it seems that HCMV developed two independent mechanisms affecting MICA whereas HIV Nef downmodulates cell surface expression of MICA, ULBP-1 and ULBP-2 [45].
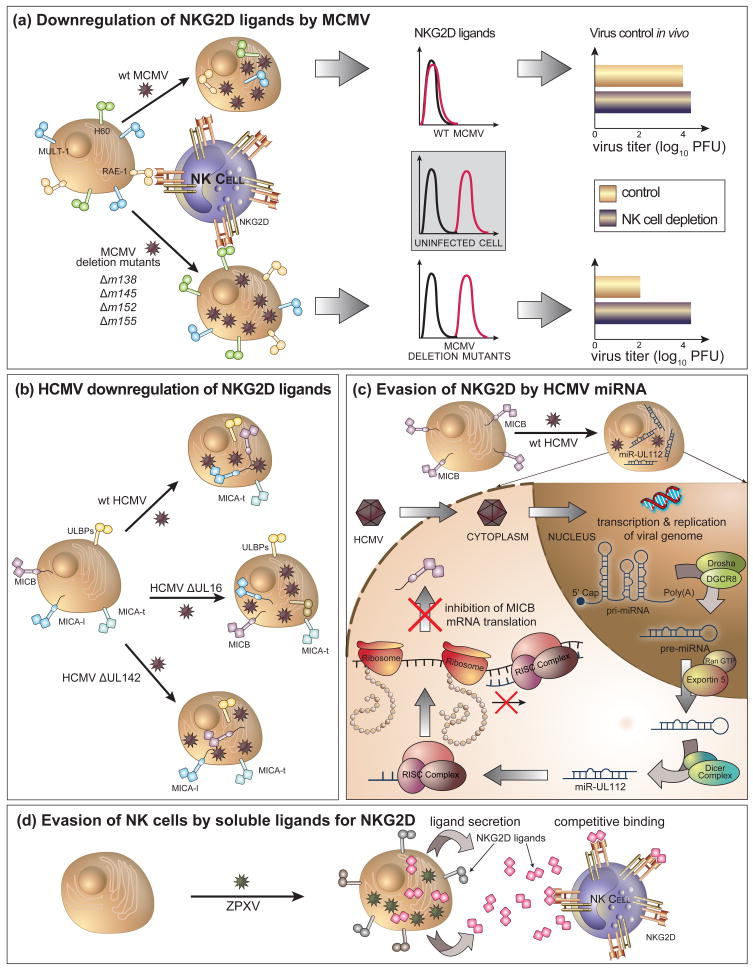
Figure 1. A schematic overview of viral intereference with NK cell activation via NKG2D.
(a)MCMV downmodulates ligands for the NKG2D receptor. m145, m152, m155 affect cell surface expression of MULT-1, RAE-1 family and H60, respectively. m138 is the first described MCMV protein with a dual function on NKG2D ligands, affecting both MULT-1 and H60. The figure shows that the deletion of either of these viral proteins results in the reconstitution of the surface expression of the respective ligands and the conversion of the virus from being NK cell-resistant to become NK cell-sensitive. (b) HCMV acts in a similar manner as MCMV, also exploiting different mechanisms. HCMV UL16 downmodulates ULBP-1 and -2, and also MICB whereas UL142 downmodulates full-length MICA. The truncated form of MICA shows resistance to downmodulation by HCMV. (c) After virus enters the cell, its genome is transcribed into mRNA but also noncoding RNA molecules. By using the cellular machinery, HCMV expresses its own miRNAs, including miR-UL112, which forms the RISC complex and binds the 3′ UTR region of MICB. In this way, miR-UL112 inhibits the translation of MICB mRNA, thus preventing it in reaching the cell surface and signaling. (d) Orthopoxviruses utilize a previously undescribed mode of interference with NKG2D signaling. They secrete a soluble protein, named OMCP, which competitively binds to NKG2D and antagonizes the function of cellularly expressed ligands.
The significance of viral immunoevasins for NKG2D has been also extensively studied in mice whose ligands for the NKG2D receptor, RAE-1 family, H60 and MULT-1, are subject for downregulation by several MCMV proteins (Table 1). Regarding susceptibility to MCMV, most laboratory mouse strains are MCMV-sensitive, characterized by the inability to mount a NK cell response against MCMV [11]. Why these mouse strains fail to generate an efficient virus control via NKG2D and other activating receptors, had been an enigma for a long time, especially keeping in mind that the cellular ligands for NKG2D are inducible by infection [9]. We hypothesized that the virus actively compromises NK cells by downmodulating NKG2D ligands. By using a systemic approach, we characterized the first MCMV gene involved in the downmodulation of NKG2D ligands [46]. Unlike cells infected with wt MCMV, cells infected with the mutant virus lacking the m152 gene expressed NKG2D ligands. The deletion of m152 converted the virus from NK cell-resistant to NK cell-sensitive in vivo (Figure 1a).
Others expanded our data showing that m152/gp40 is involved in the regulation of the members of RAE-1 family [47]. Later on, we and others characterized additional ORFs encoding proteins responsible for the downmodulation of NKG2D ligands [23,48–50]. It turned out that MCMV possesses not only efficient but also redundant mechanisms to avoid expression of NKG2D ligands, suggesting strong selective pressure enforced by NK cells. The product of m145 gene is involved in downmodulation of MULT-1, whereas the m155 protein is responsible for the regulation of H60. The deletion of either of these two MCMV genes resulted in the reconstitution of the corresponding NKG2D ligand on the surface of infected cells and increased virus sensitivity to NK cell control in vivo (Figure 1a).
The molecular mechanisms by which three MCMV immunoevasins interfere with the NKG2D ligands are so far poorly understood. However, some mechanisms have been elucidated from previous studies. For example, the m152 protein has originally been identified for its ability to retain MHC class I complexes in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC)/cis Golgi compartment [51]. Our unpublished results suggest that m152 might also cause the downmodulation of RAE-1 molecules by the same mechanism (SJ, unpublished). However, from our data it seems that RAE-1 isoforms express differential susceptibility to m152. With respect to H60, Lodoen et al. have reported that it is degraded in MCMV-infected cells in proteasome-dependent pathway via m155 [50]. We have shown that the acquisition of endo-b-N-acetylglucosaminodase H resistance and the half-life of H60 is not affected in the presence of m155, suggesting that m155 affects H60 after it exits the ERGIC/cis Golgi compartment [48]. This discrepancy should be clarified by further studies. However, H60 is not only regulated by m155, but also by m138 [49**], an originally described MCMV receptor for Fc portion of IgG [52]. We have previously shown that the virus lacking m138 is attenuated in vivo in normal as well as in immunoglobulin deficient mice, suggesting an additional immunoevasive function of the m138 protein [53]. We tested the ability of the virus lacking the m138 to downregulate ligands for the activating NK receptors and reported that two NKG2D ligands, MULT-1 and H60, are subject to downmodulation by m138 [49**]. Recently, others have demonstrated that this viral immunoevasin is able to target the costimulatory molecule B7-1 [54]. m138 interferes with the recycling pathway of surface MULT-1 and leads to its degradation in lysosomes [49**]. The ability of inhibitors of clathrin-dependent endocytosis to rescue the expression of MULT-1 in MCMV-infected cells suggests that m138 interferes with this endocytic pathway although the modes of interaction and the synergistic effect with m145 remain unanswered. Interestingly, although both MULT-1 and H60 are regulated by m138, it appears that distinct N-terminal domains within the fcr-1 ectodomain are involved in downregulation of these ligands.
Immunoevasion of NKG2D by regulatory RNA
Upon entering target cells, DNA viruses transcribe their DNA not only into protein encoding messenger RNA (mRNA), but also into noncoding regulatory RNA, such as microRNA (miRNA). Although noncoding RNAs were known to be involved in the regulation of gene expression, the potential significance of this mechanism in the regulation of immune response was unknown until recently. Stern-Ginossar et al. provided evidence that virus can use miRNA to thwart NK cells [55**]. They used an algorithm to predict miRNA targets and showed that HCMV-derived miRNA (miR-UL112) displays sequence complementarity to the 3′-UTR of mRNA encoding a human ligand for the NKG2D receptor, MICB (Figure 1c). When expressed in recombinant lentivirus vector this miRNA protects infected cells from NKG2D-dependent lysis by preventing translation of MICB mRNA. Similar observations were observed during authentic virus infection. The fact that MICB protein is also downmodulated by HCMV UL16 again indicates a strong selective pressure on the virus for the development of redundant immunoevasion mechanisms against NKG2D. As compared to the immunoevasion by viral proteins, regulatory RNA may have some advantages due to its mechanism of action involving only a small amount of nucleic acid of poor immunogenicity. This could play a role in latently infected cells undergoing reactivation in fully immunocompetent host.
Evasion of NK cells by virally encoded soluble ligands for NKG2D receptors
Unlike CMVs, zoonotic orthopoxviruses secrete a protein, distantly related to class I molecules, that serves as a competitive antagonist of the NKG2D receptor [56**] (Figure 1d). Having in mind the dual function of NKG2D for NK and T cells, one could speculate that the strategy adopted by the zoonotic orthopoxviruses is less selective as compared to the downmodulation of cellular ligands by CMVs. By downmodulating NKG2D ligands only on infected cells, the CMVs are limiting its immunomodulatory potential to infected target cells; in the case of orthopoxviruses, a secreted soluble competitive inhibitor is likely to attenuate the response to any pathogen otherwise controlled via NKG2D. Similarly, some tumors evade NK cell activation by releasing soluble MICA, which results in downmodulation and inactivation of the NKG2D receptor [57]. Thus, perhaps these different strategies have broader implications for viral immunoevasion of NKG2D.
MCMV interference with activating Ly49 NK cell receptors
Some NK cell receptors are specific for viral proteins. The first described one was the Cmv1r locus, which encodes Ly49H, an activating NK cell receptor [58–60]. Unlike most of other Ly49 receptors the ligand for Ly49H is not an MHC class I molecule but a product of the MCMV m157 gene [61,62]. However, as predicted, m157 adopts MHC class I-like protein fold and its intra- and intermolecular interactions within and between domains enable this protein to be even more compact than classical MHC class I [63**]. Recently, additional genetic loci have also been implicated in the NK cell-dependent resistance. Vidal and colleagues have shown that the genetic resistance in Cmv3+ mice maps to Ly49P in MCMV-resistant MA/My mice, and that NK cell-mediated resistance to infection in these mice involves epistatic recognition of a viral protein in the context of the MHC H-2k molecule [64]. Although the molecular nature of target recognition remains unknown, it is likely that the recognition of H-2Dk molecules depends on the viral peptide. These conclusions are corroborated by Brown and colleagues [65]. Altogether, by its specificity for MHC class I allele and viral protein, Ly49P represents another highly specific mechanism of NK cell-mediated control. An additional locus, Cmv4, is also suggested to encode an NK cell activating receptor and mediate MCMV resistance in PWK mice [66].
On the first glance, it makes no sense that viruses encode ligands for an activating NK cell receptor. Indeed, m157 is also a ligand for the inhibitory NK cell receptor Ly49I [62], which was perhaps its primary function. Two independent studies have shown that m157 is subject for mutation under selective pressure of Ly49H+ NK cells [67,68]. Therefore, for Ly49P it could be speculated that the virus would not “tolerate” the genes encoding its viral ligand, and that Ly49P must have an inhibitory counterpart with specificity for H-2Dk and a so far undefined viral peptide.
NCRs as putative targets for viral immunoevasins
NCRs are represented by activating receptors NKp30, NKp44 and NKp46 (mice express only NKp46) [69,70]. Unlike other NK cell receptor families, which include inhibitory and activating isoforms, NCRs share no such heterogeneity. While cellular ligands for NCRs remain unknown, it has been reported that viral hemaglutinin can bind NKp46 and NKp44 [71,72]. Mice lacking the NKp46 activating receptor are much more sensitive to influenza virus infection [73**]. It remains to be tested whether NKp46 receptor or its cellular ligands are subjects for viral immunoevasion. Since most mouse strains fail to mount a NK cell response to MCMV, it is possible that, similarly to NKG2D, there may be an MCMV mechanism to prevent NK cell activation via NCRs. Consistent with this speculation, it has been shown that pp65, a tegument protein of HCMV, can antagonize NKp30 activation by causing its dissociation from the adaptor molecule CD3ζ [74]. Moreover, the downmodulation of NKp30 and NKp46 ligands, but not of NKp44, on T-cell blasts, is reported in patients infected with HIV [75].
More question than answers
It is generally believed that the major function of viral NK cell immunoevasion is to enable the virus to replicate and spread before the onset of the specific immune response. However, in the case of herpesviruses such as MCMV, the most explored viral model in NK cell research, one can challenge this simplified view. In spite of a plethora of immunoregulatory functions, MCMV does not cause significant disease in immunocompetent hosts. Although data published so far indicate an important role of NK cells in shaping the CD8+ T cell response, the lessons obtained in so-called MCMV-sensitive mice, indicate that inefficient early NK cell response cannot be a decisive factor for the generation of the adaptive immunity. Namely, Ly49H− mice, such as BALB/c, can control the infection and establish latency. Does this suggest that NK cells are not essential for surveillance of the primary CMV infection? However, based on the attenuation of mutant viruses lacking NK cell immunoevasins, we can conclude that they are nevertheless important in virus control. A better understanding of the linkage between NK cell functions and viral immunoevasins might require additional considerations. Two major parameters are at the present in use for the assessment of the role of immunoevasins in vivo: (a) NK cell-dependent virus control in tissues after primary infection and (b) survival after challenge infection. Are the results obtained by these approaches really sufficient for reaching conclusions about the significance of viral immunoevasins for the pathogenesis of MCMV infection in natural conditions? We think they are not. The right question may be why it is so important to limit NK cell response and is this good for the virus or for the host, or for both. Our knowledge about the natural virus spread and the putative significance of the viral immunoregulatory mechanisms is still limited. Are these viral genes developed during the evolution to assist herpesviruses in horizontal and vertical spread? Another important role of these immunoevasins could be to allow sufficient antigenic load and optimal priming and shaping of specific immune response required for maintenance of viral latency. Altogether, new experimental approaches are needed to answer questions about the importance of viral immunoevasins for balancing the immune response, the function that is equally important for the virus and its host. Of equal importance is also the question about the putative role of immunoevasins in the prevention of immunopathology, particularly in the case of chronic infections. Although the deletion of individual viral immunoevasion genes usually results in altered sensitivity to NK cells in vivo, it is questionable whether we would be able to get the full picture by using the individual mutants. Do we need the viruses lacking sets of these immunoevasins to assess their function in the context of primary and chronic infection? We also need to understand the meaning of redundancy of viral immunoevasins. Bearing in mind that numerous NK cell receptors are also expressed on T cells and some other cells, we need to focus our efforts on the role of immunoevasins in these cells as well.
Thanks to new high-throughput technologies, we are constantly learning about new viral genes involved in the regulation of immune response, but their role in natural infection and viral pathogenesis will remain without complete answers without more sensitive and more specific approaches for monitoring immune surveillance in vivo.
Acknowledgments
Our work is supported by Croatian Ministry of Science, Education and Sport (grants 062–0621261–1263 to SJ and 062–0621261–1271 to BP), and NIH R01 HD044721–01. AK is supported by the Howard Hughes Medical Institute Scholars grant. We thank Francesco Colucci and Ofer Mandelboim for discussions about the manuscript and Vanda Juranic for help with the figure. We apologize to our colleagues whose work could not be cited owing to space limit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds. dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 2.Robbins SH, Bessou G, Cornillon A, Zucchini N, Rupp B, Ruzsics Z, Sacher T, Tomasello E, Vivier E, Koszinowski UH, et al. Natural Killer Cells Promote Early CD8 T Cell Responses against Cytomegalovirus. PLoS Pathog. 2007;3:e123. doi: 10.1371/journal.ppat.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raulet DH. Natural Killer Cells. In: Paul WE, editor. Fundamental Immunology. 5. Lippincott Williams & Wilkins; 2003. pp. 365–391. [Google Scholar]
- 4.Kirwan SE, Burshtyn DN. Regulation of natural killer cell activity. Curr Opin Immunol. 2007;19:46–54. doi: 10.1016/j.coi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006;24:249–257. doi: 10.1016/j.immuni.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 9.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 10.Orange JS, Fassett MS, Koopman LA, Boyson JE, Strominger JL. Viral evasion of natural killer cells. Nat Immunol. 2002;3:1006–1012. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- 11.Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol Cell Biol. 2007;85:46–54. doi: 10.1038/sj.icb.7100013. [DOI] [PubMed] [Google Scholar]
- 12.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Immunol Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 14.Guma M, Angulo A, Lopez-Botet M. NK cell receptors involved in the response to human cytomegalovirus infection. Curr Top Microbiol Immunol. 2006;298:207–223. doi: 10.1007/3-540-27743-9_11. [DOI] [PubMed] [Google Scholar]
- 15.Reyburn HT, Mandelboim O, Vales-Gomez M, Davis DM, Pazmany L, Strominger JL. The class I MHC homologue of human cytomegalovirus inhibits attack by natural killer cells. Nature. 1997;386:514–517. doi: 10.1038/386514a0. [DOI] [PubMed] [Google Scholar]
- 16.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 17.Leong CC, Chapman TL, Bjorkman PJ, Formankova D, Mocarski ES, Phillips JH, Lanier LL. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: the role of endogenous class I major histocompatibility complex and a viral class I homolog. J Exp Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Prod’homme V, Griffin C, Aicheler RJ, Wang EC, McSharry BP, Rickards CR, Stanton RJ, Borysiewicz LK, Lopez-Botet M, Wilkinson GW, et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J Immunol. 2007;178:4473–4481. doi: 10.4049/jimmunol.178.7.4473. These two papers [18*, 19*] demonstrate that HCMV MHC class I homolog UL18 can activate NK cells in a LIR-1 independent fashion. In addition to the previously established binding of the inhibitory receptor LIR-1 on NK cells, the results suggest the existence of additional receptor for UL18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Wagner CS, Riise GC, Bergstrom T, Karre K, Carbone E, Berg L. Increased expression of leukocyte Ig-like receptor-1 and activating role of UL18 in the response to cytomegalovirus infection. J Immunol. 2007;178:3536–3543. doi: 10.4049/jimmunol.178.6.3536. See annotation to [18*]. [DOI] [PubMed] [Google Scholar]
- 20.Cerboni C, Achour A, Warnmark A, Mousavi-Jazi M, Sandalova T, Hsu ML, Cosman D, Karre K, Carbone E. Spontaneous mutations in the human CMV HLA class I homologue UL18 affect its binding to the inhibitory receptor LIR-1/ILT2/CD85j. Eur J Immunol. 2006;36:732–741. doi: 10.1002/eji.200425220. [DOI] [PubMed] [Google Scholar]
- 21.Vales-Gomez M, Shiroishi M, Maenaka K, Reyburn HT. Genetic variability of the major histocompatibility complex class I homologue encoded by human cytomegalovirus leads to differential binding to the inhibitory receptor ILT2. J Virol. 2005;79:2251–2260. doi: 10.1128/JVI.79.4.2251-2260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell HE, Vally H, Lynch DM, Fleming P, Shellam GR, Scalzo AA, Davis-Poynter NJ. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 23.Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, Polic B, Bubic I, Kriegeskorte A, Pernjak-Pugel E, et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar V, McNerney ME. A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat Rev Immunol. 2005;5:363–374. doi: 10.1038/nri1603. [DOI] [PubMed] [Google Scholar]
- 25.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 27*.Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, Carlyle JR. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. This report describes the viral interference with the NK cell activation via Clr-b/Ocil missing self axis. While RCMV infection causes the downmodulation of cellular Clr, this virus encodes a decoy ligand RCTL that binds to Nkrp1b, thus ensuring the inhibition of NK cells. [DOI] [PubMed] [Google Scholar]
- 28.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 29.Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, Weiss EH. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol. 2000;164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Botet M, Llano M, Ortega M. Human cytomegalovirus and natural killer-mediated surveillance of HLA class I expression: a paradigm of host-pathogen adaptation. Immunol Rev. 2001;181:193–202. doi: 10.1034/j.1600-065x.2001.1810116.x. [DOI] [PubMed] [Google Scholar]
- 31.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 32.Ishido S, Choi JK, Lee BS, Wang C, DeMaria M, Johnson RP, Cohen GB, Jung JU. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi’s sarcoma-associated herpesvirus K5 protein. Immunity. 2000;13:365–374. doi: 10.1016/s1074-7613(00)00036-4. [DOI] [PubMed] [Google Scholar]
- 33.Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 34.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 35.Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, Lopez-Botet M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006;194:38–41. doi: 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 36*.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. This study reports the increase of CD94/NKG2C+ over CD94/NKG2A+ cells in peripheral blood of healthy HCMV seropositive humans, suggesting that HCMV infection may shape the NK cell repertoire. [DOI] [PubMed] [Google Scholar]
- 37.Cerwenka A, Lanier LL. NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens. 2003;61:335–343. doi: 10.1034/j.1399-0039.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 38.Kubin M, Cassiano L, Chalupny J, Chin W, Cosman D, Fanslow W, Mullberg J, Rousseau AM, Ulrich D, Armitage R. ULBP1, 2, 3: novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur J Immunol. 2001;31:1428–1437. doi: 10.1002/1521-4141(200105)31:5<1428::AID-IMMU1428>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 40.Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U, Rammensee HG, Steinle A. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol. 2003;33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
- 41.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou Y, Bresnahan W, Taylor RT, Stastny P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol. 2005;174:3098–3104. doi: 10.4049/jimmunol.174.5.3098. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Lazaro AM, Lavingia B, Stastny P. Typing for all known MICA alleles by group-specific PCR and SSOP. Hum Immunol. 2001;62:620–631. doi: 10.1016/s0198-8859(01)00241-5. [DOI] [PubMed] [Google Scholar]
- 44*.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun. 2006;346:175–181. doi: 10.1016/j.bbrc.2006.05.092. This paper extended the work described in reference [42] showing that the product of HCMV UL142 is responsible for the downregulation of full-length MICA. Since this gene is absent in AD169 strain, it has been suggested that there must be another viral inhibitor of MICA. [DOI] [PubMed] [Google Scholar]
- 45.Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol. 2007;88:242–250. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- 46.Krmpotic A, Busch DH, Bubic I, Gebhardt F, Hengel H, Hasan M, Scalzo AA, Koszinowski UH, Jonjic S. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat Immunol. 2002;3:529–535. doi: 10.1038/ni799. [DOI] [PubMed] [Google Scholar]
- 47.Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasan M, Krmpotic A, Ruzsics Z, Bubic I, Lenac T, Halenius A, Loewendorf A, Messerle M, Hengel H, Jonjic S, et al. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J Virol. 2005;79:2920–2930. doi: 10.1128/JVI.79.5.2920-2930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, et al. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J Exp Med. 2006;203:1843–1850. doi: 10.1084/jem.20060514. This paper describes the downmodulation of two NKG2D ligands, MULT-1 and H60, caused by m138, an MCMV encoded FcγR. In addition, the data provide evidence that m138 interferes with a clathrin-dependent endocytic pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med. 2004;200:1075–1081. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziegler H, Thäle R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski UH. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]
- 52.Thäle R, Lucin P, Schneider K, Eggers M, Koszinowski UH. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol. 1994;68:7757–7765. doi: 10.1128/jvi.68.12.7757-7765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crnkovic-Mertens I, Messerle M, Milotic I, Szepan U, Kucic N, Krmpotic A, Jonjic S, Koszinowski UH. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol. 1998;72:1377–1382. doi: 10.1128/jvi.72.2.1377-1382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mintern JD, Klemm EJ, Wagner M, Paquet ME, Napier MD, Kim YM, Koszinowski UH, Ploegh HL. Viral interference with B7-1 costimulation: a new role for murine cytomegalovirus fc receptor-1. J Immunol. 2006;177:8422–8431. doi: 10.4049/jimmunol.177.12.8422. [DOI] [PubMed] [Google Scholar]
- 55**.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. This is the first report on the virally encoded miRNA with the role in immunoevasion of NK cells. The HCMV encoded miR-UL112 is shown to inhibit translation of MICB mRNA by binding its 3′UTR, and thus preventing its cell surface expression and signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Campbell JA, Trossman DS, Yokoyama WM, Carayannopoulos LN. Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J Exp Med. 2007;204:1311–1317. doi: 10.1084/jem.20062026. This study demonstrated a novel mechanism of NK cell evasion utilized by zoonotic orthopoxviruses. These viruses encode a protein that is secreted by infected cells and competitively binds to NKG2D, thus blocking the recognition of infected cells by NK cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 58.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 59.Daniels KA, Devora G, Lai WC, O’Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 61.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 63**.Adams EJ, Juo ZS, Venook RT, Boulanger MJ, Arase H, Lanier LL, Garcia KC. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci U S A. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. This paper describes the crystal structure of the MCMV MHC class I-like protein m157 that serves as a ligand for Ly49H and Ly49I NK cell receptors. Although m157 does not bind peptide and beta-2-microglobulin, it possesses compact MHC class I-like fold as a result of extensive intra- and intermolecular interactions within and between domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desrosiers MP, Kielczewska A, Loredo-Osti JC, Adam SG, Makrigiannis AP, Lemieux S, Pham T, Lodoen MB, Morgan K, Lanier LL, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie X, Dighe A, Clark P, Sabastian P, Buss S, Brown MG. Deficient major histocompatibility complex-linked innate murine cytomegalovirus immunity in MA/My. L-H2b mice and viral downregulation of H-2k class I proteins. J Virol. 2007;81:229–236. doi: 10.1128/JVI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adam SG, Caraux A, Fodil-Cornu N, Loredo-Osti JC, Lesjean-Pottier S, Jaubert J, Bubic I, Jonjic S, Guenet JL, Vidal SM, et al. Cmv4, a new locus linked to the NK cell gene complex, controls innate resistance to cytomegalovirus in wild-derived mice. J Immunol. 2006;176:5478–5485. doi: 10.4049/jimmunol.176.9.5478. [DOI] [PubMed] [Google Scholar]
- 67.French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, Koszinowski U, Jonjic S, Yokoyama WM. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 2004;20:747–756. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 68.Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HR, Yokoyama WM, Scalzo AA. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A. 2003;100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnon TI, Markel G, Mandelboim O. Tumor and viral recognition by natural killer cells receptors. Semin Cancer Biol. 2006;16:348–358. doi: 10.1016/j.semcancer.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Moretta A, Biassoni R, Bottino C, Mingari MC, Moretta L. Natural cytotoxicity receptors that trigger human NK-cell-mediated cytolysis. Immunol Today. 2000;21:228–234. doi: 10.1016/s0167-5699(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 71.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 72.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 73**.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. By generating the mouse strain with the Ncr1gene replaced with GFP reporter cassette, the authors presented a mouse model for studying the role of NKp46 in vivo. They extended their own finding that NKp46 binds to influenza virus hemaglutinin, and showed enhanced susceptibility of the mutant mice to influenza virus infection. [DOI] [PubMed] [Google Scholar]
- 74.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, Gonen-Gross T, Hanna J, Nahari E, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 75.Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 2007;110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kloover JS, Grauls GE, Blok MJ, Vink C, Bruggeman CA. A rat cytomegalovirus strain with a disruption of the r144 MHC class I-like gene is attenuated in the acute phase of infection in neonatal rats. Arch Virol. 2002;147:813–824. doi: 10.1007/s007050200028. [DOI] [PubMed] [Google Scholar]
- 77.Barel MT, Ressing M, Pizzato N, van Leeuwen D, Le Bouteiller P, Lenfant F, Wiertz EJ. Human cytomegalovirus-encoded US2 differentially affects surface expression of MHC class I locus products and targets membrane-bound, but not soluble HLA-G1 for degradation. J Immunol. 2003;171:6757–6765. doi: 10.4049/jimmunol.171.12.6757. [DOI] [PubMed] [Google Scholar]
- 78.Schust DJ, Tortorella D, Seebach J, Phan C, Ploegh HL. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J Exp Med. 1998;188:497–503. doi: 10.1084/jem.188.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Llano M, Guma M, Ortega M, Angulo A, Lopez-Botet M. Differential effects of US2, US6 and US11 human cytomegalovirus proteins on HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur J Immunol. 2003;33:2744–2754. doi: 10.1002/eji.200324182. [DOI] [PubMed] [Google Scholar]
- 80.Kavanagh DG, Koszinowski UH, Hill AB. The murine cytomegalovirus immune evasion protein m4/gp34 forms biochemically distinct complexes with class I MHC at the cell surface and in a pre-Golgi compartment. J Immunol. 2001;167:3894–3902. doi: 10.4049/jimmunol.167.7.3894. [DOI] [PubMed] [Google Scholar]
- 81.Holtappels R, Gillert-Marien D, Thomas D, Podlech J, Deegen P, Herter S, Oehrlein-Karpi SA, Strand D, Wagner M, Reddehase MJ. Cytomegalovirus encodes a positive regulator of antigen presentation. J Virol. 2006;80:7613–7624. doi: 10.1128/JVI.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C, McSharry BP, Morris RJ, Llewellyn-Lacey S, Rickards C, et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6:181–188. doi: 10.1038/ni1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crotta S, Stilla A, Wack A, D’Andrea A, Nuti S, D’Oro U, Mosca M, Filliponi F, Brunetto RM, Bonino F, et al. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J Exp Med. 2002;195:35–41. doi: 10.1084/jem.20011124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee P, Feuer G, Barker E. Human T-cell leukemia virus type 1 (HTLV-1) p12I down-modulates ICAM-1 and -2 and reduces adherence of natural killer cells, thereby protecting HTLV-1-infected primary CD4+ T cells from autologous natural killer cell-mediated cytotoxicity despite the reduction of major histocompatibility complex class I molecules on infected cells. J Virol. 2007;81:9707–9717. doi: 10.1128/JVI.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]